Abstract
Drought is a serious, worldwide problem for crop production and also affects yields of barley and wheat, together with other stressors such as frost, viral diseases, or fungal pathogens. Although a number of candidate genes have been identified by transcriptome approaches in recent years, only very few have been tested in functional assays for a beneficial effect on drought tolerance. Here, a transient assay system in microprojectile-bombarded barley leaves is described that allows the functional testing of dehydration stress-related candidate genes by RNA interference (RNAi) or overexpression. Cellular stress or damage in dedydrated leaves is reported by a reduced accumulation of slowly maturing, native red-fluorescing protein DsRed that is known to be sensitive to denaturing conditions. After a dehydration-stress period of 4 d during which the relative fresh weight of leaves was kept at 60–66% of initial fresh weight, a reproducible reduction of normalized DsRed fluorescence was observed. In order to obtain proof of concept, a number of barley mRNAs homologous to drought response genes were selected and targeted by transient induced gene silencing (TIGS). TIGS of four tested genes resulted in a significantly stronger decrease of normalized DsRed fluorescence in dehydration-stressed leaves, whereas they had no effect in fully turgescent control leaves. These genes encode barley drought-responsive factor HvDRF1 (DREB2-like), dehydrin 6, late embryogenesis-abundant protein HVA1, and the vacuolar sodium/proton antiporter HvHNX1. The four targeted transcripts were also found to accumulate rapidly in dehydration-stressed barley leaf segments. The results suggest a value of the TIGS system for functional pre-screening of larger numbers of drought or dehydration stress-related candidate genes in barley.
Keywords: DsRed, particle bombardment, RNAi, single cell
Introduction
Land plants have evolved a number of strategies to cope with periods of moderate to severe drought. These include pre-formed escape strategies such as early flowering, avoidance such as deep rooting, enhanced water uptake efficiency, or reduced water loss, as well as tolerance mechanisms that include maintenance of root growth under water limitation, the accumulation of osmotically active substances, antioxidants, and proteins that protect other protein (complexes) or membrane systems in root and shoot (Ingram and Bartels, 1996). There is an ongoing debate as to whether the exploitation of avoidance or tolerance mechanisms should be the focus of plant breeding programmes. However, it appears likely that the exploitation of tolerance mechanisms may be more promising for the stabilization of crop yield under severe drought conditions as encountered in near-east or African countries, as well as in Australia, although the corresponding adapted breeding material might have a lower yield potential (Araus et al., 2002).
Transcriptome analysis, mostly in the model plant Arabidopsis thaliana, together with the phenotyping of mutated or transgenic plants has revealed a number of regulatory as well as protective proteins that are involved in the tolerance of plants to drought (Shinozaki and Yamaguchi-Shinozaki, 2007). The identified regulatory proteins can be roughly divided into the ones participating in abscisic acid (ABA)-dependent and the ones involved in ABA-independent signalling pathways (Riera et al., 2005). The former group includes transcription factors of the MYB/MYC class, the bZIP class (AREB/ABF) binding to the ABA response element (ABRE), plus NAC transcription factors, whereas the latter includes the Apetala2-like transcription factors (DREB2) that bind to drought-response elements (DREs). These transcription factors control the expression of genes involved in drought response and tolerance.
In the Triticeae cereals barley and wheat, a number of drought stress-related candidate genes have been identified, based on expression profiling. Two multigene families of barley that have been subjected to deeper analysis with respect to gene family organization and regulation of individual members encode dehydrin (Dhn) and C-repeat-binding factor (CBF) proteins (Choi et al., 1999; Skinner et al., 2005). However, only very few drought-related candidate genes have been tested directly in transgenic barley and wheat plants. These include a DREB-encoding gene from soybean, the HVA1-encoding gene of barley, the DREB1A-encoding gene of Arabidopsis, and the mtlD gene of Escherichia coli encoding a mannitol-1-phosphate dehydrogenase (Abebe et al., 2003; Pellegrineschi et al., 2004; Bahieldin et al., 2005; Gao et al., 2005). Although genetic [quantitative trait locus (QTL)] or association-genetic data related to candidate genes of the drought response in Triticeae cereals have been reported, no clear evidence for an involvement of any of the discussed genes is currently available (Cattivelli et al., 2002; Diab et al., 2004; Tondelli et al., 2006; Qian et al., 2007).
In order to assess gene function directly in barley and wheat suffering from biotic stress caused by the powdery mildew fungus Blumeria graminis, a transient assay system based on bombarded leaf epidermis was developed and proved to be useful (Panstruga, 2004; Dong et al., 2006; Trujillo et al., 2006; Zimmermann et al., 2006; Shen et al., 2007). This system, which can be used for transient overexpression of genes as well as for transient induced gene silencing (TIGS), has recently been further developed by using GATEWAY technology and microscope robotics in order to enhance throughput (Ihlow and Seiffert, 2004; Douchkov et al., 2005; Ihlow et al., 2008). The resulting phenomics tool now allows testing of hundreds to a few thousands of candidate or arbitrarily chosen genes in barley for a role in supporting or warding off powdery mildew, a model for other biotic plant–pathogen interactions.
Here, a medium- to high-throughput phenomics tool for testing dehydration stress-related genes of barley is described. Proof of concept was obtained with four candidate genes that enhanced susceptibility to dehydration stress in epidermal cells upon TIGS.
Materials and methods
Plant growth
Three different barley cultivars, cv. Golden Promise, cv. Steptoe, and cv. Morex, were used for investigations on dehydration stress response. Seeds were grown in pots of compost soil (from IPK nursery) in a growth chamber [16 h light from metal halogen lamps (120 μmol m2 s−1 at plant level), 8 h darkness, 55% (light period) to 70% (dark perdiod) relative humidity, 20 °C constant temperature].
Preparation of RNAi constructs
Sequences of the target genes were amplified from cDNA clones by PCR using a universal forward primer and a specific reverse primer (Supplementary Table S1 available at JXB online). Specific primers were designed by the software program Lasergene (DNASTAR, Madison, WI, USA). The resulting PCR fragments with an average length of ∼500 bp were purified by MinElute 96 UF PCR purification plates (Qiagen, Hilden, Germany) and ligated into the SwaI site of vector pIPKTA38 according to Douchkov et al. (2005). The ligation reactions were used for transformation of chemically competent Escherichia coli TOP10 cells (Invitrogen, Karlsruhe, Germany), and spread on LB medium containing kanamycin (50 μg ml−1). One colony of each cloning reaction was used for plasmid DNA isolation with the QIAprep® Miniprep Kit (Qiagen). Control digestion of pIPKTA38 clones was done with EcoRI to confirm the presence of inserts. PCR fragments in pIPKTA38 were recombined into the RNA interference (RNAi) destination vector pIPKTA30N as inverted repeats by using the Gateway LR clonase (Invitrogen). The entire reaction volume of 6 μl contained 1 μl of LR clonase mix II, 1 μl of pIPKTA30 destination vector (150 ng μl−1), 1 μl of the pIPKTA38 donor vector, and 3 μl of H2O. The recombination reaction was carried out overnight at room temperature (Douchkov et al., 2005), and 5 μl of the reaction were used to transform 50 μl of chemically competent E. coli TOP10 cells. The cells were spread onto LB medium containing ampicillin (100 μg ml−1) and incubated overnight at 37 °C. Plasmid DNA isolation of one clone per LR reaction was done with the QIAprep® Miniprep Kit. The presence of both inverted repeats in the final RNAi constructs was confirmed by EcoRV digestion.
Microprojectile bombardment and dehydration stress treatment
Segments of five primary leaves (with a length of ∼6 cm) were detached from 7-d-old barley seedlings and placed onto 0.5% (w/v) water–phytoagar (Duchefa) containing 10 mg l−1 benzimidazole as senescence inhibitor.
Gold particles (diameter 1 μm) were coated with plasmid DNA, consisting of three different vectors, pGFP (Schweizer et al., 1999) for normalization, pUbi-DsRed-nos for assessment of RNAi effects (Panstruga et al., 2003), and pIPKTA30_Target (RNAi vector) as described (Douchkov et al., 2005). The bombardment was performed by using a PDS-1000/He Biolistic® Particle Delivery System (BioRad) with a helium pressure of 900 psi. Bombarded leaf segments were incubated in closed Petri dishes in a climatized room [18 °C constant temperature, natural (indirect) daylight, 30 μmol m2 s−1]. These light conditions were found largely to reduce senescence of detached leaves, compared with artificial light of different types. Supplementary light from fluorescent tubes (Philips TLD 36W) in order to extend short days to 16 h also accelerated leaf senescence and was therefore omitted (data not shown). At 24 h post-bombardment, green fluorescent protein (GFP)-expressing epidermal cells were counted by using a Zeiss Axioplan 2 imaging microscope. For the subsequent exposure to dehydration stress the leaf segments were placed onto filter paper (Type 813, Macherey-Nagel, Düren, Germany) and air-dried to a fresh weight of 60%, relative to the initial fresh weight measured immediately before the dehydration treatment (RFW). Where indicated, leaf segments were exposed to more severe stress and dehydrated to 50% or 55% RFW. The dehydration treatment was carried out in the laboratory at 20 °C, 40±5% relative air humidity, daylight conditions, and lasted 90±30 min. Control leaf segments remained on water–phytoagar. After the dehydration-stressed leaves had reached the desired RFW, the filter paper was moisturized with 0.35 ml of H2O and the Petri dishes of 9 cm diameter were closed. Sealing the plates with Parafilm® (Pechiney, Chicago, IL, USA) caused constant dehydration stress conditions during the entire experiment. Dehydration-stressed and control leaf segments were incubated for 4 d as described above. At the end of the stress treatment, RFW was measured again and DsRed-expressing epidermal cells were counted in control and dehydration-stressed leaves under the microscope. The DsRed/GFP ratio was finally calculated in control and stressed leaf segments.
Fluorescence microscopy of GFP and DsRED
For GFP and DsRed detection, single-cell fluorescence was examined 24 h (GFP) or 120 h (DsRed) post-bombardment by using a Zeiss Axioplan 2 imaging microscope with the filter set BP450–490 excitation, FT510 beam splitter, BP515–565 emission for GFP fluorescence, and the filter set BP546/12 excitation, FT580 beam splitter, BP590 emission for DsRed detection. The estimation of GFP-expressing cells was done for one row of optical fields that runs from the top to the bottom of each bombarded leaf segment. The quantification of DsRed-expressing cells was done in the entire bombarded leaf segment. For RNAi silencing experiments with GFP fusion constructs, the numbers of GFP- and DsRed-expressing cells were counted in the entire bombarded leaf segments.
GFP fusion constructs
The plasmid pIPKTA40_HvDREB1 was constructed by inserting the PCR-amplified target gene sequence from cDNA clone HS08A22 as an AscI/SbfI fragment into the multiple cloning site of the vector pIPKTA40 in front of GFP (Supplementary Fig. S1 at JXB online). The PCR amplification was done with the proofreading Thermal Ace polymerase (Invitrogen, Karlsruhe, Germany) by using primers 5′-ACGAGGCGCGCCGAGATCTCTCTCTCCCTTCCTCCCTCCTC and 5′-CAAATCCTGCAGGCATTTCGGTTTCACCTTAAGGCCCACA-GT (AscI and SbfI sites underlined).
The plasmid pIPKTA40_HvDRF1 was constructed in a similar manner with the following modifications. First, site-directed mutagenesis was performed with the Quikchange® Mutagenesis Kit (Stratagene) to correct for a frameshift that occurred within the coding region of the selected cDNA clone. Secondly, a putative protein destabilization domain was deleted by combining two PCR fragments. For this purpose, two fragments of the HvDRF1 sequence with a corrected open reading frame were amplified by using the primer pair I (5′-AGAAGGCGCGCCGCGCGATTGCGAGCTCTAGATACCTC and 5′-CCATACATTGCTCTGGCTGGTCGACCATAAGC, SalI restriction site underlined) plus primer pair II (5′-CCTCTCGGTCAGATTGTCGACAGTCCACCC and 5′-GGCCACCTGCAGGCACAACCCCTCAAAGAACTCGCTCATCTC; SalI restriction site underlined). The fragments were digested with either with AscI/SalI or with SalI/SbfI and ligated together into AscI/SbfI-digested pIPKTA40 in front of GFP. Finally the GFP-coding sequence was amplified from plasmid pIPKTA40 by using the forward primer 5′-GGAGCCCGGGTCACCATGGTGAGCAAGG (adding a start ATG to the GFP sequence) and the reverse primer 5′-GCGGCGCGCCCTGCAGTAACTTGTAC. The ends of the amplicon were trimmed with XmaI/AscI and inserted into pIPKTA40 already carrying the modified HvDRF1 sequence. The resulting construct contained an internally deleted version of HvDRF1 as a translational fusion between two GFP sequences (Supplementary Fig. S1 at JXB online).
Transcript abundance
Transcripts of candidate genes were quantified by reverse transcription, real-time PCR. For this purpose, total RNA was extracted from control or treated leaves at different time points after the onset of dehydration stress. cDNA was synthesized from total RNA by using 1 μg of DNA-free RNA and the iScript Synthesis Kit (Bio-Rad, Munich, Germany) in a 20 μl reaction. A 1 μl aliquot of cDNA was used as template for real-time PCR on a 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) by using the QuantiTect SYBR Green Kit (Qiagen) according to the manufacturer's manual, except that the total reaction volume was reduced to 10 μl. Specific primer pairs were designed by the software program Lasergene (DNASTAR) (Supplementary Table S2 at JXB online). A gene encoding a ubiquitin-conjugating enzyme (NUBC, TIGR unigene TC139190 from assembly 9.0) was used for normalization of the transcript abundances. Two biological replicates with two technical replicates each were carried out.
Statistical analysis of TIGS effects
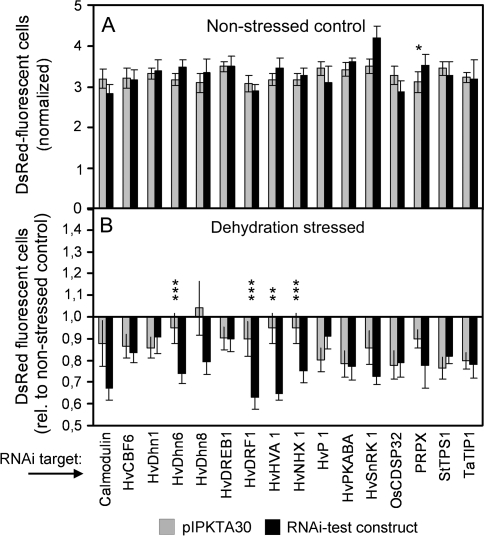
Supplementary Table S3 at JXB online shows two types of statistical analysis of TIGS effects carried out by using GraphPad InStat 3 software. First, mean numbers of DsRed-fluorescing cells in dehydration-stressed leaves (normalized to non-stressed control) were compared between RNAi test constructs and pIPKTA30N empty vector control (paired two-sample t-test). Alternatively the ratio (column_4/column_3) of the normalized number of DsRed fluorescent cells in dehydration-stressed leaves after bombardment with the RNAi test construct or the empty vector control was calculated. Statistical significance was tested by using the one-sample t-test against the hypothetical value ‘1’. This statistical test was found to be less sensitive to outliers and, therefore, less prone to false-negative results. Therefore, the one-sample t-test was used for the initial identification of candidate genes. However, for statistical analysis of the final data (usually based on 10 independent experiments), a paired two-sample t-test of mean values from the RNAi test construct versus empty vector control was applied (see Fig. 3B).
Fig. 3.
Effect of TIGS of candidate genes on the number of DsRed fluorescent cells in non-stressed control and dehydration-stressed leaf segments. Leaf segments were co-bombarded with DsRed and GFP expression plasmids, plus an RNAi construct targeting the barley genes listed below the graph. If no annotated target gene is known in barley, the name of the closest homologue from another plant species is given (see also Table 2). After a dehydration stress period of 4 d, the number of DsRed fluorescent cells was counted and normalized to GFP. During the stress period, control leaves were incubated on water–agar. (A) Number of DsRed fluorescent cells in non-stressed control leaves in the presence of the empty RNAi vector pIPKTA30 or the RNAi test construct. Mean ±SEM of five independent experiments, except for RNAi constructs targeting HvDRF1, HvDhn6, HVA1, and HvNHX1 where combined data from two series of five experiments each are shown. An asterisk indicates a statistically significant difference (P < 0.05). (B) Decrease in the number of DsRed fluorescent cells in dehydration-stressed leaf segments, compared with non-stressed control leaves, in the presence of empty RNAi vector pIPKTA30 or RNAi test construct. Mean ±SEM of five independent experiments, except for RNAi constructs targeting HvDRF1, HvDhn6, HVA1, and HvNHX1 where combined data from two series of five experiments each are shown. Statistically significant difference at the **P < 0.01 and ***P < 0.001 level (two-sample t-test, two-sided).
Results
Detached leaf assay for dehydration tolerance
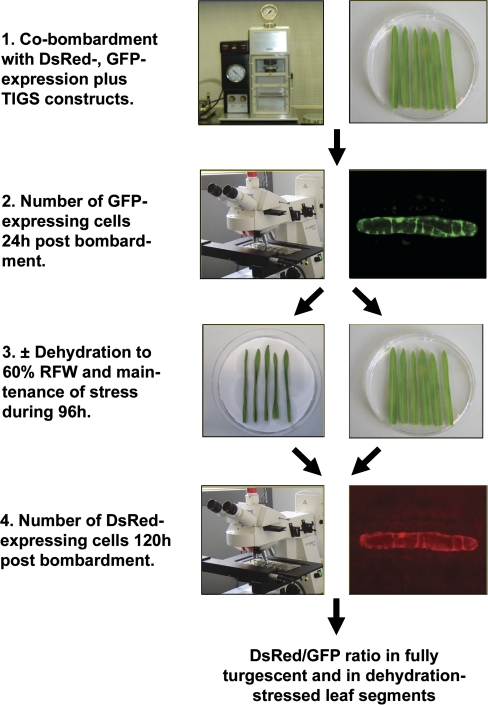
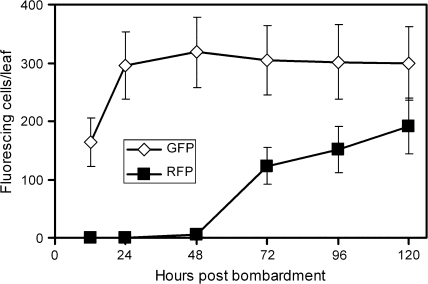
Cells suffering from drought or dehydration stress are potentially damaged in membrane integrity, protein folding, and redox status to name a few of the most prominent problems (Ingram and Bartels, 1996). The biochemical properties of the red fluorescent protein, DsRed, were exploited to report drought stress severity in bombarded epidermal cells of barley. DsRed is known to require several days for maturation into the fluorescent homo-tetrameric complex in animal and plant systems and to suffer from a shift from its red fluorescence to weak green fluorescence upon denaturation (Baird et al., 2000; Gross et al., 2000). It therefore appeared likely that DsRed fluorescence is sensitive to denaturing conditions imposed by severe drought stress over a period of 4 d. Moreover, enhanced proteolytic activity and cell death will also reduce the amount of mature, fluorescent DsRed. Figure 1 outlines the detached leaf assay for dehydration tolerance, which is based on co-bombardment of three plasmids: pGFP encoding the GFP for internal normalization of DsRed fluorescence, Ubi-DsRed-nos as reporter for cellular dehydration stress, and pIPKTA30_Target for knockdown of barley candidate genes by RNAi. Instead of the RNAi constructs, overexpression constructs could also be used. After the bombardment, GFP, immature (non-fluorescing) DsRed, as well as double-stranded RNA were allowed to accumulate in non-stressed conditions for a period of 24 h. Figure 2 shows that GFP fluorescence was fully developed 24 h post-bombardment, whereas DsRed fluorescence was still increasing after 4 d, as expected. After counting the number of GFP-expressing epidermal cells, 50% of the leaf segments were subjected to dehydration stress by partial desiccation until their RFW has reached 60% of the initial value. The other 50% of leaf segments were incubated on water–agar as fully turgescent, non-stressed controls. Over all bombardments where RFW was adjusted to 60% at the beginning of the 4 d stress period, RFW increased to 66.4±9.3% (mean ±SD, n=111) during the stress treatment in closed Petri dishes, indicating reproducible and approximately constant stress conditions. In order to compare RFW with RWC of leaves, which is a widely used parameter of drought stress (Barrs and Weatherley, 1962), RWC was calculated in a typical experiment. As shown in Supplementary Table S4 at JXB online, the experimental conditions chosen correspond to severe drought, with RWC of ∼60% after 4 d of stress (Teulat et al., 2003). At the end of the dehydration stress period, the number of DsRed-fluorescing cells was counted and normalized by the number of GFP-fluorescing cells observed in the same leaf segments before the onset of stress. In order to test whether DsRed reported dehydration stress in a reliable manner, we determined the normalized DsRed fluorescence in three dehydration-stressed barley genotypes. We found moderate reduction of the number of DsRed fluorescent cells in dehydrated leaf segments of cv Golden Promise and cv Morex, whereas the number was strongly decreased in cv Steptoe (Table 1). The stronger reduction in cv Steptoe, as compared with cv Morex, was found to be in agreement with a stronger vigour of cv Morex seedlings germinated and grown in the presence of 15% polyethylene glycol (K Neumann, IPK, personal communication). These result suggests that the decrease of the normalized number of DsRed fluorescent cells is a reliable read-out of dehydration stress in bombarded epidermal cells. However, it remains to be tested which barley genotype will be most suited for testing candidate genes by TIGS. So far, cv Golden Promise has been used, which exhibited only a moderate reduction of the number of DsRed-fluorescing cells under dehydration stress, despite its adaptedness to wet climate (UK). This suggests that TIGS of important genes for the relative robustness to dehydration of cv Golden Promise might result in a detectable decrease of DsRed fluorescence.
Fig. 1.
Flow chart of the TIGS screening system to assess gene function in dehydration-stressed barley leaves. RFW, relative fresh weight.
Fig. 2.
Kinetics of transient GFP and DsRed expression, reflected by the number of fluorescing epidermal cells per bombardment. The same bombarded leaf segments were repeatedly counted for the number of green- or red-fluorescing epidermal cells at the times indicated. Mean ±SEM of five leaf segments. The same experiment was repeated with similar results.
Table 1.
Reduction of the number of DsRed fluorescent cells in dehydration-stressed leaf segments from different barley cultivars
| Cultivar | DsRed (stressed/control)a | P (t-test)b | nc |
| Golden Promise | 0.84±0.03 | <0.0001 | 30 |
| Steptoe | 0.54±0.06 | 0.0017 | 5 |
| Morex | 0.86±0.09 | 0.18 | 5 |
Ratio of the number of DsRed fluorescent cells in dehydration-stressed leaf segments versus control leaf segments. Mean value ±SEM.
One-sample t-test versus hypothetical value ‘1’.
Number of independent bombardments.
TIGS of candidate genes
In order to obtain proof of concept for the TIGS system of dehydration-stressed leaves, a list of candidate genes were selected based on pre-existing knowledge. The list of 16 genes is shown in Table 2 and includes 14 candidates for which a role in drought tolerance has been proposed based on expression data or demonstrated by mutant or transgenic approaches (for references, see Table 2). In addition, two genes were included as negative controls that are probably not involved in abiotic stress tolerance. Expressed sequence tag (EST) clones corresponding to the selected gene candidates were identified and used for the generation of RNAi constructs (Supplementary Fig. S1 at JXB online). For the analysis of RNAi effects, two types of comparisons were made. First, the number of DsRed fluorescent cells was compared between RNAi test constructs and the empty vector control in non-stressed control leaf segments (Fig. 3A). With the exception of the construct targeting PRPX, which actually served as the control target because it encodes a pathogenesis-related protein, no significant effect was found. This indicates that silencing of drought stress-related genes in barley under non-stressed conditions does not generally impair cellular vitality. Secondly, the stress-induced decrease of the number of DsRed-fluorescing cells in the presence of empty RNAi vector was compared with the decrease in the presence of RNAi test constructs (Fig. 3B). Dehydration stress in the presence of the empty RNAi vector decreased the number of DsRed fluorescent cells in most cases, but this effect varied to a certain extent depending on the individual (five) experiments that contributed to the mean values shown. One of the major sources of variation may be leaf width, which varied to some extent from one experiment to the other and which was found to influence dehydration tolerance (data not shown). Four RNAi test constructs further decreased the number of DsRed-expressing cells in a statistically significant manner upon direct comparison with the corresponding control bombardments with empty RNAi vector. These four RNAi test constructs were targeting candidate genes HvDRF1, HvDHn6, HvHNX1, and HVA1. In order to test the initial results for reproducibility, a second set of five biological replicates was performed using the same four RNAi constructs mentioned above. In both experimental series a significant decrease of DsRed fluorescence was induced by the RNAi constructs in dehydration-stressed leaves (Supplementary Table S3 at JXB online). The available data from 10 independent experiments were used to calculate the minimum number of experiments required for TIGS effects of the four RNAi constructs to become statistically significant (P < 0.05). Supplementary Fig. S2 at JXB online shows that between three and five experiments were required to reveal the effects and that P-values usually dropped to <0.0001 with the final number of 10 experiments. It is therefore suggested to carry out at least five independent TIGS experiments in order to provide statistically robust data. Another indication of the robustness of the TIGS system comes from the series of 10 experiments with the RNAi construct that targets HvDRF1: during the first four and the remaining six experiments leaves were dehydrated to 50% and 60% initial RFW, respectively. In both series the TIGS effect of the construct was significant (DsRed ratio of test construct versus pIPKTA30 empty vector in stressed leaves below hypothetical value ‘1’ with P < 0.05). On the other hand, the mean ratio of DsRed fluorescence was not significantly different between the more and less severe stress conditions (P=0.12). This indicates that the phenotypic effect of the RNAi construct targeting HvDRF1 could be revealed over a range of stress intensity and allowed to combine data of all 10 experiments (Fig. 3). Taken together, the data presented here suggest that the TIGS system may be useful for revealing gene function in dehydration-stressed barley epidermal cells.
Table 2.
Dehydration stress-related candidate genes of barley tested by transient induced gene silencing (TIGS) in barley cv Golden Promise
| Clone ID | Function | BlastX hit (NCBI) | E-value | Identity | Accession no. | Remark | Reference |
| HO33D09 | Calmodulin | H. vulgare | 0 | 99% | M27303 | BlastN | Perruc et al. (2004) |
| HI09P16 | HvCBF6 (CBF2/DREB1-like) | H. vulgare | 4.0E-50 | 99% | AAX23701 | Oh et al. (2007) | |
| HV04I10 | HvDhn1 (Dehydrin 1) | H. vulgare | 6,0E-23 | 100% | AAF01689 | Suprunova et al. (2004) | |
| HC02P10 | HvDhn6 (Dehydrin 6) | H. vulgare | 4.0E-19 | 97% | AAF01694 | Suprunova et al. (2004) | |
| HO01L05 | HvDhn8 (Dehydrin 8) | H. vulgare | 2,0E-52 | 98% | AAD02259 | Choi et al. (1999) | |
| HS08A22 | HvDREB1 (DREB2-like) | H. vulgare | 2.0E-23 | 100% | AAY25517 | Sakuma et al. (2006a) | |
| HU05J23 | HvDRF1 (DREB2-like) | H. vulgare | 3.0E-51 | 100% | AAO38211 | Xue and Loveridge (2004) | |
| HV09A17 | HVA1 | H. vulgare | 2.0E-48 | 100% | CAA55041 | Bahieldin et al. (2005) | |
| HG01K10 | PRPX | H. vulgare | 2.0E-70 | 99% | CAA34641 | Negative control | Unpublished |
| HO16L13 | HvPKABA1 (SAPK1-like kinase) | H. vulgare | 2,0E-112 | 99% | BAB61736 | Yamauchi et al. (2002) | |
| HS03E24 | SnRK1-type protein kinase | H. vulgare | 2.0E-85 | 83% | CAA07813 | Negative control | Radchuk et al. (2006) |
| HQ01K06 | Sodium/proton antiporter HvHNX1 | H. vulgare | 1.0E-79 | 100% | BAC56698 | Fukuda et al. (2004) | |
| HO36E09 | Thioredoxin CDSP32 | O. sativa | 2.0E-90 | 87% | BAC75581 | Broin and Rey (2003) | |
| HR01K17 | Tonoplast intrinsic protein | T. aestivum | 2.0E-64 | 95% | ABI96816 | Peng et al. (2007) | |
| HZ41I22 | Trehalose-6-phosphate synthase 1 | S. lycopersicum | 1e-62 | 90% | ABO61742 | Karim et al. (2007) | |
| HO10E23 | Vacuolar pyrophosphatase | H.vulgare | 9E-76 | 90% | BAB18681 | (Park et al. 2005) |
TIGS of DREB2-like genes
Two DREB2-like genes were included in the list of 16 candidates: HvDRF1 and HvDREB1. As shown in Fig. 3B, TIGS of HvDRF1 clearly reduced DsRed fluorescence in dehydration-stressed leaves, whereas RNAi of HvDREB1 had no effect. These two genes were selected in order to test whether the difference in TIGS effects was due to specific silencing of two genes with different cellular functions or whether the RNAi construct targeting HvDREB1 was simply not efficient. For this aim, two translational Target:GFP fusion constructs were constructed for transient expression in barley epidermal cells, as shown in Supplementary Fig. S1 at JXB online. However, initial attempts to express a HvDRF1:GFP fusion construct resulted in no detectable GFP fluorescence. A previously identified destabilization domain lying immediately upstream from the AP2 domain of the transcription factor was therefore deleted (Sakuma et al., 2006b). Still, the fusion construct did not give rise to a fluorescent protein. Finally, a ‘sandwich’ construct with the engineered HvDRF1 cDNA between two GFP open reading frames yielded a green-fluorescing fusion product. Supplementary Fig. S3 shows an example of GFP fluorescence of both constructs. The HvDREB1:GFP fusion protein was localized in epidermal nuclei, as expected (Supplementary Fig. S3C, D). In contrast, no preferred nuclear localization of the GFP:HvDRF1:GFP fusion protein was found (Supplementary Fig. S3E, F), which was probably due to masking of potential nuclear localization signal(s) by the GFP ‘sandwich’. Each of the two DREB2-like:GFP fusion constructs was then co-bombarded with either of the two RNAi constructs targeting one of the DREB2-like genes, plus with pUbi-DsRed-nos, followed by counting the number of GFP-fluorescing cells (normalized to DsRed expression). A highly significant and target-specific reduction of GFP-fluorescing cells was observed, indicating that both RNAi constructs were causing efficient silencing and that no cross-silencing between the two RNAi targets occurred (Table 3). The fact that GFP fluorescence did not drop to zero in the presence of a matching RNAi construct may reflect some lagging of RNAi onset behind the accumulation of the GFP fusion proteins, which resulted in a higher background GFP fluorescence.
Table 3.
Efficiency and specificity of silencing of two members of the DREB2-like gene family of barley
| RNAi construct | HvDRF1:GFP expressiona | nb | HvDREB1:GFP expressiona | nb |
| pIPKTA30 | 100 | 20 | 100 | 30 |
| pIPKTA30_HvDRF1 | 60.0±5.8 (P < 0.0001)c | 20 | 130.5±15.4 (P=0.058)c | 30 |
| pIPKTA30_HvDREB1 | 105.3±9.8 (P=0.598)c | 20 | 56.8±7.7 (P < 0.0001)c | 30 |
Fusion:GFP expression, normalized to co-bombarded pUbi-DsRed-Nos, was compared with the expression in the presence of the empty RNAi vector pIPKTA30 (set to 100%).
Number of analysed leaf segments.
One-sample t-test versus hypothetical value 100.
RNAi constructs may produce off-target effects, which will lead to false conclusions about gene function. This question was addressed with additional constructs targeting different regions of the HvDRF1 mRNA. Figure 4 shows that all three tested constructs produced a very similar, statistically significant reduction of the number of DsRed-fluorescing cells in dehydrated leaf segments compared with the empty vector control. It was therefore concluded that the observed RNAi effect was due to reduced HvDRF1 mRNA in the transformed cells.
Fig. 4.
Similar effect of RNAi constructs targeting different regions of the HvDRF1 mRNA. The horizontal lines below the mRNA indicate the RNAi target region of the corresponding RNAi construct. Numbers below the lines indicate the ratio of the number of DsRed fluorescent cells in dehydration-stressed leaves after bombardment with RNAi test construct or the empty vector control (see also Supplementary Table S1 at JXB online). Mean values ±SEM from 9–10 independent experiments. P-values, two-sample t-test, two-sided.
Gene regulation
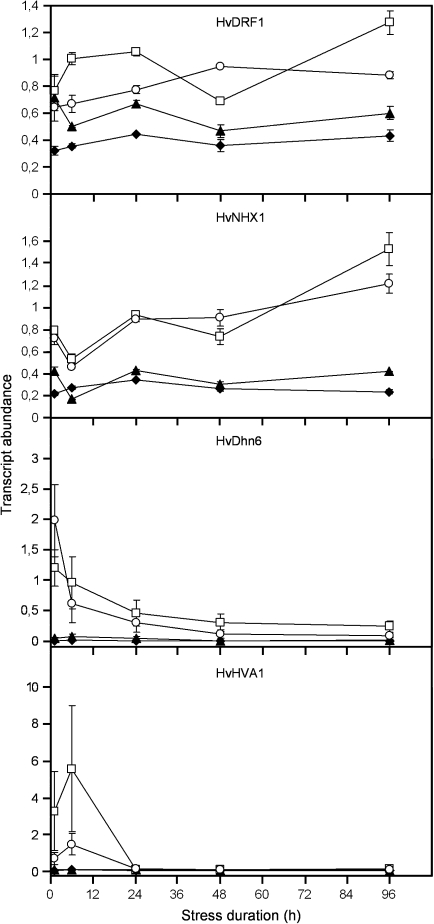
The four candidate genes reducing the number of DsRed-fluorescent cells upon TIGS in a dehydration-specific manner might be important for drought tolerance and, therefore, differentially regulated by the dehydration stress. To test this hypothesis, quantitative reverse transcription real-time PCR (RT-qPCR) was carried out in detached leaf segments. As shown in Fig. 5, mRNAs of all four functionally validated genes were up-regulated during the dehydration treatment in non-bombarded as well as in bombarded leaf segments. However, the kinetics of induction were clearly different between the four genes. The mRNA of HvDRF1 and HvNHX1 accumulated during the entire stress period, whereas the mRNA of HVA1 and HvDhn6 reached a maximum already at 6 h and 1 h after the onset of dehydration, respectively, and had returned to basal levels by the end of the experiment. The reproducibility of the regulation and normalized transcript levels of the four selected candidate genes during dehydration stress is shown in Supplementary Fig. S4 at JXB online. Together with the data shown in Supplementary Table S4, it is assumed that the stress conditions of bombarded leaf segments were sufficiently standardized to allow reproducible phenotypic effects of TIGS. In conclusion, although transcript accumulation was probably not restricted to the epidermis where TIGS phenotypes were observed, the combination of gene regulation and TIGS data does provide convergent evidence and might be used as filter set for longer lists of candidate genes possibly related to dehydration stress.
Fig. 5.
Transcript profiles of candidate genes in dehydration-stressed leaf segments. Transcripts were quantified by reverse transcription, real-time PCR of leaf RNA isolated during the dehydration-stress period (open symbols) or during control incubation on water–agar (filled symbols). RNA was extracted from either non-bombarded (filled triangles, open squares) or bombarded (filled diamonds, open circles) leaf segments. The time at which the relative fresh weight of detached, air-dried leaf segments had reached 55% was set to zero. Transcript abundance is expressed as the ratio between the gene of interest and the control gene NUBC. Mean values ±range from two independent PCR experiments using the same cDNA samples are shown.
Discussion
Functional genomic approaches require the availability of phenomics tools such as high-throughput assessment of quantitative phenotypes depending on natural or induced genetic variation. The TIGS system described here for testing dehydration stress-related genes represents a second phenomics tool in barley, after the establishment of a transient assay system for testing defence-related genes against the powdery mildew pathogen (Douchkov et al., 2005). The availability of the two TIGS screening tools now allows for testing of hundreds up to a few thousand genes for a potential role in dehydration tolerance or pathogen resistance. Importantly, this can be done directly in relevant cultivars of the target crop without the need to refer to model systems such as Nicotiana benthamiana, Arabidopsis thaliana, mammalian cell lines, or yeast, where high throughput phenomics has been demonstrated to be feasible (Warringer et al., 2003; Brigneti et al., 2004; Silva et al., 2004; Devi et al., 2006). The current throughput of the TIGS system for testing genes of dehydration tolerance is ∼40 (candidate) genes per person month. However, the system has the potential for throughput enhancement by robotizing the microscopic analysis, as has been achieved in the TIGS screening system for pathogen resistance (Ihlow et al., 2008). A throughput of 80–100 genes per person month is therefore estimated to be a realistic goal, from the moment where libraries of either RNAi or (over)expression constructs are ready for bombardments. In summary, the TIGS system for dehydration-related genes may be termed medium to high throughput, especially when compared with the other functional genomics tools in barley such as TILLING or transgenic plants that have an estimated throughput of one gene per person month including the phenotyping (Caldwell et al., 2004; Zhao et al., 2006). Moreover, complementation or transient overexpression assays can also be performed in the transient single-cell system.
The TIGS system for dehydration stress tolerance can address gene function with respect to a few aspects of the overall drought-related problem. First of all, genes involved in avoidance traits such as cuticle or epicuticular wax thickness and composition, or stomatal opening cannot be tested. Because target genes are only silenced in single epidermal cells, genes that are involved in the mobility of compatible solutes also cannot be tested. What our test system should be able to reveal is cellular survival, protection from protein denaturation and damage, plus enzymatic protein degradation. Most probably, both water loss and abundance of reactive oxygen species such as H2O2 can inhibit DsRed maturation. Which of these stress consequences are more detrimental for DsRed fluorescence cannot be judged at the moment, due to a lack of a larger amount of tested protective genes against either of these consequences.
By using a limited number of well described candidate genes based on pre-existing knowledge, proof of concept of the TIGS system in dehydration-stressed barley could be obtained. These genes encode the DREB2-like transcription factor HvDRF1, the dehydrin HvDhn6, the HVA1 protein belonging to the LEA family, and the vacuolar sodium–proton antiporter HvNHX1. Interestingly, silencing of HVA1, one of the direct downstream targets of HvDRF1 (Xue and Loveridge, 2004), also caused a significant reduction of DsRed maturation, indicating that the TIGS system is sensitive enough to reveal gene function not only for key regulators (HvDRF1) with a predicted epistatic effect but also for important downstream protective genes. HvDFR1 mRNA was present at a quite high basal level but TIGS did not reduce DsRed fluorescence in non-stressed leaves. This suggests that HvDRF1 protein undergoes post-translational activation upon stress or that the probably pre-existing, functional protein is not involved in housekeeping cellular functions. The effect of HVA silencing does not come as a surprise because this gene has been shown to mediate drought tolerance in a series of transgenic approaches in different plant species including wheat (Xu et al., 1996; Bahieldin et al., 2005; Fu et al., 2007). For HvNHX1, a beneficial role in drought stress tolerance has also been shown in transgenic plants (Brini et al., 2007). The effect of HvDRF1 and HvDhn6 silencing shown here is the first direct demonstration of a protective function of the encoded proteins, although a protective effect of other dehydrins on enzymatic activity in vitro has been previously reported (Reyes et al., 2005). Taken together, these four functionally validated gene candidates represent leads for molecular marker development in barley, in order to test co-segregation and association with QTLs for drought tolerance in barley mapping and association genetic populations, respectively. Another potentially promising follow-up approach to TIGS would be the generation of transgenic barley plants carrying either corresponding RNAi constructs or introgressed gene haplotypes of drought-tolerant genotypes.
The small set of candidate genes tested here included two and three members of the DREB2-like and Dhn multigene families, respectively. However, only one RNAi construct per gene family exhibited a significant phenotypic effect. In the case of HvDRF1 and HvDREB1 (both DREB2-like), it is shown that the construct specificity of reduced numbers of DsRed fluorescent cells was indeed due to target-specific silencing because both constructs caused only degradation of the corresponding target:GFP fusion RNA. This opens up the possibility to carry out systematic TIGS of multigene families in barley such as Dhn or CBF, in order to obtain a clearer picture of the physiological role of their individual members. However, family-wide TIGS approaches would require a more sophisticated construct design than the one applied here because all three RNAi constructs targeting Dhn genes were found to have largely overlapping specificities, as determined by the software tool ‘siRNA Scan’ (Xu et al., 2006). It appears therefore more likely that differences in phenotypic effects of the three Dhn–RNAi constructs was due to different RNAi efficiency, which appears possible because target sequences were largely non-overlapping (data not shown). The RNAi construct targeting HvDhn8 also showed an effect that was not statistically significant due to a higher variability of empty vector control data and that therefore might be a false-negative result. The throughput of the TIGS system will allow the testing of lists of candidate genes that have been published to be up-regulated in dehydrated or drought-stressed barley to come up with a first functional inventory of the genes underlying dehydration tolerance and, therefore, tolerance to extreme drought, in barley (Talame et al., 2007). The extension of the DsRed-based TIGS system to cold tolerance in barley might be a further option worth testing.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of PCR primers for the generation of inverted repeat RNAi constructs.
Table S2. List of PCR primers for reverse transcription, real-time PCR.
Table S3. Reproducible phenotypic effect of RNAi constructs targeting four candidate genes.
Table S4. Relationship between relative fresh weight (RFW) and relative water content (RWC) of bombarded barley leaf segments.
Fig. S1. Schematic representation of constructs for transient expression of GFP fusion proteins and for TIGS.
Fig. S2. Minimum number of independent TIGS experiments required for statistically significant effects of four candidate genes.
Fig. S3. Fluorescence of GFP wild-type and DREB:GFP fusion proteins upon transient expression in barley epidermal cells.
Fig. S4. Reproducibility of drought-induced gene regulation in bombarded leaf segments.
Supplementary Material
Acknowledgments
This work was supported by WTZ grant HUN 04/A01 from the German Ministry of Education and Research (BMBF) and by Grant 3593A/0405T from the state of Saxonia-Anhalt, Germany. We would like to acknowledge valuable comments made by two anonymous reviewers.
Glossary
Abbreviations
- ABA
abscisic acid
- bZIP
basic leucine zipper factor
- DREB
drought response element-binding
- DsRed
red fluorescent protein from Discosoma sp.
- Hv
Hordeum vulgare
- MYB
transcription factor related to v-Myb from the avian myeloblastosis virus
- MYC
transcription factor related to v-Myc from the avian myelocytomatosis
- RFW
relative fresh weight
- RWC
relative water content
- TIGS
transient induced gene silencing
References
- Abebe T, Guenzi AC, Martin B, Cushman JC. Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiology. 2003;131:1748–1755. doi: 10.1104/pp.102.003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus JL, Slafer GA, Reynolds MP, Royo C. Plant breeding and drought in C-3 cereals: what should we breed for? Annals of Botany. 2002;89:925–940. doi: 10.1093/aob/mcf049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahieldin A, Mahfouz HT, Eissa HF, Saleh OM, Ramadan AM, Ahmed IA, Dyer WE, El-Itriby HA, Madkour MA. Field evaluation of transgenic wheat plants stably expressing the HVA1 gene for drought tolerance. Physiologia Plantarum. 2005;123:421–427. [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proceedings of the National Academy of Sciences, USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrs HD, Weatherley PE. A re-examination of relative turgidity technique for estimating water deficits in leaves. Australian Journal of Biological Sciences. 1962;15:413. [Google Scholar]
- Brigneti G, Martin-Hernandez AM, Jin HL, Chen J, Baulcombe DC, Baker B, Jones JDG. Virus-induced gene silencing in Solanum species. The Plant Journal. 2004;39:264–272. doi: 10.1111/j.1365-313X.2004.02122.x. [DOI] [PubMed] [Google Scholar]
- Brini F, Hanin M, Mezghani I, Berkowitz GA, Masmoudi K. Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. Journal of Experimental Botany. 2007;58:301–308. doi: 10.1093/jxb/erl251. [DOI] [PubMed] [Google Scholar]
- Broin M, Rey P. Potato plants lacking the CDSP32 plastidic thioredoxin exhibit overoxidation of the BAS1 2-cysteine peroxiredoxin and increased lipid peroxidation in thylakoids under photooxidative stress. Plant Physiology. 2003;132:1335–1343. doi: 10.1104/pp.103.021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.) The Plant Journal. 2004;40:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- Cattivelli L, Baldi P, Crosatti C, Di Fonzo N, Faccioli P, Grossi M, Mastrangelo AM, Pecchioni N, Stanca AM. Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Molecular Biology. 2002;48:649–665. doi: 10.1023/a:1014824404623. [DOI] [PubMed] [Google Scholar]
- Choi DW, Zhu B, Close TJ. The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theoretical and Applied Genetics. 1999;98:1234–1247. [Google Scholar]
- Devi SR, Chen X, Oliver DJ, Xiang CB. A novel high-throughput genetic screen for stress-responsive mutants of Arabidopsis thaliana reveals new loci involving stress responses. The Plant Journal. 2006;47:652–663. doi: 10.1111/j.1365-313X.2006.02814.x. [DOI] [PubMed] [Google Scholar]
- Diab AA, Teulat-Merah B, This D, Ozturk NZ, Benscher D, Sorrells ME. Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theoretical and Applied Genetics. 2004;109:1417–1425. doi: 10.1007/s00122-004-1755-0. [DOI] [PubMed] [Google Scholar]
- Dong WB, Nowara D, Schweizer P. Protein polyubiquitination plays a role in basal host resistance of barley. The Plant Cell. 2006;18:3321–3331. doi: 10.1105/tpc.106.046326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Molecular Plant-Microbe Interactions. 2005;18:755–761. doi: 10.1094/MPMI-18-0755. [DOI] [PubMed] [Google Scholar]
- Fu DL, Huang BR, Xiao YM, Muthukrishnan S, Liang GH. Overexpression of barley hva1 gene in creeping bentgrass for improving drought tolerance. Plant Cell Reports. 2007;26:467–477. doi: 10.1007/s00299-006-0258-7. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Chiba K, Maeda M, Nakamura A, Maeshima M, Tanaka Y. Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter from barley. Journal of Experimental Botany. 2004;55:585–594. doi: 10.1093/jxb/erh070. [DOI] [PubMed] [Google Scholar]
- Gao SQ, Xu HJ, Cheng XG, Chen M, Xu ZS, Li LC, Ye XG, Du LP, Hao XY, Ma YZ. Improvement of wheat drought and salt tolerance by expression of a stress-inducible transcription factor GmDREB of soybean (Glycine max) Chinese Science Bulletin. 2005;50:2714–2723. [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proceedings of the National Academy of Sciences, USA. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihlow A, Schweizer P, Seiffert U. A high-throughput screening system for barley/powdery mildew interactions based on automated analysis of light micrographs. BMC Plant Biology. 2008 doi: 10.1186/1471-2229-8-6. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Karim S, Aronsson H, Ericson H, Pirhonen M, Leyman B, Welin B, Mantyla E, Palva ET, Van Dijck P, Holmstrom KO. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Molecular Biology. 2007;64:371–386. doi: 10.1007/s11103-007-9159-6. [DOI] [PubMed] [Google Scholar]
- Oh SJ, Kwon CW, Choi DW, Song SI, Kim JK. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnology Journal. 2007;5:646–656. doi: 10.1111/j.1467-7652.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- Panstruga R. A golden shot: how ballistic single cell transformation boosts the molecular analysis of cereal–mildew interactions. Molecular Plant Pathology. 2004;5:141–148. doi: 10.1111/j.1364-3703.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- Panstruga R, Kim MC, Cho MJ, Schulze-Lefert P. Testing the efficiency of dsRNAi constructs in vivo: a transient expression assay based on two fluorescent proteins. Molecular Biology Reports. 2003;30:135–140. doi: 10.1023/a:1024945920331. [DOI] [PubMed] [Google Scholar]
- Park S, Li JS, Pittman JK, Berkowitz GA, Yang HB, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proceedings of the National Academy of Sciences, USA. 2005;102:18830–18835. doi: 10.1073/pnas.0509512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- Peng YH, Lin WL, Cai WM, Arora R. Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta. 2007;226:729–740. doi: 10.1007/s00425-007-0520-4. [DOI] [PubMed] [Google Scholar]
- Perruc E, Charpenteau M, Ramirez BC, Jauneau A, Galaud JP, Ranjeva R, Ranty B. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. The Plant Journal. 2004;38:410–420. doi: 10.1111/j.1365-313X.2004.02062.x. [DOI] [PubMed] [Google Scholar]
- Qian G, Han ZX, Zhao T, Deng GB, Pan ZF, Yu MQ. Genotypic variability in sequence and expression of HVA1 gene in Tibetan hulless barley, Hordeum vulgare ssp vulgare, associated with resistance to water deficit. Australian Journal of Agricultural Research. 2007;58:425–431. [Google Scholar]
- Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiology. 2006;140:263–278. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Rodrigo MJ, Colmenero-Flores JM, Gil JV, Garay-Arroyo A, Campos F, Salamini F, Bartels D, Covarrubias AA. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant, Cell and Environment. 2005;28:709–718. [Google Scholar]
- Riera M, Valon C, Fenzi F, Giraudat J, Leung J. The genetics of adaptive responses to drought stress: abscisic acid-dependent and abscisic acid-independent signalling components. Physiologia Plantarum. 2005;123:111–119. [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell. 2006a;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proceedings of the National Academy of Sciences, USA. 2006b;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Abderhalden O, Dudler R. A transient assay system for the functional assessment of defense-related genes in wheat. Molecular Plant-Microbe Interactions. 1999;12:647–654. [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proceedings of the National Academy of Sciences, USA. 2004;101:6548–6552. doi: 10.1073/pnas.0400165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Molecular Biology. 2005;59:533–551. doi: 10.1007/s11103-005-2498-2. [DOI] [PubMed] [Google Scholar]
- Suprunova T, Krugman T, Fahima T, Chen G, Shams I, Korol A, Nevo E. Differential expression of dehydrin genes in wild barley, Hordeum spontaneum, associated with resistance to water deficit. Plant, Cell and Environment. 2004;27:1297–1308. [Google Scholar]
- Talame V, Ozturk NZ, Bohnert HJ, Tuberosa R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. Journal of Experimental Botany. 2007;58:229–240. doi: 10.1093/jxb/erl163. [DOI] [PubMed] [Google Scholar]
- Teulat B, Zoumarou-Wallis N, Rotter B, Ben Salem M, Bahri H, This D. QTL for relative water content in field-grown barley and their stability across Mediterranean environments. Theoretical and Applied Genetics. 2003;108:181–188. doi: 10.1007/s00122-003-1417-7. [DOI] [PubMed] [Google Scholar]
- Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca AM, Pecchioni N. Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theoretical and Applied Genetics. 2006;112:445–454. doi: 10.1007/s00122-005-0144-7. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Altschmied L, Schweizer P, Kogel KH, Huckelhoven R. Respiratory burst oxidase homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp hordei. Journal of Experimental Botany. 2006;57:3781–3791. doi: 10.1093/jxb/erl191. [DOI] [PubMed] [Google Scholar]
- Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proceedings of the National Academy of Sciences, USA. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DP, Duan XL, Wang BY, Hong BM, Ho THD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiology. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiology. 2006;142:429–440. doi: 10.1104/pp.106.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Loveridge CW. HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. The Plant Journal. 2004;37:326–339. doi: 10.1046/j.1365-313x.2003.01963.x. [DOI] [PubMed] [Google Scholar]
- Yamauchi D, Zentella R, Ho TH. Molecular analysis of the barley (Hordeum vulgare L.) gene encoding the protein kinase PKABA1 capable of suppressing gibberellin action in aleurone layers. Planta. 2002;215:319–326. doi: 10.1007/s00425-002-0740-6. [DOI] [PubMed] [Google Scholar]
- Zhao T, Palotta M, Langridge P, Prasad M, Graner A, Schulze-Lefert P, Koprek T. Mapped Ds/T-DNA launch pads for functional genomics in barley. The Plant Journal. 2006;47:811–826. doi: 10.1111/j.1365-313X.2006.02831.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Baumlein H, Mock HP, Himmelbach A, Schweizer P. The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiology. 2006;142:181–192. doi: 10.1104/pp.106.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.