Abstract
Mass spectrometry (MS) has become increasingly important for tissue specific protein quantification at the isoform level, as well as for the analysis of protein post-translational regulation mechanisms and turnover rates. Thanks to the development of high accuracy mass spectrometers, peptide sequencing without prior knowledge of the amino acid sequence—de novo sequencing—can be performed. In this work, absolute quantification of a set of key enzymes involved in carbon and nitrogen metabolism in Medicago truncatula ‘Jemalong A17’ root nodules is presented. Among them, sucrose synthase (SuSy; EC 2.4.1.13), one of the central enzymes in sucrose cleavage in root nodules, has been further characterized and the relative phosphorylation state of the three most abundant isoforms has been quantified. De novo sequencing provided sequence information of a so far unidentified peptide, most probably belonging to SuSy2, the second most abundant isoform in M. truncatula root nodules. TiO2-phosphopeptide enrichment led to the identification of not only a phosphorylation site at Ser11 in SuSy1, but also of several novel phosphorylation sites present in other root nodule proteins such as alkaline invertase (AI; EC 3.2.1.26) and an RNA-binding protein.
Keywords: Absolute quantification, alkaline invertase, asparagine synthetase, de novo sequencing, mass western, Medicago truncatula, phosphoproteomics, plant proteomics, root nodules, stable isotope labelling, sucrose synthase
Introduction
Legumes are able to establish nitrogen-fixing symbiotic relations with soil bacteria generally termed rhizobia. This symbiosis is beneficial for both partners: plants provide a carbon source and nutrients for the microsymbiont which, in return, produces a reduced form of nitrogen directly assimilatable by the plant. Sucrose is the main carbohydrate transported via phloem to sink tissues like nodules, where it is metabolized by either SuSy or AI (Morell and Copeland, 1984; Flemetakis et al., 2006). SuSy is a cytosolic enzyme, which catalyses the cleavage of sucrose to fructose and UDP-glucose (Okamoto and Akazawa, 1980). This enzyme is considered primarily responsible for sucrose metabolism in mature nitrogen-fixing nodules, as it has been shown to be essential for symbiotic nitrogen fixation in a variety of legume plants (Gordon et al., 1999; Horst et al., 2007; Kuster et al., 2007). In addition, it has been demonstrated that SuSy is involved in the regulation of nitrogen fixation under different abiotic stresses, both at the transcriptional and post-transcriptional level (Gonzalez et al., 1995; Gordon et al., 1997; Zhang et al., 1999).
SuSy has been well-characterized at the molecular level in the model legume Medicago truncatula (Hohnjec et al., 1999, 2003). Based on transcriptional profiles, five isoforms have been described that are expressed in root nodules. However, a recent multi-dimensional LC-MS/MS-based proteomic profiling study which identified around 400 proteins from M. truncatula root nodules was only able to identify SuSy1, the most abundant isoform (Larrainzar et al., 2007). In contrast to shotgun proteomic techniques, targeted MS approaches allow for the detection and quantification of low abundance proteins present in complex samples (Wienkoop and Weckwerth, 2006; Mallick et al., 2007). While western blots are very sensitive and have been widely used for semi-quantitative protein analysis, absolute quantification can only be achieved when used in conjunction isoform specific antibodies and is subsequently scarcely used (Duncan et al., 2006). Thus, the fact that protein quantification by western blots is only relative, the requirement of specific antibodies, together with the limitation in the number of proteins detectable per analysis, has led to the development of alternative LC/MS-based techniques such as the so-called ‘mass western’ (Lehmann et al., 2008).
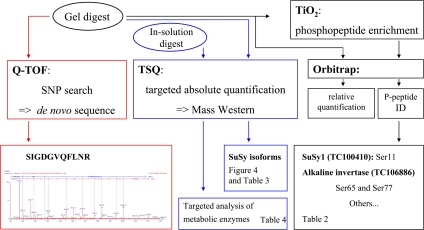
In this current work, a selective LC-MS/MS-based method using synthetic isotope-labelled peptides (Wienkoop and Weckwerth, 2006) has been applied for accurate quantification of SuSy proteins and other nodule metabolic enzymes (Fig. 1). Within a single analysis, several proteins involved in root nodule N metabolism as well as SuSy were detected and quantified with high sensitivity in absolute terms. Specific identification of different SuSy isoforms sharing high sequence similarity is presented. Furthermore, phosphopeptide-enrichment techniques have been applied to analyse the state of SuSy1 phosphorylation as well as for the identification of new phosphorylation sites in several nodule proteins. A schematic overview of the workflow can be found in Fig. 1.
Fig. 1.
Schematic workflow of the different MS analysis carried out. ID, identification.
Materials and methods
Plant growth conditions and nodule harvest
Medicago truncatula ‘Jemalong A17’ plants were inoculated with Sinorhizobium meliloti strain 2011. Plants were grown in 1.0 l pots with a mixture of vermiculite:perlite (5:2 v:v) as substrate under controlled environment conditions (14/10 h day/night, 22/16 °C temperature, and 70/60% relative humidity). The photosynthetic photon flux was 600 μmol m−2 s−1. When plants were 10 weeks old, root nodules were collected, frozen in liquid nitrogen and stored at –80 °C for further analysis.
Protein extraction
Nodules (0.1 g fresh weight) were homogenized in a mortar and pestle with 0.5 ml ice-cold homogenization buffer (50 mM HEPES, pH 7.8). Homogenates were centrifuged at 2000 g at 4 °C for 15 min and supernatants were collected and kept as nodule plant fractions.
Generation of stable isotope-labelled peptides
For absolute quantification of SuSy and proteins involved in nodule N assimilation, isoform-specific 13C/15N leucine-labelled peptides were synthesized (Thermo Electron, Ulm, Germany) and employed as internal standard peptides. Two approaches were used for the selection of peptides, empirical (i) and theoretical (ii).
(i) Many of the proteins that were to be targeted for quantitation had already been previously identified by MS (Larrainzar et al., 2007). Consequently, peptides derived from these proteins were already known and can be found in the ProMEX spectral library (Hummel et al., 2007). From this list of peptides, isoform-specific peptides were selected according to the recommendations of Thermo Electron in the instructions for the design of heavy peptide standards. The guidelines advise on sequence length and on the avoidance of certain amino acids that are common targets of uncontrolled modifications and in some cases were limiting factors for the successful selection of candidate peptides.
(ii) Peptides selected for quantitation that had not been identified by MS in previous studies were designed based on proteotypic peptide prediction. In the case of the SuSy isoforms where peptides from SuSy1 have previously been identified, the peptide sequences were aligned and proteotypic peptides from the other isoforms selected based on similarity to a known peptide from SuSy1. These peptides displayed changes in amino acid composition that resulted in significant mass differences and thus a specific proteotypic peptide for each isoform (Fig. 2, framed segment).
Fig. 2.
Alignment of SuSy isoforms sequences of M. truncatula. The de novo peptide sequence is included in bold as part of SuSy2 isoform. Framed segment indicates the proteotypic peptides from the SuSy isofroms selected for absolute quantification.
Detailed information about peptide sequences and specific single reaction monitoring transition data can be found in Table 1. Peptide mass shifts caused by methionine oxidation reactions were below 1% and, therefore, did not significantly influence the quantification.
Table 1.
Triple quadrupole tune settings for the specific standard peptides used for absolute quantification
| TC code | Name | Peptide sequence | Standard peptide |
Native peptide |
||||
| Precursor ion (m/z) | Product ions (m/z) | Collision energy | Precursor ion (m/z) | Productions (m/z) | Collision energy | |||
| TC100391 | AS | STYAWGLEAR | 580.7 | 738.3 | 19 | 577.2 | 731.3 | 19 |
| TC100393 | AS | STFAWGLEAR | 572.8 | 552.2 | 18 | 569.3 | 545.2 | 18 |
| TC106729 | GS1a | ITEIAGVVVSFDPK | 741.4 | 243.9 | 36 | 737.9 | 243.9 | 36 |
| TC106808 | GS1b | HETADINTFLWGVANR | 617.6 | 702.3 | 19 | 615.3 | 702.3 | 19 |
| TC106913 | GS2 | IHIEAYGEGNER | 465.5 | 475.1 | 14 | 463.2 | 475.1 | 14 |
| TC94780 | GOGAT | EVLVDFDNLLPK | 704.9 | 1166 | 17 | 701.4 | 1159 | 17 |
| TC94631 | AAT2 | ATAELLLGADNPAIK | 752.4 | 785.4 | 22 | 748.9 | 785.4 | 22 |
| TC106918 | AAT1 | TEEGKPLVLDVVR | 487.9 | 917.6 | 19 | 485.6 | 910.6 | 19 |
| TC94623 | AAT | IPSGHGYDDFEVVR | 534.2 | 744.3 | 18 | 530.9 | 739.3 | 18 |
| TC94704 | GDH | GLDIPSLLK | 481.8 | 564.3 | 15 | 478.3 | 557.3 | 15 |
| TC100410 | SuSy1 | VHSLKER | 438.2 | 639.3 | 18 | 434.7 | 632.3 | 18 |
| TC100410 | SuSy1 | ALESEMLSR | 521.7 | 851.4 | 18 | 518.2 | 851.4 | 18 |
| TC95820 | SuSy2 | ALENEMLAR | 527.2 | 862.4 | 18 | 523.7 | 862.4 | 18 |
| TC94447 | SuSy3 | ALENEMLR | 491.7 | 791.3 | 17 | 488.2 | 791.3 | 17 |
| TC99016 | SuSy5 | ALEEELLQK | 540.2 | 888.5 | 18 | 536.7 | 888.5 | 18 |
Peptide sequences employed in this work, as well as their corresponding precursor masses and product ions are given. Stable isotope-labelled leucine residues are marked in bold.
SDS-PAGE and in-gel protein digestion
Prior to mass western analyses, plant nodule protein extracts (80 μg of protein per sample) were separated by SDS-PAGE on 6% (w/v) polyacrylamide gels. After electrophoresis, proteins were visualized using Gel-Code Blue Stain reagent (Pierce Biotechnology, Rockford, USA). For the specific analysis of SuSy isoforms and determination of phosphorylation levels, gel bands corresponding to a molecular weight of approximately 90 kDa were cut out and standard synthetic peptides were added. For the identification of novel phosphoproteins, bands in the range between 25–60 kDa were also excised from the gels. A total number of four gel band replicates were analysed. In-gel digestion was carried out as previously described by Shevchenko et al. (1996).
In-solution protein digestion
Aliquots containing 50 μg of plant nodule proteins were digested overnight at 37 °C with Porosyzme immobilized trypsin beads (1:10, v/v, Applied Biosystems, Darmstadt, Germany). After centrifugation to remove beads the peptide mixtures were desalted using SPEC C18 columns according to the manufacturer's instructions (Varian, Darmstadt, Germany). Desalted digest solutions were dried and pellets stored at –20 °C until use. Four biological replicates were analysed.
Phosphopeptide enrichment
For the identification of novel nodule phosphopeptides, gel digests corresponding to protein bands within the range of 25–60 kDa were loaded onto titanium dioxide columns (TiO2, TopTips) purchased from SunChrom (Friedrichsdorf, Germany). Phosphopeptide enrichment was carried out as previously described by Mazanek et al. (2007).
Determination of phosphorylation stoichiometry
To determine the relative phosphorylation states of proteins, corresponding bands were excised from SDS-PAGE gels, digested, and analysed by MS. Relative abundance was assessed by comparing peak areas for the phosphorylated and non-phosphorylated tryptic peptides (see also ‘Orbitrap settings for the identification of phosphopeptides and relative phosphorylation state analysis’, and the workflow diagram in Fig. 1).
Q-TOF settings for de novo sequencing analysis
For de novo sequencing, gel digests corresponding to SuSy were loaded onto a HPLC system (Waters 2695 Alliance, Waters GmbH, Eschborn, Germany) directly coupled to a Q-TOF mass spectrometer (Waters GmbH, Eschborn, Germany). Peptides were eluted using a 15-min gradient from 0–80% (v/v) acetonitrile (ACN)/0.1% (v/v) formic acid (FA) using a C18 column (150×1 mm, Jupiter 4μ Proteo 90A, Phenomenex, Aschaffenburg, Germany). De novo sequencing was performed using Mass Lynx software (Waters GmbH, Eschborn, Germany). The software was programmed to screen for the doubly charged precursor masses 521.7 and 602.8 of the known SuSy peptides (ALESEMLSR and SIGNGVQFLNR, corresponding to SuSy1; TC100410), a mass 527.2 for ALENEMLAR (corresponding to isoform SuSy2; TC95820) and a mass of 602.8 plus a mass shift of 0.5, in an attempt to identify a possible sequence variation due to amino acid differences encoded by single nucleotide polymorphisms (SNPs), for example, glutamine (Q)/glutamic acid (E) or asparagine (N)/aspartic acid (D) exchange.
Triple quadrupole settings for absolute quantification
For the identification and absolute quantification of different SuSy isoforms, a one-dimensional (1D) nano-flow LC-MS/MS system equipped with a pre-column (Agilent, Zeesen, Germany) was employed. Peptides were eluted using a monolithic column (Merck, Darmstadt, Germany) of 15 cm length and 0.1 mm internal diameter during a 30 min gradient from 5% to 100% (v/v) methanol/0.1% (v/v) FA with a controlled flow rate of 0.7 μl min−1. 1 pmol of each internal standard peptide was added to the protein sample prior to analysis, as peptides at this concentration have been shown to lie within the linear range of detection (Wienkoop and Weckwerth, 2006). MS analysis was performed on a TSQ Quantum Discovery MAX mass spectrometer (Thermo Electron, Ulm, Germany) operated in the positive mode and tuned to its optimum sensitivity for each standard peptide as previously described (Glinski and Weckwerth, 2005). Scan width for all MRMs (multiple reaction monitoring) was 0.7 mass units, and resolution was set to 0.25 and 0.7 mass units for Q1 and Q3, respectively. The dwell time per transition was 50 ms, spray voltage was set to 1.8 kV and temperature of heated transfer capillary was set to 150 °C. Collision energies used for recorded transitions are summarized in Table 1. Absolute quantification was performed based on peak integration.
Orbitrap settings for the identification of phosphopeptides and relative phosphorylation state analysis
After phosphopeptide enrichment, TiO2-peptide eluates were loaded on a 1D nano-flow LC-MS/MS system equipped with pre-column (Agilent, Zeesen, Germany). A monolithic column (Merck, Darmstadt, Germany) of 15 cm length and 0.1 mm internal diameter was coupled to an Orbitrap LTQ XL mass spectrometer (Thermo Electron, Ulm, Germany). Peptides were eluted during a 50-min gradient from 5–100% (v/v) methanol/0.1% (v/v) FA with a controlled flow rate of 0.5 μl min−1. For precursor masses determinations, resolution and accuracy of the Orbitrap were set to 3000 ppm and 2 ppm, respectively. MS/MS measurements were performed on wideband mode. Spray voltage was set to 1.8 kV and temperature of the heated transfer capillary was set to 150 °C. Potential phosphopeptide candidates were evaluated using MSQuant 1.4 software. A post-translational modification (PTM) score for localization, which estimates the probability to find an actual phosphorylation site based on fragment ions (Olsen et al., 2006), is shown in Table 2. For the relative quantification of phosphorylation levels, gel digests were directly analysed using the same settings as described for phosphopeptide identification.
Table 2.
Phosphopeptides found in M. truncatula root nodules after TiO2-enrichment and MSQuant validation (PTM: post-translational modification)
| Protein | Peptide sequence | Phospho site | Mascot score | MSQuant PTM score |
| TC106886 | AGLDNYDNYS(p)PGGR | Ser65 | 69 | 114 |
| SGFNTPAS(p)SAR | Ser77 | 48 | 73 | |
| TC101080 | KHASS(p)PPPDSPSQDSVEKPTYVR | Ser21 | 53 | 60 |
| TC97295 | HQSAAT(p)PTPTAGAR | Thr33 | 40 | 70 |
Results and discussion
Detection of different SuSy isoforms in M. truncatula root nodules
Although detection of three SuSy isoforms using isoform-specific antibodies has been reported in maize (Duncan et al., 2006), multi-dimensional LC-MS/MS and western blot analysis did not allow detection of SuSy isoforms other than SuSy1 in M. truncatula root nodules (Larrainzar et al., 2007). This is probably due to the relatively low abundance of the other isoforms, together with the similarity they share at the amino acid sequence level, which limits an MS/MS-based discrimination. Figure 2 represents an alignment of the N-terminal region of the five described SuSy isoform sequences. Sequences for SuSy1 and SuSy3 (TC94447) appear to be complete, whereas the known sequences for SuSy2 (TC95820), SuSy5 (TC99016), and SuSy4 (TC98648) are estimated to be 72, 40, and 19%, respectively. To improve the detection of different SuSy isoforms in root nodules further, two approaches were carried out: (i) targeted screening for peptide sequence variation using a Q-TOF mass spectrometer with high mass accuracy; and (ii) multiple reaction monitoring on a triple quadrupole based on predicted proteotypic peptides.
Identification of a new SuSy peptide sequence
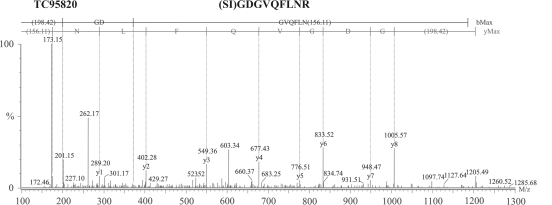
When comparing sequences of protein isoforms, it is common to find certain amino acid differences encoded by SNPs [i.e. glutamine (Q) for glutamic acid (E) or asparagine (N) for aspartic acid (D)]. These sequence variations are translated into mass differences in peptides enabling isoform discrimination using MS. Looking for this type of polymorphism, the predicted amino acid sequences of the SuSy isoforms were aligned (Fig. 2) and examined for potential candidates with this type of polymorphism. A SuSy1 tryptic peptide containing some of the above-mentioned residues was subsequently chosen (SIGNGVQFLNR). After screening the gel sample for masses corresponding to the doubly charged peptide, two ions were selected: one corresponding to a mass-to-charge ratio (m/z) of 602.8 derived from a SuSy1 tryptic peptide, and the other peptide of unknown sequence with an m/z of 603.3. De novo sequencing was performed on the unknown ion and was identified as the peptide (SI)GDGVQFLNR (Fig. 3), which did not match any of the previously reported sequences (Fig. 2). The observed mass difference corresponds to a shift starting from ion y7, explained by a replacement of D instead of the original N residue present in SuSy1. The only other possible combination producing a 603.3 m/z peptide would be the replacement of isoleucine (I) for leucine (L) in the N-terminal region. However, as this region appears to be well-conserved in the amino acid sequence of SuSy in different legume plants (data not shown), this alternative appears to be less likely. Furthermore, peptides derived from both SuSy1 and SuSy2 were detected during the analysis of 1D-PAGE samples (see Materials and methods and Fig. 2), further supporting the hypothesis that the new peptide is driven from SuSy2 (Fig. 2). Interestingly, SuSy2 is the second most abundant isoform in M. truncatula root nodules at the transcript level (Hohnjec et al., 2003).
Fig. 3.
Q-TOF driven MS/MS spectrum of the de novo sequence for a tryptic peptide with m/z 603.3 (SIGDGVQFLNR).
Mass western of different SuSy isoforms in root nodules
For the specific absolute quantification of protein isoforms, a technique based on high resolution multiple reaction monitoring in a triple quadrupole mass spectrometer, called mass western, has recently been established (Lehmann et al., 2008). Instead of antibodies, isoform-specific stable isotope-labelled peptides are synthesized. These are then employed as internal standards and added to protein samples, which are subsequently digested either in gel or directly in solution (Wienkoop and Weckwerth, 2006). As the concentration of the standard peptides is known, absolute quantification of native proteins is achievable in fmol μg−1 of total protein.
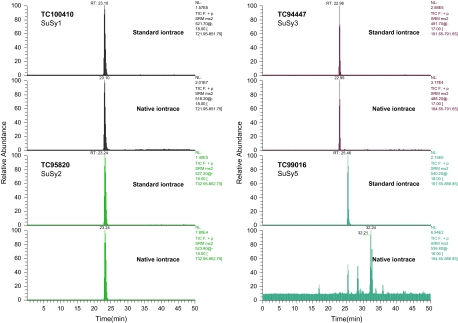
In the present study, four standard peptides specific for each of the isoforms SuSy1, 2, 3, and 5 were selected (Table 1). As only 19% of the SuSy4 sequence is available (Fig. 2), this isoform was excluded from the analysis. Out of the four isoforms analysed, absolute quantification was only possible for three of them (Table 3; Fig. 4). SuSy5 appeared either to be below the detection limit or not present in nodules under the conditions investigated. Results show that SuSy2 is the second most abundant isoform present in M. truncatula root nodules. This finding further supports the hypothesis that the de novo peptide sequence belongs to this second isoform. Due to the high sensitivity of this technique, SuSy3 was also successfully identified, despite its much weaker signal (Fig. 4). Absolute quantification of the different isoforms shows that the protein ratios for SuSy1:SuSy2:SuSy3 are 1:0.013:0.0018 (Table 3). These results are in agreement with a recent transcript analysis in pea nodules (Marino et al., 2008), where the ratio for the three isoforms was 1:0.002:1.7 × 10−5. From a technical point of view, the four orders of magnitude range determined in a single analysis corresponds to the current maximum capacity for a MS-based analysis (Wienkoop et al., 2006; Wienkoop and Weckwerth, 2006). This limitation is mainly due to the limit of detection of each peptide and the loading capacity of the columns. Nevertheless, the sensitivity (2 ng) in the present study has been shown to be comparable to those of western blots (Lehmann et al., 2008). A significant advantage of the mass western is that it allows for absolute quantification and for multiple targets to be analysed in a single run. Furthermore, mass western provides a direct measurement of protein concentration, not available by transcript analysis.
Table 3.
Absolute quantification of three different SuSy isoforms in M. truncatula root nodules after in-gel digestion (n=4)
| TC Code | Name | Specific amount a |
||
| fmol μg−1 protein | pmol per gel slide | ng per gel slide | ||
| TC100410 | SuSy1 | 160±2.21 | 12.8 | 1000 |
| TC95820 | SuSy2 | 2.1±0.03 | 0.170 | 16 |
| TC94447 | SuSy3 | 0.3±0.25 | 0.023 | 2 |
| TC99016 | SuSy5 | n.d. | n.d. | n.d. |
n.d., Not detected.
Fig. 4.
Mass western of four SuSy isoforms detected in M. truncatula root nodule protein extract using a triple quadrupole mass spectrometer and specific stable isotope-labelled standard peptides (see Table 1).
Absolute quantification of proteins involved in N metabolism present in complex nodule protein samples
Absolute quantification enables the analysis of complete metabolic pathways in complex protein samples in a single analysis. In contrast to the previous analysis, where the starting materials employed were in-gel digests, absolute quantification was also carried out using nodule extract protein samples as the starting material. Internal standard peptides for glutamine synthetase (GS), glutamate dehydrogenase (GDH), asparagine synthetase (AS), and aspartate aminotransferase (AAT) were synthesized (peptide sequences shown in Table 1), and these metabolic enzymes, together with SuSy, were subjected to absolute quantification (Table 4). Regarding SuSy, only the most abundant isoform, SuSy1, representing about 1% of the total nodule proteins, was detected. The reason for this might be a higher signal-to-noise ratio of a complex protein sample compared to the pre-fractionated gel sample used in the previous analysis. However, the absolute amount of SuSy1 appears to be slightly higher in the complex nodule protein analysis indicating that the in-gel analysis may be even more sensitive.
Table 4.
Absolute quantification of several proteins involved in C and N metabolism in M. truncatula root nodules after gel free shotgun analysis
| TC Code | Name | fmol mg−1 proteina |
| TC100410 | SuSy1 | 195.2±23.7 |
| TC95820 | SuSy2 | n.d. |
| TC94447 | SuSy3 | n.d. |
| TC100391 | AS | 76.5±2.5 |
| TC100393 | AS | 90.5±8.5 |
| TC106729 | GS1a | 98.9±1.0 |
| TC106808 | GS1b | 29.6±8.6 |
| TC106913 | GS2 | 23.8±5.8 |
| TC94631 | AAT2 | 44.3±0.9 |
| TC106918 | AAT1 | 5.1±0.9 |
| TC94623 | AAT | 20.7±4.1 |
| TC94704 | GDH | 51.1±9.3 |
n.d., Not detected.
Most of these enzymes involved in nodule N metabolism (GS, GDH, AAT) are well characterized and their activities are routinely assessed (Vance, 2008). Although most of the N exported from temperate climate nodules is in the form of asparagine, detection of AS activity in plant tissues is extremely difficult (Lea et al., 2007). The reasons for this are not clear, but they may be related to the presence of inhibitors (Joy et al., 1983), asparaginase activity (Streeter, 1977) or protein instability (Huber and Streeter, 1985). To overcome these limitations, quantification of AS gene expression has been suggested to be a good semi-quantitative indicator of its activity levels, showing a good correlation with asparagine:aspartate ratios transported to the shoot via the xylem (Antunes et al., 2008). The strategy shown in the present paper may provide a more accurate alternative for the quantification of AS in nodules.
Identification of phosphorylation sites and determination of the phosphorylation state of SuSy1 in nodules under normal growth conditions
SuSy has been shown to be phosphorylated in a number of tissues in different plants. While initial evidence that SuSy was phosphorylated came from work on maize cell cultures (Shaw et al., 1994), it was Huber et al. (1996) who first characterized a phosphorylation site at Ser15 in SuSy isolated from maize leaves. Further studies have described other phosphorylation sites in maize (Hardin et al., 2003) and legume plants like soybean (Zhang and Chollet, 1997; Zhang et al., 1999; Komina et al., 2002). In particular, Zhang et al. (1999) described a putative seryl-phosphorylation domain near the N-terminus of the protein (LTRVHS11LRER) in soybean. This motif, which was reported to be the primary phosphorylation site of nodule soybean SuSy in planta, is present in practically all the SuSy sequences reported to date.
In the present work, MS analysis revealed that SuSy1 from the nodules of M. truncatula is also phosphorylated at Ser11, the same residue identified as phosphorylated in soybean and homologous to Ser15 in maize SuSy1 (VHS11LKER; spectra can be found in ProMEX database http://promex.mpimp-golm.mpg.de/home.shtml). In addition, the relative phosphorylation state of the protein was analysed and quantified by comparing peak areas of the phosphorylated and non-phosphorylated tryptic peptides derived from the same analysis (see also ‘Determination of phosphorylation stoichiometry’, and the workflow diagram in Fig. 1).
M. truncatula nodule SuSy1 was found to be hyperphosphorylated, as the phosphopeptide was 1.7-fold more abundant than the non-phosphorylated peptide. The meaning of this hyperphosphorylation state remains the subject of much debate. In soybean, in vitro studies suggest that nodule SuSy sucrose-cleavage activity is not dependent on the phosphorylation state of the protein, but rather influences the attachment of this enzyme to the membrane (Zhang et al., 1999). However, studies in maize have shown that leaf SuSy is activated when phosphorylated at its primary site (Ser15), while phosphorylation at Ser170 would trigger degradation of the enzyme (Hardin et al., 2003, 2004). More recently, it has been suggested that sucrose concentration and phosphorylation may regulate SuSy oligomerization and F-actin association in maize kernels and seedlings (Duncan and Huber, 2007). Regarding membrane attachment, Komina et al. (2002) investigated the phosphorylation state of membrane and soluble SuSy forms of nodules in soybean. Using several non-MS-based approaches, the microsomal SuSy fraction was found to be hypophosphorylated relative to the soluble form (on an equivalent SuSy protein base). Although abiotic stresses seem to have a negative impact on SuSy abundance, as well as on its phosphorylation levels (data not shown), the physiological meaning of a state of hyper- or hypophosphorylation remains to be elucidated.
Other phosphoproteins found in nodules
SuSy is one of the most characterized nodule proteins in legumes. Due to its relatively high abundance in root nodules and, specifically with regard to M. truncatula, its hyperphosphorylation state, it is understandable that the conserved Ser11 phosphopeptide has been described several times. In order to identify novel putative phosphorylated proteins in M. truncatula root nodules, a phosphopeptide enrichment strategy based on TiO2-affinity chromatography was carried out. As a result, several other phosphopetides were identified (Table 2). Among them, two phosphopeptides (AGLDNYDNYS65PGGR and SGFNTPAS77SAR) belonging to AI (TC106886), the other enzyme with a sucrose-cleavage activity in nodules. In contrast to SuSy, relative peak area quantification of both peptides indicated that AI is mostly hypophosporylated in nodules. The non-phosphorylated to the phosphorylated forms of Ser65 and Ser77 were determined to be in a ratio of 1:0.01:0.03, respectively. It has been suggested that AI is post-transcriptionally regulated through an interaction with a phospho-binding 14-3-3 protein in developing barley grains (Alexander and Morris, 2006). However, to our knowledge this is the first report where the actual phosphorylation sites of the protein are described. Several other phosphopeptides were found that belong to a putative RNA-binding protein (TC101080) and a hypothetical protein (TC97295), which is similar to a universal stress response protein described in Arabidopsis thaliana (At4g27320) (Table 2). Although, thus far, little is known about the function of these proteins in nodules, they may be considered interesting candidates for further studies given the roles of phosphorylation in metabolic regulation.
Nevertheless, several other nodule proteins have been suggested to be phosphorylated and were not identified in this present study. This is the case for both GS (Lima et al., 2006) and phosphoenolpyruvate decarboxylase (Zhang et al., 1995) from M. truncatula. Previously reported phosphopeptides from GS did not appear to be detectable after tryptic digestion, but may be detected using different proteolytic enzymes. In the case of phosphoenolpyruvate decarboxylase, phosphopeptides were not detected using the current approach as its molecular weight was out of range of the analysed bands subjected to TiO2 enrichment. Therefore, future analysis may include a wider range of molecular weights to identify new nodule protein subject to protein phosphorylation.
Conclusion
This study has demonstrated for the first time the absolute quantification of three distinct M. truncatula SuSy isoforms in an independent tissue-specific manner. It is shown that, besides the nodule-enhanced SuSy1, two other protein isoforms are present in root nodules at quantifiable amounts. De novo sequencing allowed for the identification of a new peptide sequence, most probably driven from the second most abundant isoform SuSy2, which complements the existing sequence information based on transcript data. Absolute quantification of N metabolism enzymes was carried out, enabling the discrimination of different protein isoforms from a single analysis, even when those isoforms were present at very low amounts. Nodule SuSy1 was shown to be hyperphosphorylated at Ser11 in M. truncatula and novel phosphoproteins present in nodules were identified. All peptides found in this study can be viewed in the ProMEX spectral library (Hummel et al., 2007).
Acknowledgments
We thank Joshua L Heazlewood and Waltraud Schulze for helpful discussion. A special thank you to Wolfgang Höhenwarter and Joshua L Heazlewood for proof reading. EL is the holder of a Formación de Profesorado Universitario PhD fellowship from the Spanish Ministry of Education and Science. EL, EMG, and CA-I acknowledge financial support from DGI-MEC (Spain) grant AGL2005-0274/AGR and its associated FEDER funding. CA-I also wishes to acknowledge the support provided by the Mobility Programme of the Spanish Ministry of Education and Science.
Glossary
Abbreviations
- AAT
aspartate aminotransferase
- AI
alkaline invertase
- AS
asparagine synthetase
- GDH
glutamate dehydrogenase
- GOGAT
glutamate synthase
- GS
glutamine synthetase
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- Q-TOF
quadrupole time of flight
- SuSy
sucrose synthase
References
- Alexander RD, Morris PC. A proteomic analysis of 14-3-3 binding proteins from developing barley grains. Proteomics. 2006;6:1886–1896. doi: 10.1002/pmic.200500548. [DOI] [PubMed] [Google Scholar]
- Antunes F, Aguilar M, Pineda M, Sodek L. Nitrogen stress and the expression of asparagine synthetase in roots and nodules of soybean (Glycine max L. Merr.) Physiologia Plantarum. 2008;133:736–743. doi: 10.1111/j.1399-3054.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- Duncan KA, Hardin SC, Huber SC. The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant and Cell Physiology. 2006;47:959–971. doi: 10.1093/pcp/pcj068. [DOI] [PubMed] [Google Scholar]
- Duncan KA, Huber SC. Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Plant and Cell Physiology. 2007;48:1612–1623. doi: 10.1093/pcp/pcm133. [DOI] [PubMed] [Google Scholar]
- Flemetakis E, Efrose RC, Ott T, Stedel C, Aivalakis G, Udvardi MK, Katinakis P. Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Molecular Biology. 2006;62:53–69. doi: 10.1007/s11103-006-9003-4. [DOI] [PubMed] [Google Scholar]
- Glinski M, Weckwerth W. Differential multisite phosphorylation of the trehalose-6-phosphate synthase gene family in Arabidopsis thaliana: a mass spectrometry-based process for multiparallel peptide library phosphorylation analysis. Molecular and Cellular Proteomics. 2005;4:1614–1625. doi: 10.1074/mcp.M500134-MCP200. [DOI] [PubMed] [Google Scholar]
- Gonzalez EM, Gordon AJ, James CL, Arrese-Igor C. The role of sucrose synthase in the response of soybean nodules to drought. Journal of Experimental Botany. 1995;46:1515–1523. [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O. Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiology. 1999;120:867–877. doi: 10.1104/pp.120.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, Skot L, James CL. Stress-induced declines in soybean N2 fixation are related to nodule sucrose synthase activity. Plant Physiology. 1997;114:937–946. doi: 10.1104/pp.114.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin SC, Tang GQ, Scholz A, Holtgraewe D, Winter H, Huber SC. Phosphorylation of sucrose synthase at serine 170: occurrence and possible role as a signal for proteolysis. The Plant Journal. 2003;35:588–603. doi: 10.1046/j.1365-313x.2003.01831.x. [DOI] [PubMed] [Google Scholar]
- Hardin SC, Winter H, Huber SC. Phosphorylation of the amino terminus of maize sucrose synthase in relation to membrane association and enzyme activity. Plant Physiology. 2004;134:1427–1438. doi: 10.1104/pp.103.036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N, Becker JD, Puhler A, Perlick AM, Küster H. Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Molecular and General Genetics. 1999;261:514–522. doi: 10.1007/s004380050995. [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Puhler A, Küster H. The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Molecular Plant–Microbe Interactions. 2003;16:903–915. doi: 10.1094/MPMI.2003.16.10.903. [DOI] [PubMed] [Google Scholar]
- Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang TL. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiology. 2007;144:806–820. doi: 10.1104/pp.107.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL, Liao PC, Gage DA, McMichael RW, Chourey PS, Hannah LC, Koch K. Phosphorylation of serine-15 of maize leaf sucrose synthase: occurrence in vivo and possible regulatory significance. Plant Physiology. 1996;112:793–802. doi: 10.1104/pp.112.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TA, Streeter JG. Purification and properties of asparagine synthetase from soybean root-nodules. Plant Science. 1985;42:9–17. [Google Scholar]
- Hummel J, Niemann M, Wienkoop S, Schulze W, Steinhauser D, Selbig J, Walther D, Weckwerth W. ProMEX: a mass spectral reference database for proteins and protein phosphorylation sites. BMC Bioinformatics. 2007;8:216. doi: 10.1186/1471-2105-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy KW, Ireland RJ, Lea PJ. Asparagine synthesis in pea leaves and the occurrence of an asparagine synthetase inhibitor. Plant Physiology. 1983;73:165–168. doi: 10.1104/pp.73.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komina O, Zhou Y, Sarath G, Chollet R. In vivo and in vitro phosphorylation of membrane and soluble forms of soybean nodule sucrose synthase. Plant Physiology. 2002;129:1664–1673. doi: 10.1104/pp.002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster H, Vieweg MF, Manthey K, Baier MC, Hohnjec N, Perlick AM. Identification and expression regulation of symbiotically activated legume genes. Phytochemistry. 2007;68:8–18. doi: 10.1016/j.phytochem.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Weckwerth W, Ladrera R, Arrese-Igor C, Gonzalez EM. Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiology. 2007;144:1495–1507. doi: 10.1104/pp.107.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Sodek L, Parry MAJ, Shewry PR, Halford NG. Asparagine in plants. Annals of Applied Biology. 2007;150:1–26. [Google Scholar]
- Lehmann U, Wienkoop S, Weckwerth W. If the antibody fails: a mass western approach. The Plant Journal. 2008 doi: 10.1111/j.1365-313X.2008.03554.x. doi: 10.1111/j.1365-313X.2008.03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. Journal of Experimental Botany. 2006;57:2751–2761. doi: 10.1093/jxb/erl036. [DOI] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. eComputational prediction of proteotypic peptides for quantitative proteomics. Nature Biotechnology. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- Marino D, Hohnjec N, Küster H, Moran JF, Gonzalez EM, Arrese-Igor C. Evidence for transcriptional and post-transcriptional regulation of sucrose synthase in pea nodules by the cellular redox state. Molecular Plant–Microbe Interactions. 2008;21:622–630. doi: 10.1094/MPMI-21-5-0622. [DOI] [PubMed] [Google Scholar]
- Mazanek M, Mituloviae G, Herzog F, Stingl C, Hutchins JR, Peters JM, Mechtler K. Titanium dioxide as a chemo-affinity solid phase in offline phosphopeptide chromatography prior to HPLC-MS/MS analysis. Nature Protocols. 2007;2:1059–1069. doi: 10.1038/nprot.2006.280. [DOI] [PubMed] [Google Scholar]
- Morell M, Copeland L. Enzymes of sucrose breakdown in soybean nodules: alkaline invertase. Plant Physiology. 1984;74:1030–1034. doi: 10.1104/pp.74.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Akazawa T. Enzymic mechanism of starch breakdown in germinating rice seeds. 9. De novo synthesis of β-amylase. Plant Physiology. 1980;65:81–84. doi: 10.1104/pp.65.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Shaw JR, Ferl RJ, Baier J, St Clair D, Carson C, McCarty DR, Hannah LC. Structural features of the maize sus1 gene and protein. Plant Physiology. 1994;106:1659–1665. doi: 10.1104/pp.106.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Streeter JG. Asparaginase and asparagine transaminase in soybean leaves and root nodules. Plant Physiology. 1977;60:235–239. doi: 10.1104/pp.60.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP. Carbon and nitrogen metabolism in legume nodules. In: Dilworth MJ, James EK, Sprent JI, Newton WE, editors. Nitrogen-fixing leguminous symbioses. New York: Springer-Verlag; 2008. pp. 293–320. [Google Scholar]
- Wienkoop S, Larrainzar E, Niemann M, Gonzalez EM, Lehmann U, Weckwerth W. Stable isotope-free quantitative shotgun proteomics combined with sample pattern recognition for rapid diagnostics. Journal of Separation Science. 2006;29:2793–2801. doi: 10.1002/jssc.200600290. [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Weckwerth W. Relative and absolute quantitative shotgun proteomics: targeting low-abundance proteins in Arabidopsis thaliana. Journal of Experimental Botany. 2006;57:1529–1535. doi: 10.1093/jxb/erj157. [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Chollet R. Seryl-phosphorylation of soybean nodule sucrose synthase (nodulin-100) by a Ca2+-dependent protein kinase. FEBS Letters. 1997;410:126–130. doi: 10.1016/s0014-5793(97)00537-1. [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Li B, Chollet R. In vivo regulatory phosphorylation of soybean-nodule phosphoenolpyruvate carboxylase. Plant Physiology. 1995;108:1561–1568. doi: 10.1104/pp.108.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Lund AA, Sarath G, Cerny RL, Roberts DM, Chollet R. Soybean nodule sucrose synthase (Nodulin-100): further analysis of its phosphorylation using recombinant and authentic root-nodule enzymes. Archives of Biochemistry and Biophysics. 1999;371:70–82. doi: 10.1006/abbi.1999.1415. [DOI] [PubMed] [Google Scholar]