Abstract
The Arabidopsis thaliana sterol carrier protein-2 (AtSCP2) is a small, basic and peroxisomal protein that in vitro enhances the transfer of lipids between membranes. AtSCP2 and all other plant SCP-2 that have been identified are single-domain polypeptides, whereas in many other eukaryotes SCP-2 domains are expressed in the terminus of multidomain polypeptides. The AtSCP2 transcript is expressed in all analysed tissues and developmental stages, with the highest levels in floral tissues and in maturing seeds. The expression of AtSCP2 is highly correlated with the multifunctional protein-2 (MFP2) involved in β-oxidation. A. thaliana Atscp2-1 plants deficient in AtSCP2 show altered seed morphology, a delayed germination, and are dependent on an exogenous carbon source to avoid a delayed seedling establishment. Metabolomic investigations revealed 110 variables (putative metabolites) that differed in relative concentration between Atscp2-1 and normal A. thaliana wild-type seedlings. Microarray analysis revealed that many genes whose expression is altered in mutants with a deficiency in the glyoxylate pathway, also have a changed expression level in Atscp2-1.
Keywords: Arabidopsis, β-oxidation, germination, glyoxylate cycle, lipid, lipid transport, metabolomics, microarray, peroxisomes, SCP-2
Introduction
Sterol carrier protein-2 (SCP-2) is an intracellular, small, basic protein domain that in vitro stimulates the transfer of lipids between membranes (Ritter et al., 1971; Bloj et al., 1978). In mammals, SCP-2 is implicated to have role in peroxisomal β-oxidation. The exact function of SCP-2 in β-oxidation is unclear, but it might facilitate the presentation and solubilization of the substrates or stabilizing the enzymes involved in catalysing the reaction cycles (Seedorf et al., 2000). Such suggestions are mainly based on studies of the mammalian peroxisomal proteins sterol carrier protein-X (SCP-X) and D-bifunctional protein (DBP), which both contain C-terminal SCP-2 domains. The human gene SCPX (also known as SCP2) encodes SCP-X, which consists of a 3-ketoacyl-CoA thiolase domain connected to a C-terminal SCP-2 domain (Ohba et al., 1994). Due to the existence of dual promoters, as well as proteolytic cleavage sites the SCPX encoded SCP-2 domain is also expressed as a single-domain protein (Ohba et al., 1995). SCP-X and the single-domain SCP-2 are both predominantly located to peroxisomes. Gene targeting in mice revealed that complete deficiency of SCPX resulted in an impaired catabolism of 2-methyl branched-chain fatty acyl CoAs as shown by a 10-fold accumulation of phytanic acid in SCPX(–/–) mice (Seedorf et al., 1998). Further, it has been demonstrated with FRET microscopy that the single-domain SCP-2 interacts in peroxisomes with enzymes involved in β-oxidation such as acyl-CoA oxidase, bifunctional protein, and 3-ketoacyl-CoA thiolase (Wouters et al., 1998). It may be noteworthy that plants are not encoding any gene orthologous to the 3-ketoacyl-CoA thiolase in SCP-X (Edqvist and Blomqvist, 2006).
A SCP-2 domain is also present in the C-terminus of the mammalian DBP (also referred to as MFE-2), which in the human genome is encoded from HSD17B4. DBP has domains for D-3 (equivalent to 3R)-hydroxyacyl-CoA dehydrogenase, 2-enoyl-CoA hydratase, and SCP-2 (Adamski et al., 1995; Leenders et al., 1998) and catalyses the second and third steps of the β-oxidation pathway. Reactions catalysed by the mammalian DBP proceed through D-3-hydroxyl-CoA esters. DBP is suggested to be required for the peroxisomal β-oxidation of the enoyl-CoA esters of very long chain fatty acids, pristanic acid, and of dihydroxycholestanoic acid (DHCA) (Wanders, 2004). The role of the SCP-2 domain in DBP is unclear, but it contains a peroxisomal targeting signal (PTS1) that locates DBP to the peroxisomes. The structural and functional conservation may indicate that the SCP-2 domain in DBP also has additional functions, such as to interact with enzymatic domains of DBP to form an extended hydrophobic cavity for the hydrophobic tails of some β-oxidation substrates (Haapalainen et al., 2001).
A. thaliana do not encode DBP, and there are no plant genes identified orthologous to the D-3-hydroxyacyl-CoA dehydrogenase domain of mammalian DBP (Edqvist and Blomqvist, 2006). Rather, the A. thaliana multifunctional proteins AIM1 and MFP2 each share domain structure and approximately 50% amino acid sequence similarity to the human peroxisomal L-bifunctional protein (LBP) (also referred to as MFE-1) (Kiema et al., 2002). LBP and DBP lack significant sequence similarity and LBP, as well as AIM1 and MFP2, are not carrying any C-terminal SCP-2 domain. Furthermore, reactions catalysed by LBP, AIM1, and MFP2 proceeds through L-3-hydroxyl-CoA esters.
Thus, plants do not encode DBP or SCP-X, but it has recently been shown that plants also encode and express SCP-2 (Eklund and Edqvist, 2003; Edqvist et al., 2004; Edqvist and Blomqvist, 2006). The gene AtSCP2 (At5g42890) on chromosome 5 encodes the sole SCP-2 domain in the A. thaliana genome. AtSCP2 is a 13.6 kDa protein with a pI of 9.2, which localizes to peroxisomes through its C-terminal PST1 targeting signal. It has lipid transfer activity in vitro (Edqvist et al., 2004). The known crystal structures of SCP-2 from rabbit (Choinowski et al., 2000), of the SCP-2 domain of the human DBP (Haapalainen et al., 2001) and of yellow fever mosquito SCP-2 (Dyer et al., 2003) have an α/β-fold consisting of a five stranded β-sheet and four or five α-helices. A C-terminal segment, together with part of the β-sheet and four α-helices form a hydrophobic tunnel, which is very suitable for binding of lipids or other hydrophobic ligands. According to generated models, AtSCP2 has a similar α/β-fold forming a hydrophobic tunnel (Edqvist et al., 2004). AtSCP2 and also all other plant SCP-2 that have been identified are single-domain polypeptides (Edqvist and Blomqvist, 2006; Viitanen et al., 2006), whereas, as indicated above, SCP-2 domains in animals and many other eukaryotes are often present in the terminal of polypeptides which carry multiple protein domains.
The only SCP-2 domain encoded in A. thaliana is the single-domain protein AtSCP-2. As described above and in Edqvist and Blomqvist (2006), the situation is more complex in animals, with larger SCP-2 gene families and often quite complicated arrays of protein domain fusions. We reason that this turns A. thaliana into a very suitable model organism for studying the function of the still enigmatic SCP-2 domain. Here, an initial investigation on the biological function of AtSCP2 is presented. It is shown that the activity of the peroxisomal protein AtSCP2 is important for the metabolism in A. thaliana seeds and seedlings.
Materials and methods
Plant materials and growth conditions
A. thaliana ecotype Columbia (Col-0) was used as the wild-type plant. Seeds of the T-DNA insertion lines Sail_1231_F11 were purchased from the European Arabidopsis Stock Centre (NASC) (Loughborough, UK). The Sail_1231_F11 line is referred to as Atscp2-1. Seeds were surface-sterilized (washed in 70% ethanol for 1 min and in 15% chlorine and 0.5% SDS for 10 min followed by at least four washes in sterile distilled water) and sown on half-strength Murashige and Skoog medium (1/2 MS). Before cultivation, seed dormancy was broken by 72 h of cold treatment (4 °C). The synthetic auxin 2,4-dichlorophenoxybutyric acid (2,4-DB) (0.1 μM, 4 μM) and indole-3-butyric acid (IBA) (3 μM, 30 μM) were added to the autoclaved medium where indicated. Ten-day-old plants grown under sterile conditions were transplanted on soil mixed with vermiculite (2:1 v/v). The plants were cultivated under controlled conditions in environmental chambers at 20–22 °C under long day (16/8 h light/dark) conditions. The Atscp2-1 mutant was back-crossed to wild-type A. thaliana Col-0.
For expression of AtSCP2 in Atscp2-1 under the control of its own promoter, a DNA fragment carrying the AtSCP2 gene including the promoter was obtained through amplification of A. thaliana genomic DNA with primers ATSCP2promattB1F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCACACCTCCTATTTATCGGACAT-3′) and AtSCP2attB2R (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCACAACTTTGAAGGTTTACGGAAGAT-3′). The PCR fragment was recombined into the destination vector pMDC99 (Curtis and Grossniklaus, 2003) resulting in the plasmid pJE602. For expression of AtSCP2 cDNA under control of the cauliflower mosaic virus (CaMV) 35S promoter, a fragment carrying a cDNA copy of AtSCP2 was amplified from A. thaliana cDNA with ATSCP2attB1F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCGAATACCCAACTCAAATC-3′) and ATSCP2attB2R. The PCR fragment was recombined into destination vector pMDC32 (Curtis and Grossniklaus, 2003) yielding plasmid pJE601. Recombination events were done with the Gateway technology from Invitrogen (Carlsbad, CA, USA). pJE601 and pJE602 were transformed into Agrobacterium tumefaciens C58. The floral dip method (Clough and Bent, 1998) was used to transform A. thaliana Atscp2-1 with A. tumefaciens C58 carrying pJE601 or pJE602. Transformations and selection of transformants were done at the Uppsala Transgenic Arabidopsis Facility. The transformants obtained were denoted Atscp2-1(35S::AtSCP2) for transformation with pJE601 and Atscp2-1(AtSCP2::AtSCP2) for transformation with pJE602.
Phenotypic assays
The hypocotyl and root lengths were measured on 3-d-old and 7-d-old seedlings grown on ½ MS medium with 0% or 1% sucrose. At least 20 seedlings were measured from each growth condition and time point. All experiments were performed in triplicate. Rosette diameter was measured at the widest point of the plant without disturbing any leaves under light. The developing seeds were counted after self-pollination. The percentage of seed germination was scored every 12 h after transferring stratified seeds on the MS media to a growth chamber. Germination was defined as an obvious protrusion of the radicle through the seed coat. Several seed lots were tested with similar results.
Histochemical and quantitative GUS activity assays
A DNA fragment carrying the AtSCP2 promoter was amplified from the A. thaliana Col-0 genome by the use of primers SCPPrU2 (5′-CACACCTCCTATTTATCGGACAT-3′) and SCPPrN2 (5′-GATTTTTGTTAGAGACTGGCACG-3′). The PCR primers were designed such that a fragment was amplified stretching from the untranslated region of the nearest gene upstream of AtSCP2 to the 5’ untranslated region of AtSCP2. The obtained 1.4 kb AtSCP2 promoter fragment was inserted into vector PCR2.1-TOPO (Invitrogen) to yield the plasmid pER2. The AtSCP2 promoter fragment was released from pER2 by restriction enzymes XbaI+BamH1, and subsequently fused to the β-glucuronidase (GUS) reporter gene by ligation to the XbaI+BamH1 sites of vector pBI101 resulting in plasmid pER1. The plasmid pER1 was transformed to A. tumefaciens C58.
Histochemical GUS-assays were performed as described by Jefferson et al. (1987). Plant tissues were incubated in a substrate solution containing 50 mM Na-phosphate buffer (pH 7.0), 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid cyclohexyl ammonium salt (X-GlcA CHA) (Duchefa Biochemie, Haarlem, The Netherlands), 0.5 mM K4Fe(CN)6, 0.5 mM K3Fe(CN)6, and 0.01% (w/v) Triton X-100 at 37 °C overnight. Stained samples were incubated in 95% ethanol at room temperature to extract the chlorophyll.
Quantitative real-time reverse transcriptase-PCR, reverse transcriptase-PCR and genomic PCR
RNA was extracted from A. thaliana using the Qiagen RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Five μg RNA was used for cDNA synthesis using oligo dT-primer and Superscript II Rnase-Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Amplification of the cDNA was performed in the presence of gene-specific primers and the SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) in MicroAmp Optical 96-well reaction plates with optical covers using an ABI Prism 7000 Sequence Detector (Applied Biosystems). Reaction conditions were 50 °C for 2 min, 94 °C for 10 min, followed by 40 cycles of 94 °C for 15 s and 60 °C for 1 min. All cDNA-samples were included in triplicate in all assays. Primers were designed using Primer express software (Applied Biosystems). Relative quantification of gene expression data was carried out with the 2–ΔΔCT or comparative CT method (Livak and Schmittgen, 2001). The threshold cycle (CT) indicates the cycle number at which the amount of amplified transcript reaches a fixed threshold. Expression levels were normalized with the CT values obtained for the A. thaliana tubulin β-2/β-3 chain (At5g62690). The following gene-specific primers were used: AtSCP2: SCPF1 (5′-GCCGGGAAAGAGGTCACA-3′) and SCPR1 (5′-TGTAAGTAACCTCTTCAAACCCTAATTTCT-3); A. thaliana tubulin β-2/β-3 chain (At5g62690): Tub1 (5′-ACACCAGACATAGTAGCAGAAATCAAG-3′) and Tub2 (5′-GAGCCTTACAACGCTACTCTGTCTGTC-3′); At3g21720: 3g21720F (5′-TTGCACAGATCGCAGACATCA-3′) and 3g21720R (5′-GAATTGGGTGCATTCATTGAGA-3′); At1g06570: 1g06570F (5′-CTCCGCCAAATCCGATCTT-3′) and 1g06570R (5′-GGAGGTCACCGGAGGTGAGT-3′); At3g47340: 3g47340F (5′-AACGTCTTGCCGTCATCGAT-3′) and 3g47340R (5′-CAATGGTCTTGTCCTCGTTGAA-3′); At1g08630: 1g08630F (5′-CGATGCGAGAAGCAATGTGTA-3′) and 1g08630R (5′-CGTCTAGCCGTTGGGTCATATC-3′); At1g21400: 1g21400F (5′-AGACACAGGCTGATCATTTGGTT-3′) and 1g21400R (5′-TTTGCCTCCTGGGAAATCC-3′); At3g45140: 3g45140F (5′-AGCCCCAATGGAAACAAGTCT-3′) and 3g45140R (5′-AGCAAGATTCCATAGCCAGCA-3′); At5g55730: 5g55730F (5′-GGCACCAGAGGATGGTGATG-3′) and 5g55730R (5′-TCTTTCCTTTCGCTTTCCCTTT-3′); At5g04960: 5g04960F (5′-AAACGCGAGCCGATCAAG-3′) and 5g04960R (5′-AGAGCTCCGTGATGGTGACTTT-3′); At3g14210: 3g14210F (5′-CAGGAGGAAATGGCTCATCTTCTA-3′) and 3g14210R (5′-AGCTCACGGACCGTCATTG-3′); At1g52050: 1g52050F (5′-TTCCTAAGCTACAGAAGTTTGTTCATGCATG-3′) and 1g52050R (5′-CATAGTTGACAACATCGGAATCG-3′).
Reverse transcriptase-PCR (RT-PCR) was done as described previously (Edqvist et al., 2004). The oligonucleotides ATSCPRT1 and ATSCPRT2 were used for expression analysis of AtSCP2 cDNA, while UBL1 and UBL2 were used for amplification of cDNA for the ubiquitin-conjugating enzyme E2-21 kD (At5g41340) (Edqvist et al., 2004).
For isolation of A. thaliana genomic DNA, 500 μl of an extraction buffer consisting of 200 mM TRIS, 250 mM NaCl, and 25 mM EDTA was added to plant tissues. After addition of six glass beads (3 mm in diameter), the tissues were disrupted by incubation in a FAST-prep instrument (MP Biomedicals, Irvine, CA, USA) for 30 s. 25 μl of 10% SDS was added, and the samples were incubated at 65 °C for 10 min and then centrifuged for 5 min. 450 μl of the supernatant was removed and precipitated with an equal volume of cold isopropanol. After incubation for 2 min at room temperature, samples were centrifuged for 5 min. The supernatant was discarded, and the DNA pellet obtained was dried, dissolved in water, and used for PCR. The following primers were used for analysing genomic DNA from Atscp2-1: SAIL_1231_F11_RP (5′-AACATTGCTCCAAAGGTTGGT-3′), SAIL_1231_F11_LP (5′-GGACCAAATCCAAGTCACACA-3′) and SAIL_LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGC-3′).
Expression profiling through microarray analysis
Two-day-old seedlings of the Atscp2-1 and wild-type plants grown under light conditions on 1/2 MS without sucrose were harvested and frozen quickly in liquid nitrogen. Two-day-old seedlings were selected for the expression analysis due to the high AtSCP-2 expression levels and the manifested phenotype of young Atscp2-1 seedlings when grown on 1/2 MS without exogenous carbon source. RNA samples were extracted using RNeasy Plant Mini Kit (Qiagen). Five μg of total RNA was converted to double-stranded cDNA using the SuperScript polymerase II (Invitrogen) with a T7-dT primer incorporating a T7 RNA polymerase promoter (Ambion, Austin, TX, USA). The double-stranded cDNA was amplified to cRNA through in vitro transcription using Megascript T7 kit (Ambion). To obtain aminoallyl-labelled cDNA the amplified cRNA was used as template for cDNA synthesis performed with SuperScript III (Invitrogen) in the presence of dATP, dCTP, dGTP, and aminoallyl-dUTP (aa-dUTP). After completion of cDNA synthesis unincorporated aa-dUTP was removed by using Qiagen QIAquick PCR purification kit. The purified aminoallyl-labelled cDNA (aa-cDNA) was lyophilized and subsequently dissolved in 4.5 μl of 0.1 M Na2CO3-buffer, pH 9.0. To label the aa-cDNA, 4.5 μl of the NHS-ester Cy3 or Cy5 dye, prepared in DMSO, was added and the mixture was incubated at room temperature for 1 h in darkness. The two resulting dye-labelled cDNA pools were mixed and then co-hybridized to the same A. thaliana CATMA microarray slide (Allemeersch et al., 2005) for 2 d at 42 °C in a water bath. After that, the arrays were washed and then scanned using a GenePix 4000B array scanner (Molecular Devices, Sunnyvale, CA, USA). The raw data were stored and analysed using BASE (https://base.lcb.uu.se) and the Linnaeus Centre for Bioinformatics (LCB) Data Warehouse (https://dw.lcb.uu.se) at LCB and the WCN Expression Array Facility at Uppsala University and The Swedish University of Agricultural Sciences, Uppsala, Sweden. Four replicates were made. The empirical Bayes methods B-statistics (Filtering Bstat1 >7.21) (Lönnstedt and Speed, 2002; Smyth et al., 2003; Lönnstedt and Britton, 2005) was applied to rank the genes in order of evidence for differential expression, from the strongest to the weakest evidence.
Metabolomics
Metabolites were extracted from 3-d-old seedlings of A. thaliana wild-type and Atscp2-1. Seedlings were grown on 1/2 MS without sucrose. The young seedlings were selected for metabolome profiling due to the high AtSCP-2 expression levels in wild-type and the manifested phenotype in young Atscp2-1 seedlings when grown on 1/2 MS without sucrose. From each genotype, 10 samples of 20 mg fresh weight were used for the analysis. Extraction of metabolites, GC/MS analysis, and data processing were done as described previously (Gullberg et al., 2004; Jonsson et al., 2005). All multivariate statistical investigations (PCA, PLS-DA) were performed using Simca software 10.5.0.0 (Umetrics, Umeå, Sweden). The following statistics for the PLS-DA models are discussed throughout this paper: R2X is the cumulative modelled variation in X, R2Y is the cumulative modelled variation in Y and Q2Y is the cumulative predicted variation in Y, according to cross-validation. The range of these parameters is 0–1, where 1 indicates a perfect fit.
Docking of auxin precursors into the homology model of Arabidopsis thaliana SCP-2
The docking program GOLD 3.2 (Jones et al., 1995, 1997) was used to dock the auxin precursors IBA and 2,4-DB into the previously created homology model of AtSCP-2 (Edqvist et al., 2004), which is based on the crystal structure of the SCP-2-like domain of human DBP (Haapalainen et al., 2001). The IBA and 2,4-DB structures were generated using the program SYBYL 8.0 (Tripos International, St Louis, MO, USA). Ten independent genetic algorithm runs with the default docking parameters were made in GOLD for the ligands. The binding site was restricted within a 15 Å radius of the side-chain hydrogen (HZ) of Phe112 in the AtSCP2 model. The docking was stopped if the three best scoring solutions were within 1.5 Å rmsd of each other. The cavity in the AtSCP2 model was identified using the program SURFNET (Laskowski, 1995). The docking results were visualized and examined in the BODIL modelling environment (Lehtonen et al., 2004).
Results
Expression of AtSCP2 during development
To gain insight on the expression pattern of AtSCP2, we analyzed large amount of data accessible in public databases (i.e. www.weigelworld.org, www.arabidopsis.org, and www.genevestigator.ethz.ch) from microarray analysis of gene expression during A. thaliana development. Figure 1 shows the expression of AtSCP2 in 63 samples from different tissues or stages of development. The data are from the AtGenExpress expression atlas (www.weigelworld.org) (Schmid et al., 2005) and shows that the AtSCP2 mRNA is present in all tissues and at all stages of the life of the plant. The highest levels of the AtSCP2 transcript was found in seeds, such as in green cotyledons of maturating seeds (Fig. 1, sample 63), and in floral tissues, such as the stamens of stage 12 flowers (Fig. 1, sample 51) and petals (Fig. 1, sample 50) and stamens of stage 15 flowers (Fig. 1, sample 52). The accumulation of the AtSCP2 transcript in roots, stems, buds, siliques, inflorescences, and leaves of mature A. thaliana plants was analysed with quantitative real-time RT-PCR (data not shown). The results obtained confirmed that the AtSCP2 mRNA is abundant in most A. thaliana tissues.
Fig. 1.
Expression of AtSCP2 in A. thaliana tissues. The data are collected from the microarray experiment AtGenExpress: Expression Atlas of A. thaliana (Schmid et al., 2005) obtained from www.weigelworld.org. The investigated tissue samples are from roots (RO) (samples 1–7), stems (ST) (samples 8–10), leaves (LE) (samples 11–25), whole plants (WP) (samples 26–36), shoot apex (SA) (samples 37–40), floral organs (FL) (samples 41–55), and seeds (samples 56–63) of A. thaliana Col-0. Samples referred to in the text (4, 5, 27, 28, 50, 51, and 63) are indicated with sample numbers. Samples 10, 20, 30, 40, 50, and 60 are indicated with arrows to simplify for the reader. Plants were grown on soil, unless growth substrate is indicated. (1) Root, 7 d, (2) Root, 17 d, (3) Root, 1×MS agar, 1% sucrose, 15 d, (4) Root, 8 d, 1×MS, (5) Root, 8 d, 1×MS agar, 1% sucrose, (6) Root, 1× MS agar, 21 d, (7) Root, 1×MS agar, 1% sucrose, 21 d, (8) Hypocotyl, 7 d, (9) 1st node, 21+ d, (10) 2nd internode, 21+ d, (11) Cotyledons, 7 d, (12) Leaves no. 1+2, 7 d, (13) Rosette leaf no. 4, 10 d, (14) Rosette leaf no. 2, 17 d, (15) Rosette leaf no. 4, 17 d, (16) Rosette leaf no. 6, 17 d, (17) Rosette leaf no. 8, 17 d, (18) Rosette leaf no. 10, 17 d, (19) Rosette leaf no. 12, 17 d, (20) Petiole leaf no. 7, 17 d, (21) Proximal half leaf no. 7, 17 d, (22) Distal half leaf no. 7, 17 d, (23) Leaf, 1× MS agar, 1% sucrose, 15 d, (24) Senescing leaves, 35 d, (25) Cauline leaves, 21+ d, (26) Seedling, green parts, 7 d, (27) Seedling, green parts, 1× MS agar, 8 d, (28) Seedling, green parts, 1× MS agar, 1% sucrose, 8 d, (29) Seedling, green parts, 1× MS agar, 21 d, (30) Seedling, green parts, 1× MS agar, 1% sucrose, 21 d, (31) Rosette after transition to flowering, but before bolting, 21 d, (32) Rosette after transition to flowering, but before bolting, 22 d, (33) Rosette after transition to flowering, but before bolting 23 d, (34) Vegetative rosette, 7 d, (35) Vegetative rosette, 14 d, (36) Vegetative rosette, 21 d, (37) Shoot apex, vegetative+young leaves, 7 d, (38) Shoot apex, vegetative, 7 d, (39) Shoot apex, transition (before bolting), 14 d, (40) Shoot apex, inflorescence (after bolting), 21 d, (41) Flower, stage 9, (42) Flower, stage 10–11, (43) Flower, stage 12, (44) Flower, stage 15, (45) Flower, 28 d, (46) Pedicel, stage 15, (47) Sepal, stage 12, (48) Sepal, stage 15, (49) Petal, stage 12, (50) Petal, stage 15, (51) Stamen, stage 12, (52) Stamen, stage 15, (53) Pollen, 6 weeks, (54) Carpel, stage 12, (55) Carpel, stage 15, (56) Siliques, with seeds stage 3; mid-globular to early heart embryos globular embryo, (57) Siliques, with seeds stage 4; early to late heart embryos, (58) Siliques, with seeds stage 5; late heart to mid-torpedo embryos triangle embryo, (59) Seeds, stage 6, w/o siliques; mid to late torpedo embryos torpedo embryo, (60) Seeds, stage 7, w/o siliques; late torpedo to early walking-stick embryos walking stick seed, (61) Seeds, stage 8, w/o siliques; walking-stick to early curled cotyledons embryos, (62) Seeds, stage 9, w/o siliques; curled cotyledons to early green cotyledons embryos, (63) Seeds, stage 10, w/o siliques; green cotyledons embryos.
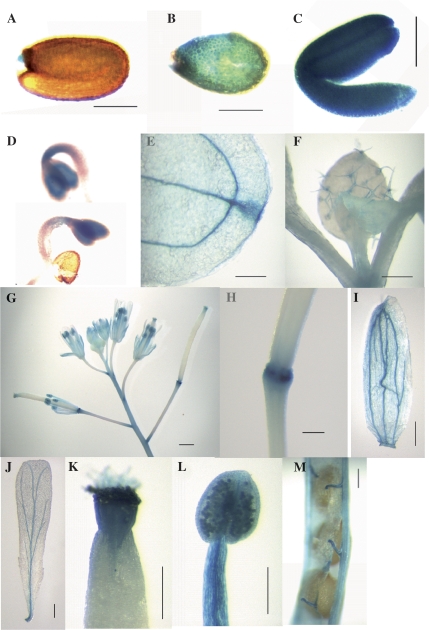
The A. thaliana AtSCP2 promoter was fused to the β-glucuronidase (GUS) reporter gene and the temporal and spatial patterns of expression were assessed during plant growth and development (Fig. 2). Endosperm (Fig. 2B), embryo (Fig. 2C), and 2-d-old seedlings (Fig. 2D) showed very high GUS activity. Staining was also detected in vascular tissues and hydathodes of cotyledons (Fig. 2E), in trichomes of the rosette leaves (Fig. 2F), in the receptacles (Fig. 2G, H), in vascular tissues of sepals and petals (Fig. 2G, I, J), in the style and stigma of the carpels (Fig. 2G, K), and in anthers, filaments, and pollen (Fig. 2G, L). The funiculi of the siliques were also shown to contain GUS activity (Fig. 2M).
Fig. 2.
Localization of GUS protein in transgenic A. thaliana plants expressing GUS from the AtSCP2 promoter. (A) Seed, (B) endosperm, (C) embryo, (D) 2-d-old seedlings, (E) cotyledon hydathode, (F) trichomes, (G) flowers, (H) receptacle, (I) sepal, (J) petal, (K) style, (L) pollen grains, anther and filament, (M) funiculus. GUS activity is visualized by the blue colour. The bar correspond to 200 μm (H, K), 250 μm (A–C, I, J, L), 500 μm (M), 1 mm (F, G) or 2 mm (E).
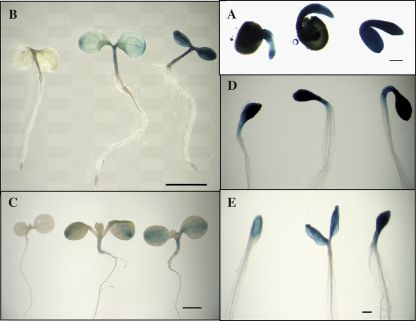
The performance of the AtSCP2 promoter-GUS fusion was also assayed in response to light, darkness, and sucrose in 1–7-d-old seedlings (Fig. 3). The 1-d-old seedlings showed high levels of GUS-activity, indicating high levels of activity of the AtSCP2 promoter early during germination. There was a decrease in expression from the AtSCP2 promoter with time, as 7-d-old seedlings showed lower GUS activity compared with 1-d-old and 3-d-old seedlings. Dark-grown seedlings showed a more intense staining, indicating a higher activity of the AtSCP2 promoter. Interestingly, the GUS activity increased with increasing amounts of sucrose when grown under constant illumination as well as in darkness. The microarray data compiled in Fig. 1 also revealed a slight increase in the accumulation of the AtSCP2 transcript in seedlings in response to sucrose (compare samples 4 and 5, and samples 27 and 28).
Fig. 3.
Sucrose and darkness stimulates expression of GUS from the AtSCP2 promoter. Seedlings were grown for 1 d (A), 3 d (B, D), or 7 d (C, E) under constant illumination (A–C) or in darkness (D, E) on 1/2 MS agar supplemented with 0% sucrose (leftmost seedling in A–E), 1% sucrose (central seedling in A–E) or 3% sucrose (rightmost seedling in A–E).
Correlated expression patterns of AtSCP2 to other genes
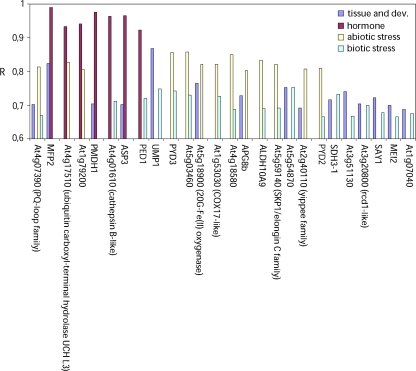
To establish a functional context of the expression pattern of AtSCP2, it was analysed whether its expression is correlated to that of other genes. To calculate Pearson correlation coefficients (R) from microarray analysis of gene expression, the gene correlator function of Genevestigator (www.genevestigator.ethz.ch) (Zimmermann et al., 2004, 2005) and the ExpressionAngler function of the Botany Array Resource in the University of Toronto (bbc.botany.utoronto.ca) (Toufighi et al., 2005) were used. Results from such investigations using ExpressionAngler on data from the AtGenExpress experiments are shown in Fig. 4. At the ExpressionAngler, the data are divided into four groups: tissue, hormone, stress, and pathogen for samples collected from various tissues and developmental stages, after hormone treatments, after abiotic stresses or biotic stresses, respectively. The 100 genes from each group giving the highest R to AtSCP2 were listed. The R cut-off values required for genes to appear on the TOP100-lists were R=0.923 (hormone), R=0.800 (stress), R=0.656 (pathogen), and R=0.686 (tissue). R >0.7 is generally considered a rule-of-thumb threshold for true correlation and used in various analysis (Lee et al., 2004; Ren et al., 2005). Figure 4 shows the R of the 26 genes that were identified in the lists from at least two of the four groups. Firstly, it can be noted that no gene was found in all the lists, and that only one gene (At4g07390 encoding a PQ-loop repeat family protein) appeared in three of the lists. Of the 25 remaining genes there are three genes (MFP2, PED1, and PMDH1) which encode proteins involved in the β-oxidation cycle (Hayashi et al., 1998; Rylott et al., 2006; Pracharoenwattana et al., 2007). The correlation of expression between MFP2 and AtSCP2 is particularly strong, with R=0.82 for tissue samples and R=0.99 for the hormone samples.
Fig. 4.
The expression of AtSCP2 is correlated to the expression of genes involved in β-oxidation. The bar graph shows the Pearson correlation coefficients (R) calculated on data from microarray analysis of gene expression. Samples are grouped in tissue and development, hormone treatments, biotic stress, and abiotic stress. Only genes, which were present among the 100 genes with the highest R values in at least two sample groups, are included in the graph. MFP2: MULTIFUNCTIONAL PROTEIN 2; PMDH1: PEROXISOMAL MALATE DEHYDROGENASE 1; ASP3: ASPARTATE AMINOTRANSFERASE 3; PED1: PEROXISOME DEFECTIVE 1; UMP1: UBIQUITIN-MEDIATED PROTEOLYSIS 1; PYD3: PYRIMIDINE DEGRADATION STEP 3; APG8b: AUTOPHAGY-RELATED UBIQUITIN-LIKE MODIFIER 8b; ALDH1: ALDEHYDE DEHYDROGENASE 1, PYD2: PYRIMIDINE DEGRADATION STEP 2; SDH3-1: SUCCINATE DEHYDROGENASE 3–1; SAY1: STERYL DEACETYLASE 1; MEI2: MEIOSIS 2; COX17: Copper chaperone for cytochrome c oxidase; SKP1: S-PHASE KINASE-ASSOCIATED PROTEIN; rcd1: required for cell differentiation 1.
Characterization of an AtSCP2 T-DNA insertion mutant
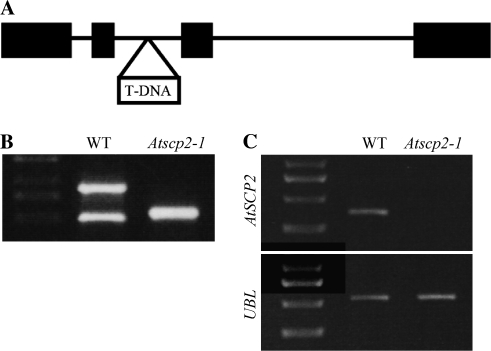
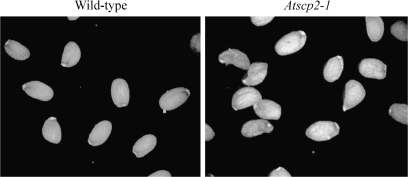
To elucidate the biological role of AtSCP2 further, the T-DNA insertion line Sail_1231_F11 was obtained from NASC (Fig. 5A). The Sail_1231_F11 line is referred to as Atscp2-1. In Atscp2-1 the T-DNA insertion is in the second intron of AtSCP2. A homozygous insertion line was obtained as confirmed by PCR analysis (Fig. 5B). Transcript analysis using RT-PCR showed that no AtSCP2 transcript was detected in Atscp2-1 seedlings (Fig. 5C). Adult plants of Atscp2-1 were normal in appearance (data not shown). Since the expression analysis revealed the highest levels of the AtSCP2 transcript in floral tissues and during seed maturation, seed development in Atscp2-1 was investigated. Thirty-three siliques were removed from wild-type and Atscp2-1 plants, and the developing seeds were examined under a microscope and counted. For wild-type plants, the average silique contained 56 green seeds, whereas the average Atscp2-1 silique contained 49 green seeds. According to the performed t test, the seed per silique numbers of Atscp2-1 were significantly different from wild-type at a P-value of 0.000155. When the morphology of the seeds was examined after harvest, it was revealed that seeds from the Atscp2-1 plants had an imperfect appearance (Fig. 6). Seeds from Atscp2-1 plants complemented with wild-type AtSCP-2 from either pJE602 containing a genomic fragment carrying AtSCP2 including the AtSCP2 promoter [Atscp2-1(AtSCP2::AtSCP2)] or pJE601 carrying a cDNA copy of AtSCP2 connected to the CaMV 35S promoter [Atscp2-1(35S::AtSCP2)] showed wild-type morphology (data not shown).
Fig. 5.
Characterization of the Atscp2-1 mutant. In (A) is the genomic structure of the AtSCP2 loci on chromosome 5 of the A. thaliana genome. Exons are indicated as bars. The location of the T-DNA insertion in Atscp2-1 is shown. (B) PCR analysis of the AtSCP2 loci in genomic DNA from A. thaliana wild-type and Atscp2-1. The absence of the AtSCP2 specific (arrow) PCR product indicates that the Atscp2-1 line is homozygous for the T-DNA insertion. (C) Analysis of the expression of AtSCP2 mRNA in wild-type Col-0 and the Atscp2-1 line. The absence of a PCR product from RT-PCR analysis of AtSCP2 cDNA shows that AtSCP2 is not expressed in the Atscp2-1 line. Information regarding primers is provided in the Materials and methods.
Fig. 6.
Seeds from Atscp2-1 have an aberrant morphology. The picture shows a random sample of seeds from one wild-type plant and one Atscp2-1 plant.
The germination of Atscp2-1 seeds was tested by scoring the radicle emergence every 12 h after transferring stratified seeds on 1/2 MS media to a growth chamber. The germination kinetics was significantly slower for Atscp2-1 seeds compared to wild-type seeds (Fig. 7). 75% of the Atscp2-1 seeds were germinated after 72 h incubation showing that the mutant seeds also had a lower germination frequency than the wild-type. Neither the lowered frequency nor the slower kinetics of germination could be rescued by the addition of 1% sucrose to the media. Germination kinetics resembling A. thaliana wild-type were obtained when Atscp2-1 was complemented with wild-type AtSCP-2 (Fig. 7).
Fig. 7.
Seeds from Atscp2-1 show slower germination kinetics and lower germination frequency. Seeds from wild-type (red lines, triangles), Atscp2-1 (35S::AtSCP2) (black lines, circles), Atscp2-1 (AtSCP2::AtSCP2) (blue lines, diamonds), and Atscp2-1 (green lines, squares) were surface-sterilized and placed on 1/2 MS-media with 1% sucrose (filled symbols) or 0% sucrose (open symbols). Seeds were then stratified for 72 h at 4 °C, and placed in growth cabinet at 22 °C. The time scale measures the time in the growth cabinet. Germination was scored as radicle protrusion. Each time point contains the scoring information from 200–300 seeds.
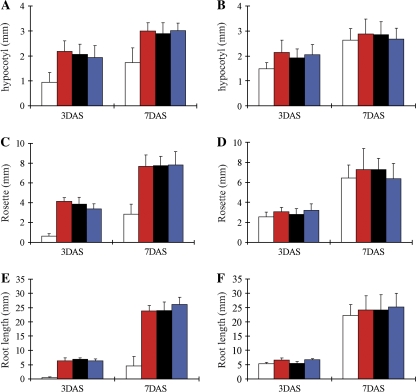
In order to investigate whether the lack of SCP-2 had an effect on seedling establishment, seeds from Atscp2-1 and wild-type were placed on agar media with or without sucrose and germinated under light or in darkness (Fig. 8). After 3 d on media without sucrose, the Atscp2-1 seedlings were clearly distinguishable from the wild-type. The insertion line showed smaller cotyledon rosettes and shorter hypocotyls than wild-type and the root elongation was also retarded (Fig. 8A–F). Seven days after sowing the root length was still retarded compared with the wild-type when grown without a carbohydrate supplement (Fig. 8E, F). Also in the dark, root elongation was inhibited on media lacking sucrose as the length of the primary root in Atscp2-1 was reduced on average to 67% and 48% of the wild-type length when assayed after 3 d and 7 d, respectively. The length of the mutant hypocotyls were slightly reduced in darkness on media lacking sucrose, as they showed a 20% and 14% reduction in length after 3 d and 7 d, respectively. Growth and development of Atscp2-1 on 1% sucrose closely matched the wild-type control, in light (Fig. 8A–F) as well as in darkness (data not shown).
Fig. 8.
AtSCP2 is important for root growth during seedling establishment. The hypocotyl length (A, B), size of the rosette (C, D) and root length (E, F) of 3-d-old and 7-d-old seedlings were measured after growing the plants on 1/2 MS without sucrose (A, C, E) or with 1% sucrose (B, D, F) under constant illumination. At least 20 seedlings were measured from each growth condition and time point. White bars: Atscp2-1, red bars: wild-type, black bars: Atscp2-1(35S::AtSCP2), blue bars: Atscp2-1(AtSCP2::AtSCP2). Error bars show standard deviation.
Effect of 2,4-DB and IBA on germination of AtSCP2 mutants
2,4-DB is β-oxidized to 2,4-dichlorophenoxyacetic acid, which inhibits root elongation. Several A. thaliana mutants with deficiencies in β-oxidation have been identified in screens for plants that elongated roots on normally inhibitory concentrations of 2,4-DB (Hayashi et al., 1998). To investigate whether the AtSCP2 protein may have a role in β-oxidation of auxin precursors and analogues, seeds of Atscp2-1 were germinated on media containing 0.1 μM and 4 μM 2,4-DB and root elongation was followed. The growth inhibition of root elongation of Atscp2-1 was indistinguishable from the control wild-type plants on any tested concentration of 2,4-DB indicating that the AtSCP2 is not involved in catabolism of 2,4-DB (data not shown). Several β-oxidation mutants also show resistance to the growth inhibition caused by IBA (Zolman et al., 2001; Adham et al., 2005). However, Atscp2-1 also showed wild-type sensitivity to IBA (data not shown).
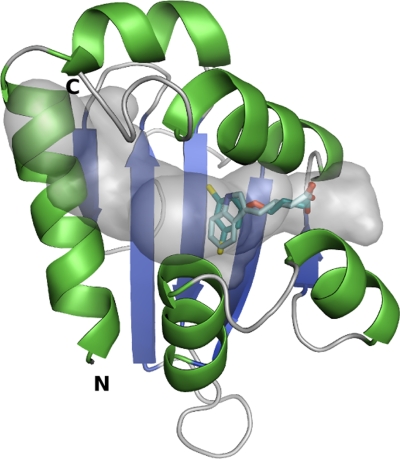
The binding mode of IBA and 2,4-DB into the homology model of AtSCP2 was studied by the automated docking program GOLD, which gave four docking results for IBA and three results for 2,4-DB. The ligands were positioned quite similarly in all the dockings and only the results with the highest fitness are presented in Fig. 9. Both IBA and 2,4-DB were positioned with their carboxyl groups in one of the cavity openings (near Gln92, Gln110, Ile102, and Leu106). The indole of IBA and the dichlorophenyl group of 2,4-DB were positioned towards the hydrophobic cavity (near Ile37, Phe74, Phe76, Phe81, Met100, and Phe112). The SURFNET plot shows that the AtSCP2 model has a very large tunnel-like cavity. Based on the docking results, the IBA and 2,4-DB molecules take up only a part of the cavity volume and, thus, do not extensively interact with the large hydrophobic cavity, indicating that they are not optimal AtSCP2 ligands (Fig. 9).
Fig. 9.
Docking of IBA and 2,4-DB into the homology model of AtSCP2. The AtSCP2 cavity is shown in transparent grey. IBA and 2,4-DB are shown in light cyan and cyan, respectively. The IBA and 2,4-DB molecules lie near one of the cavity openings and leave a large volume of the binding cavity empty, which suggests that they are not the best possible ligands for AtSCP2.
Expression profiling of Atscp2-1
To reveal the molecular mechanisms underlying the delayed root elongation of Atscp2-1 seedlings, expression profiling was performed using the CATMA microarray slides (Allemeersch et al., 2005). RNA was isolated from 2-d-old seedlings of wild-type and Atscp2-1 germinated in the light on media lacking sucrose. The RNA was used as the template for the synthesis of cDNA, which subsequently was labelled with Cy5 and Cy3 and hybridized to the gene-specific tags of 150–500 bp on the microarray slide. The empirical Bayes methods B-statistics (Lönnstedt and Speed, 2002; Smyth et al., 2003; Lönnstedt and Britton, 2005) was applied to identify and rank the 100 reporters that, among about 25 000 reporters on the slide, showed the most significant changes in expression pattern in Atscp2-1. These 100 reporters corresponded to 94 different genes from the A. thaliana genome of which 54 were up-regulated and 40 were down-regulated (including AtSCP2) in Atscp2-1 (see Supplementary Table S1 at JXB online). The expression pattern of 10 genes identified as up- or down-regulated in the microarray analysis was tested with quantitative real-time RT-PCR using gene specific primers. The results confirmed that these genes had a modified expression in Atscp2-1 (see Supplementary Table S2 at JXB online).
Those genes with a significantly altered expression represent a large range of functional categories, such as carbohydrate and amino acid metabolism, transport, and stress response. Interestingly, several genes that previously were shown to have an altered expression in seedlings of A. thaliana mutants icl-2 and mls-2 with mutations in genes encoding the key enzymes in the glyoxylate cycle, isocitrate lyase (ICL), and malate synthase (MLS) (Cornah et al., 2004), also showed an altered expression in Atscp2-1 (Table 1). In fact, of the 20 genes listed showing the most significant overexpression in icl-2 mutants, six were among the 54 genes that were up-regulated on our Top 100 list. Two out of 12 genes that showed a 2-fold repression in mls-2, and 2 out of 10 genes showing a 2-fold overexpression in mls-2 were also identified in our Top 100 list. Three genes, ASN1 (At3g47340), At2g05540, and THA1 (At1g08630), are overexpressed in Atscp2-1, icl-2, and mls-2. ASN1 is a glutamine-dependent asparagine synthase (Lam et al., 1994), At2g055490 is a Gly-rich protein, and THA1 is a Thr aldolase, which catalyse the formation of Gly from Thr (Joshi et al., 2006).
Table 1.
Atscp2-1 shows a similar pattern of gene expression as icl-2 and mls-1
| Gene | Descriptiona | Atscp2-1b | icl-2bc | mls-2bc |
| THA1 | Threonine aldolase | + | + | + |
| (At1g08630) | ||||
| At1g21400 | 2-Oxoisovalerate dehydrogenase | + | + | NA |
| At2g05540 | Gly-rich protein | + | + | + |
| ASN1 | Gln-dependent Asn-synthetase | + | + | + |
| (At3g47340) | ||||
| At5g20230 | Blue-copper binding protein | + | + | NA |
| At5g50600 | Hydroxysteroid dehydrogenase 1 | + | + | NA |
| At3g14210 | Myrosinase-associated protein | – | NA | – |
| At1g52060 | Jasmonate-inducible protein | – | NA | – |
The data are from microarray experiments, where the gene expression in Atscp2-1, icl-2, and mls-2 seedlings were compared to wild-type seedlings. Only genes ranked to be among the 100 genes showing the most significantly altered expression pattern in Atscp2-1 were included in the comparison to icl-2 and mls-2. NA, not applicable (see Cornah et al., 2004).
Descriptions are according to The Arabidopsis Information Resource (TAIR) at www.arabidopsis.org.
+ indicates that the gene is overexpressed in the mutant compared to wild-type; – indicates that the gene is expressed at lower levels in the mutant compared to in wild-type.
Data are from Cornah et al. (2004).
Metabolome analysis of Atscp2-1
We were interested in identifying metabolites that showed either significantly increased or decreased levels in Atscp2-1. Metabolome analyses were performed on 3-d-old seedlings of Atscp2-1 and A. thaliana wild-type. The samples were extracted, derivatized, and analysed by GC-MS according to Gullberg et al. (2004). The GC-MS data were analysed using hierarchical multivariate curve resolution (H-MCR; (Jonsson et al., 2005)). The data were centred and scaled to unit variance prior to partial least squares discriminant analysis (PLS-DA) classification of the genotypes. The obtained PLS-DA model (three components; R2X=0.48; R2Y=0.99; Q2Y=0.93) shows a clear separation of the genotypes for the first two components (data not shown). The identification of differences was performed by interpretation of the loadings (as described in Trygg and Wold, 2002) from the PLS-DA model together with the 99% confidence intervals calculated using jack-knifing. In seedlings, 421 variables were detected, 110 of those showed a significant difference between wild-type and Atscp2-1 according to PLS-DA analysis (one component; R2X=0.20; R2Y=0.91; Q2Y=0.81) and interpretation of first loading vector as described above. The significant metabolites were identified by comparison of retention index and mass spectra with retention index and mass spectra libraries (Schauer et al., 2005). The 20 metabolites that showed the most significant difference in accumulation between Atscp2-1 and wild-type were ranked. Twelve different metabolites were identified on the ranking list (Table 2). There were higher levels of Gln, pyroglutamic acid (a putative derivatization artefact from glutamate), Asp, β-Ala, fumaric acid, glyceraldehyde, and ribose in wild-type seedlings compared to Atscp2-1 (Table 2). The Atscp2-1 seedlings contained enhanced levels of Ser, Gly, Asn, 3-cyanoalanine, and 5-methylthiopentanitrile (tentative identification). Comparison with the previously published metabolome analyses of icl-2 and mls-2 seedlings (Cornah et al., 2004) revealed several similarities as these mutants were also shown to have lowered amounts of Gln, and that mls-2 and Atscp2-1 seedlings both contained increased levels of Ser and Gly. The increased levels of Asn in Atscp2-1 coincide with increased expression of the asparagine synthase ASN1, as shown by the microarray analysis of Atscp2-1 seedlings. Elevated levels of Gly correlated with increased expression of the Thr aldolase THA1 in Atscp2-1.
Table 2.
Compounds showing significantly different levels in 3-d-old seedlings of wild-type and Atscp2-1
| Compound | Wild-type Relativea mean area ±99% confidence interval | Atscp2-1 Relativea mean area ±99% confidence interval |
| Asp | 14 122±3572 | 8808±2170 |
| β-Ala | 798±374 | 396±65 |
| Fumaric acid | 1582±412 | 887±163 |
| Gln | 174 860±37561 | 89 282±10191 |
| Glyceraldehyde | 815±266 | 323±71 |
| Pyroglutamic acid | 312 447±55162 | 182 199±18855 |
| Ribose | 884±265 | 458±54 |
| Asn | 6477±1874 | 11 380±1560 |
| 3-Cyanoalanine | 752±100 | 1171±97 |
| Gly | 22 289±3940 | 50 760±11217 |
| 5-Methylthiopentanenitrileb | 0±0 | 876±136 |
| Ser | 27 879±9060 | 58 189±5825 |
Corrected for internal standards and weight.
Tentative identification.
Discussion
It has been shown that AtSCP2 is ubiquitously expressed in maturing seeds, young seedlings, and floral tissues, although the transcript is present throughout all developmental stages of A. thaliana. The expression of AtSCP2 is correlated to the expression of enzymes of the β-oxidation machinery, such as MFP2, PED1, and PMDH1. The AtSCP2-deficient Atscp2-1 show altered seed morphology, compromised germination, and require exogenous carbohydrates to avoid delayed seedling establishment. Transcriptome and metabolome analysis of AtSCP2-1 revealed similarities to the glyoxylate cycle mutants icl-2 and mls-2, such as increased expression of the genes ASN1 and THA1, decreased levels of Gln, and elevated levels of Gly and Ser.
According to current knowledge on β-oxidation and the glyoxylate cycle in A. thaliana (reviewed in Baker et al., 2006), fatty acids are delivered into the peroxisomes by the peroxisomal ABC-transporter CTS (also referred to as PED3 or PXA1) (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002). The delivered fatty acids are activated to acyl-CoA esters through the activity of peroxisomal acyl-CoA synthetases such as the long-chain acyl-CoA synthetases LACS6 and LACS7 (Fulda et al., 2004). The acyl-CoA esters then go through the repeated cleavage of acetate units from the thiol end through the activities of the β-oxidation enzymes: acyl-CoA oxidases ACX1-6, the multifunctional proteins MFP2 or AIM1, and the 3-ketoacyl-CoA thiolases (PED1, KAT1 and PKT2) (Hayashi et al, 1998, 1999; Richmond and Bleecker, 1999; Germain et al., 2001; Adham et al., 2005; Rylott et al., 2006). To allow for conversion of lipids into sugar via gluconeogenesis, the acetate obtained may be fed into the glyoxylate cycle for the synthesis of succinate, malate, and oxaloacetate. The reactions in the glyoxylate cycle are catalysed by citrate synthase (CSY), aconitase, ICL, MLS, and malate dehydrogenase.
Seedling establishment of the Atscp2-1 mutant is not dependent on an exogenous supply of sucrose, but sucrose is required to avoid a delay. Sucrose-dependent seedling establishment has been shown for many A. thaliana mutants deficient in proteins with roles in β-oxidation or the glyoxylate cycle, such as the acx1acx2 mutant deficient in the ACX1 long chain acyl-CoA oxidase and the ACX2 very long chain acyl CoA oxidase (Adham et al., 2005), the aim1 mutant (Richmond and Bleecker, 1999), the mfp2 mutant (Rylott et al., 2006), the pex6 mutant deficient in a putative peroxisomal ATPase (Zolman and Bartel, 2004), the pxa1/ped3/cts mutant (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002), the lacs6lacs7 mutant (Fulda et al., 2004), the ped1/kat2 mutant (Hayashi et al., 1998; Germain et al., 2001), the csy2csy3 mutant (Pracharoenwattana et al., 2005), the mls-2 mutant (Cornah et al., 2004), and the sdp2 mutant deficient in peroxisomal monohydroascorbate reductase (Eastmond, 2007). The sucrose dependency of these mutants is considered to reflect a lack of gluconeogenesis as a consequence of reduced fatty acid β-oxidation or a blocked glyoxylate cycle.
Many of the above sucrose-dependent mutants with defects in β-oxidation are able to grow in toxic levels of 2,4-DB or IBA. The phenotype of the Atscp2-1 mutant resembles that of the mfp2, lacs6lacs7, acx1acx2, and sdp2 mutants, which are sucrose-dependent but still sensitive to 2,4-DB and IBA (Fulda et al., 2004; Pinfield-Wells et al., 2005; Rylott et al., 2006; Eastmond, 2007). The phenotype of the lacs6lacs7 mutant may be explained by the fact that probably another CoA-synthetase is capable of activating 2,4-DB and IBA to the corresponding CoA-thioester before β-oxidation. Moreover, IBA and 2,4-DB sensitivity shows that the deficiency of the two long chain acyl-CoA synthetases does not cause a general block of the β-oxidation cycle in the lacs6lacs7 mutant. The 2,4-DB sensitivity of mfp2 mutants is probably due to that the other A. thaliana multifunctional protein AIM1 is active on 2,4-DB in a mfp2 background (Rylott et al., 2006). In the case of the acx1acx2 mutants, the 2,4-DB sensitivity could be due to ACX1 and ACX2 showing specificity for medium to very long chain acyl-CoA species, whereas 2,4-DB is metabolized by ACXs with short-chain specificity as suggested by Pinfield-Wells et al. (2005). The 2,4-DB sensitivity shown for sdp2 was hypothesized to be due to many peroxisomal matrix and membrane proteins remaining functional in the mutant (Eastmond, 2007). The IBA and 2,4-DB sensitivity of the Atscp2-1 mutant show that AtSCP2 is not required for β-oxidation of those compounds, and that the β-oxidation cycle is not blocked in the mutant. Furthermore, the docking analysis of IBA and 2,4-DB shows that they are not the most favourable ligands for AtSCP2, indicating that AtSCP2 would not be involved in the binding or transfer of the auxin precursors.
The Atscp2-1 mutant shows a compromised germination, with slower germination kinetics as well as a lower germination frequency. Unlike the post-germinative growth phenotype, the germination phenotype of Atscp2-1 is not rescued by the addition of exogenous sugar. The slow germination phenotype of Atscp2-1 resembles the phenotype of the lac6lac7 double mutant which reached 50% germination in 4.5 d compared to 1.5 d for wild-type (Footitt et al., 2006). Sucrose-independent germination phenotypes have also been shown for acx1acx2, ped1/kat2, and cts mutants (Pinfield-Wells et al., 2005). Seeds from these mutants showed germination frequencies below 30%, indicating a more severe block in germination compared to Atscp2-1. Seeds from ped1/kat2 and cts mutants contain significant amounts of sucrose (Footitt et al., 2002; Pritchard et al., 2002) suggesting that the germination phenotypes are not due to a limited supply of soluble sugars. Rather, as suggested recently (Pinfield-Wells et al., 2005) the β-oxidation pathway may, during germination, be utilized for the synthesis of a specific signal molecule promoting germination, or to degrade a molecule that inhibits germination. This capacity may then be compromised in Atscp2-1, lac6lac7, acx1acx2, ped1/kat2, and cts mutants leading to an inhibition of germination.
During development and maturation of A. thaliana seeds, the total fatty acid levels in the seeds increase, until the later stages of seed maturation when the levels drop to close to 30% relative to the peak levels (Baud et al, 2002). Metabolic studies of developing embryos of Brassica napus showed that at least 10% of the fatty acids stored as triacylglycerol was lost during the desiccation phase of seed development. Interestingly, metabolic labelling of the embryos revealed that β-oxidation was not associated with net gluconeogenic activity (Chia et al., 2005). The function of fatty acid breakdown during seed development remains unclear, nevertheless, the activity during seed maturation may explain the altered morphology of the Atscp2-1 seeds if AtSCP2 is involved in peroxisomal lipid utilization. We were somewhat surprised to find that sucrose stimulated the expression of AtSCP2. Rather, it had been expected that sucrose would repress the expression, as increased levels of sucrose could be signalling a limited need for lipid utilization. Further investigations of the function of AtSCP2 may possibly reveal the significance of this observation.
The Atscp2-1 seedlings had elevated levels of Asn, Ser, and Gly, and decreased levels of Asp, Glu, and Gln. Changed levels of Gln, Ser, and Gly were previously reported for icl-2 and mls-2 seedlings (Cornah et al., 2004). Asn accumulates in response to sugar starvation, probably due to an increase in expression of Asn synthase (Lam et al., 1994; Azevedo et al., 2006), which catalyses the formation of Asn through the transfer of an amide group from Gln to Asp. Our microarray experiments revealed overaccumulation of the ASN1 transcript in Atscp2-1 seedlings (see Supplementary Table S1 at JXB online) which suggests that the modified levels of Asn, Asp, Gln, and Glu in the Atscp2-1 mutant are consequences of increased accumulation of Asn synthase. Interestingly, ASN1 had elevated expression levels also in icl-2 and mls-2 seedlings (Cornah et al., 2004). Why are deficiencies in AtSCP2, ICL, and MLS leading to increased expression of ASN1? Possibly, when an exogenous carbon source is lacking, these mutant seedlings are in a condition suggestive of sugar starvation. Sugar starvation triggers ASN1 expression and leads to increased levels of Asn. Atscp2-1 also had increased levels of Gly and Ser. This was also seen for the mls-2 seedlings. In the case of mls-2, Cornah et al. (2004) hypothesized that when the glyoxylate cycle is blocked at MLS, glyoxylate feeds into the photorespiratory pathway. The increased activity of the photorespiratory pathway then leads to elevated levels of Gly and Ser. The elevated levels of Gly and Ser in Atscp2-1 possibly indicate that AtSCP2 also functions in the glyoxylate cycle. This hypothesis may also be supported by the decreased levels of fumarate in Atscp2-1. An underperforming glyoxylate cycle in Atscp2-1 may lead to lowered levels of fumarate, as fumarate is produced in the citric acid cycle from succinate, which is a product of the glyoxylate cycle. The elevated Gly levels may also, at least partially, be due to the demonstrated overexpression of THA1 in Atscp2-1. THA1, which is mainly expressed in seeds and young seedlings, encodes a Thr aldolase functioning in Thr catabolism where it is catalysing the conversion of Thr to Gly (Joshi et al., 2006). A tha1 mutant has a 50% decrease in Gly content (Joshi et al., 2006), which indicates that it is not unlikely that the overexpression of THA1 in Atscp2-1 would result in elevated Gly levels. It is interesting that the expression of THA1 was also induced in mls-2 and icl-2 seedlings (Cornah et al, 2004).Thus, it can not be excluded that the elevated Gly levels in mls-2 seedlings is, to some extent, related to increased THA1 activity.
We have presented data here showing that the activity of the peroxisomal protein AtSCP2 is important for metabolism in A. thaliana. A function in β-oxidation is supported from the co-expression with MFP2, PED1, and PMDH1. The common pattern seen in animals, fungi, and protists with fusions of SCP-2 domains to catalytic domains involved in β-oxidation could also support a direct involvement in β-oxidation for AtSCP2. However, the 2,4-DB-sensitivity shown for Atscp2-1 may suggest that AtSCP2 is not required for β-oxidation, at least not for all compounds. A function related to the glyoxylate cycle is supported by the similarities between Atscp2-1 and the glyoxylate cycle mutants icl-2 and mls-2, as revealed by transcriptome and metabolome analyses. Another possibility that cannot be excluded is that the function of AtSCP2 is not directly connected to a specific metabolic pathway. Rather, AtSCP2 could be involved in the transport, solubilization, and presentation of hydrophobic reaction intermediates from several metabolic pathways. Further detailed investigations, including lipid profiling of Atscp2-1 seedlings, as well as AtSCP2-protein and AtSCP2-ligand interactions may provide us with additional clues to the biological function of AtSCP2.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Ranking of genes with significantly altered expression in Atscp2-1 relative to wild-type.
Table S2. Comparison of results from microarray and quantitative real-time reverse transcriptase PCR (qRT-PCR) analysis of gene expression in wild-type and Atscp2-1 seedlings.
Supplementary Material
Acknowledgments
We thank Maria Andersson, Gunilla Swärd, and Gun Rönnqvist for technical assistance, Kristina Blomqvist for comments on the manuscript, and Jens Sundström for tubulin primers and advice on real-time PCR. We thank Anette Hagberg for help with designing and analysing microarray experiments. This work was supported by The Swedish Research Council (JE), Carl Trygger Foundation (JE), China Scholarship Council (BSZ), and the Natural Science Foundation of Zhejiang Province (Y305314) (BSZ).
References
- Adamski J, Normand T, Leenders F, Monte D, Begue A, Stehelin D, Jungblut PW, de Launoit Y. Molecular cloning of a novel widely expressed human 80 kDa 17 β-hydroxysteroid dehydrogenase IV. Biochemical Journal. 1995;311:437–443. doi: 10.1042/bj3110437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adham AR, Zolman BK, Millius A, Bartel B. Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. The Plant Journal. 2005;41:859–874. doi: 10.1111/j.1365-313X.2005.02343.x. [DOI] [PubMed] [Google Scholar]
- Allemeersch J, Durinck S, Vanderhaeghen R, et al. Benchmarking the CATMA microarray. A novel tool for Arabidopsis transcriptome analysis. Plant Physiology. 2005;137:588–601. doi: 10.1104/pp.104.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids. 2006;30:143–162. doi: 10.1007/s00726-005-0245-2. [DOI] [PubMed] [Google Scholar]
- Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. Chewing the fat: β-oxidation in signalling and development. Trends in Plant Science. 2006;11:124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiology and Biochemistry. 2002;40:151–160. [Google Scholar]
- Bloj B, Hughes ME, Wilson DB, Zilversmit DB. Isolation and amino acid analysis of a nonspecific phospholipid transfer protein from rat liver. FEBS Letters. 1978;96:87–89. doi: 10.1016/0014-5793(78)81068-0. [DOI] [PubMed] [Google Scholar]
- Chia TY, Pike MJ, Rawsthorne S. Storage oil breakdown during embryo development of Brassica napus (L.) Journal of Experimental Botany. 2005;56:1285–1296. doi: 10.1093/jxb/eri129. [DOI] [PubMed] [Google Scholar]
- Choinowski T, Hauser H, Piontek K. Structure of sterol carrier protein 2 at 1.8 Å resolution reveals a hydrophobic tunnel suitable for lipid binding. Biochemistry. 2000;39:1897–1902. doi: 10.1021/bi992742e. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cornah JE, Germain V, Ward JL, Beale MH, Smith SM. Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. Journal of Biological Chemistry. 2004;279:42916–42923. doi: 10.1074/jbc.M407380200. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer DH, Lovell S, Thoden JB, Holden HM, Rayment I, Lan Q. The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35A resolution. Journal of Biological Chemistry. 2003;278:39085–39091. doi: 10.1074/jbc.M306214200. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. The Plant Cell. 2007;19:1376–1387. doi: 10.1105/tpc.106.043992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist J, Blomqvist K. Fusion and fission, the evolution of sterol carrier protein-2. Journal of Molecular Evolution. 2006;62:292–306. doi: 10.1007/s00239-005-0086-3. [DOI] [PubMed] [Google Scholar]
- Edqvist J, Rönnberg E, Rosenquist S, Blomqvist K, Viitanen L, Salminen TA, Nylund M, Tuuf J, Mattjus P. Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein-2. Journal of Biological Chemistry. 2004;279:53544–53553. doi: 10.1074/jbc.M405099200. [DOI] [PubMed] [Google Scholar]
- Eklund DM, Edqvist J. Localization of non-specific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings. Plant Physiology. 2003;132:1249–1259. doi: 10.1104/pp.103.020875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO Journal. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Marquez J, Schmuths H, Baker A, Theodoulou FL, Holdsworth M. Analysis of the role of COMATOSE and peroxisomal β-oxidation in the determination of germination potential in Arabidopsis. Journal of Experimental Botany. 2006;57:2805–2814. doi: 10.1093/jxb/erl045. [DOI] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J. Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. The Plant Cell. 2004;16:394–405. doi: 10.1105/tpc.019646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM. Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. The Plant Journal. 2001;28:1–12. doi: 10.1046/j.1365-313x.2001.01095.x. [DOI] [PubMed] [Google Scholar]
- Gullberg J, Jonsson P, Nordström A, Sjöström M, Moritz T. A strategy for identifying differences in large series of metabolomic samples analysed by GC/MS. Analytical Chemistry. 2004;76:1738–1745. doi: 10.1021/ac0352427. [DOI] [PubMed] [Google Scholar]
- Haapalainen AM, van Aalten DM, Merilainen G, Jalonen JE, Pirila P, Wierenga RK, Hiltunen JK, Glumoff T. Crystal structure of the liganded SCP-2-like domain of human peroxisomal multifunctional enzyme type 2 at 1.75 Å resolution. Journal of Molecular Biology. 2001;313:1127–1138. doi: 10.1006/jmbi.2001.5084. [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M. A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. Journal of Biological Chemistry. 1999;274:12715–12721. doi: 10.1074/jbc.274.18.12715. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant and Cell Physiology. 2002;43:1–11. doi: 10.1093/pcp/pcf023. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. The Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. Journal of Molecular Biology. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G. Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. The Plant Cell. 2006;18:3564–3575. doi: 10.1105/tpc.106.044958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P, Johansson A, Gullberg J, Trygg J, Jiye A, Grung B, Marklund S, Sjöström M, Antti H, Moritz T. High through-put data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Analytical Chemistry. 2005;77:5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- Kiema TR, Taskinen JP, Pirila PL, Koivuranta KT, Wierenga RK, Hiltunen JK. Organization of the multifunctional enzyme type 1: interaction between N- and C-terminal domains is required for the hydratase-1/isomerase activity. Biochemical Journal. 2002;367:433–441. doi: 10.1042/BJ20020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Peng SS, Coruzzi GM. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiology. 1994;106:1347–1357. doi: 10.1104/pp.106.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. Journal of Molecular Graphics. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Co-expression analysis of human genes across many micro array data sets. Genome Research. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenders F, Dolez V, Begue A, Moller G, Gloeckner JC, de Launoit Y, Adamski J. Structure of the gene for the human 17β-hydroxysteroid dehydrogenase type IV. Mammalian Genome. 1998;9:1036–1041. doi: 10.1007/s003359900921. [DOI] [PubMed] [Google Scholar]
- Lehtonen JV, Still DJ, Rantanen VV, et al. BODIL: a molecular modelling environment for structure-function analysis and drug design. Journal of Computer-Aided Molecular Design. 2004;18:401–419. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lönnstedt I, Britton T. Hierarchical Bayes models for cDNA microarray gene expression. Biostatistics. 2005;6:279–291. doi: 10.1093/biostatistics/kxi009. [DOI] [PubMed] [Google Scholar]
- Lönnstedt I, Speed TP. Replicated microarray data. Statistica Sinica. 2002;12:31–46. [Google Scholar]
- Ohba T, Holt JA, Billheimer JT, Strauss JF., 3rd Human sterol carrier protein x/sterol carrier protein 2 gene has two promoters. Biochemistry. 1995;34:10660–10668. doi: 10.1021/bi00033a042. [DOI] [PubMed] [Google Scholar]
- Ohba T, Rennert H, Pfeifer SM, He Z, Yamamoto R, Holt JA, Billheimer JT, Strauss JF., 3rd The structure of the human sterol carrier protein X/sterol carrier protein 2 gene (SCP2) Genomics. 1994;24:370–374. doi: 10.1006/geno.1994.1630. [DOI] [PubMed] [Google Scholar]
- Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson TR, Graham IA. Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. The Plant Journal. 2005;43:861–872. doi: 10.1111/j.1365-313X.2005.02498.x. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. The Plant Cell. 2005;17:2037–2048. doi: 10.1105/tpc.105.031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. The Plant Journal. 2007;50:381–390. doi: 10.1111/j.1365-313X.2007.03055.x. [DOI] [PubMed] [Google Scholar]
- Pritchard SL, Charlton WL, Baker A, Graham IA. Germination and storage reserve mobilization are regulated independently in Arabidopsis. The Plant Journal. 2002;31:639–647. doi: 10.1046/j.1365-313x.2002.01376.x. [DOI] [PubMed] [Google Scholar]
- Ren XY, Fiers MW, Stiekema WJ, Nap JP. Local coexpression domains of two to four genes in the genome of Arabidopsis. Plant Physiology. 2005;138:923–934. doi: 10.1104/pp.104.055673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. The Plant Cell. 1999;11:1911–1924. doi: 10.1105/tpc.11.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter MC, Dempsey ME. Specificity and role in cholesterol biosynthesis of a squalene and sterol carrier protein. Journal of Biological Chemistry. 1971;246:1536–1539. [PubMed] [Google Scholar]
- Rylott EL, Eastmond PJ, Gilday AD, Slocombe SP, Larson TR, Baker A, Graham IA. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment. The Plant Journal. 2006;45:930–941. doi: 10.1111/j.1365-313X.2005.02650.x. [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Seedorf U, Ellinghaus P, Nofer JR. Sterol carrier protein-2. Biochimica et Biophysica Acta. 2000;1486:45–54. doi: 10.1016/s1388-1981(00)00047-0. [DOI] [PubMed] [Google Scholar]
- Seedorf U, Raabe M, Ellinghaus P, et al. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-X gene function. Genes and Development. 1998;12:1189–1201. doi: 10.1101/gad.12.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods in Molecular Biology. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. The Plant Journal. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) Journal of Chemometrics. 2002;16:119–128. [Google Scholar]
- Viitanen L, Nylund M, Eklund DM, Alm C, Eriksson AK, Tuuf J, Salminen TA, Mattjus P, Edqvist J. Characterization of SCP-2 from Euphorbia lagascae reveals that a single Leu/Met exchange enhances sterol transfer activity. FEBS Journal. 2006;273:5641–5655. doi: 10.1111/j.1742-4658.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- Wanders RJ. Peroxisomes, lipid metabolism, and peroxisomal disorders. Molecular Genetics and Metabolism. 2004;83:16–27. doi: 10.1016/j.ymgme.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Wouters FS, Bastiaens PIH, Wirtz KWA, Jovin TM. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes. EMBO Journal. 1998;17:7179–7189. doi: 10.1093/emboj/17.24.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends in Plant Science. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proceedings of the National Academy of Sciences, USA. 2004;101:1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiology. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.