Abstract
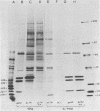
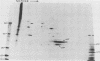
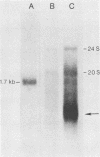
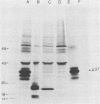
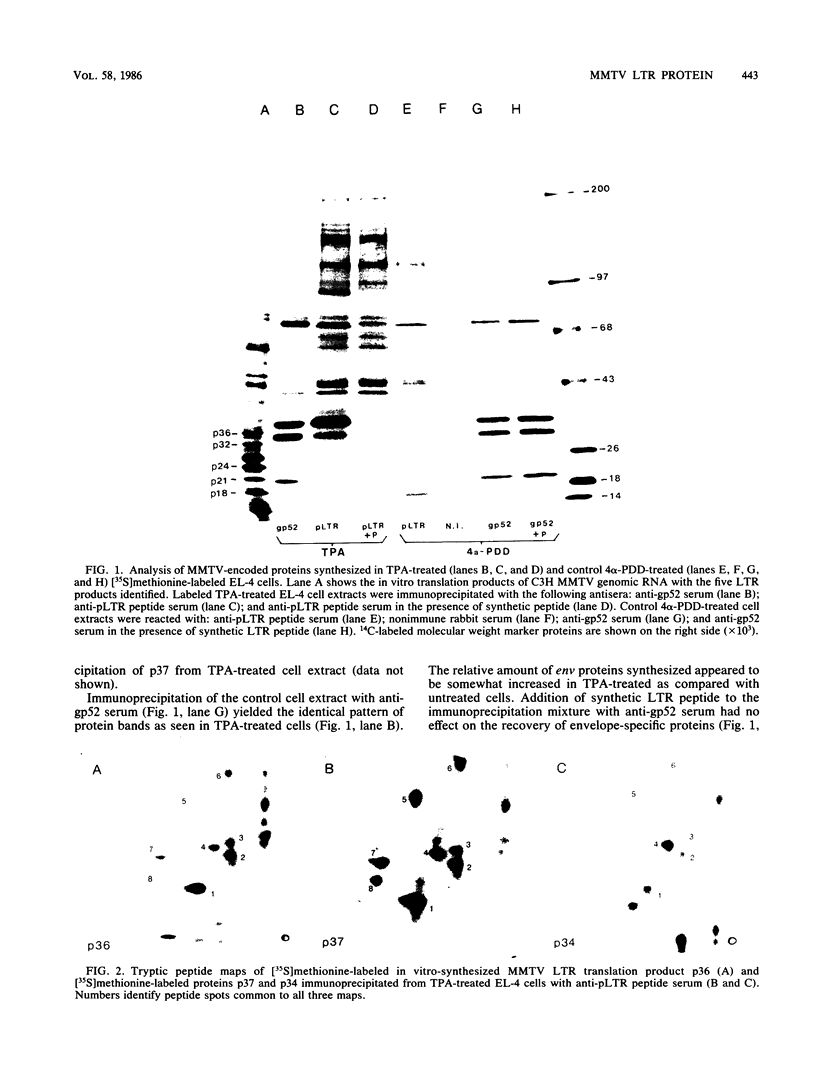
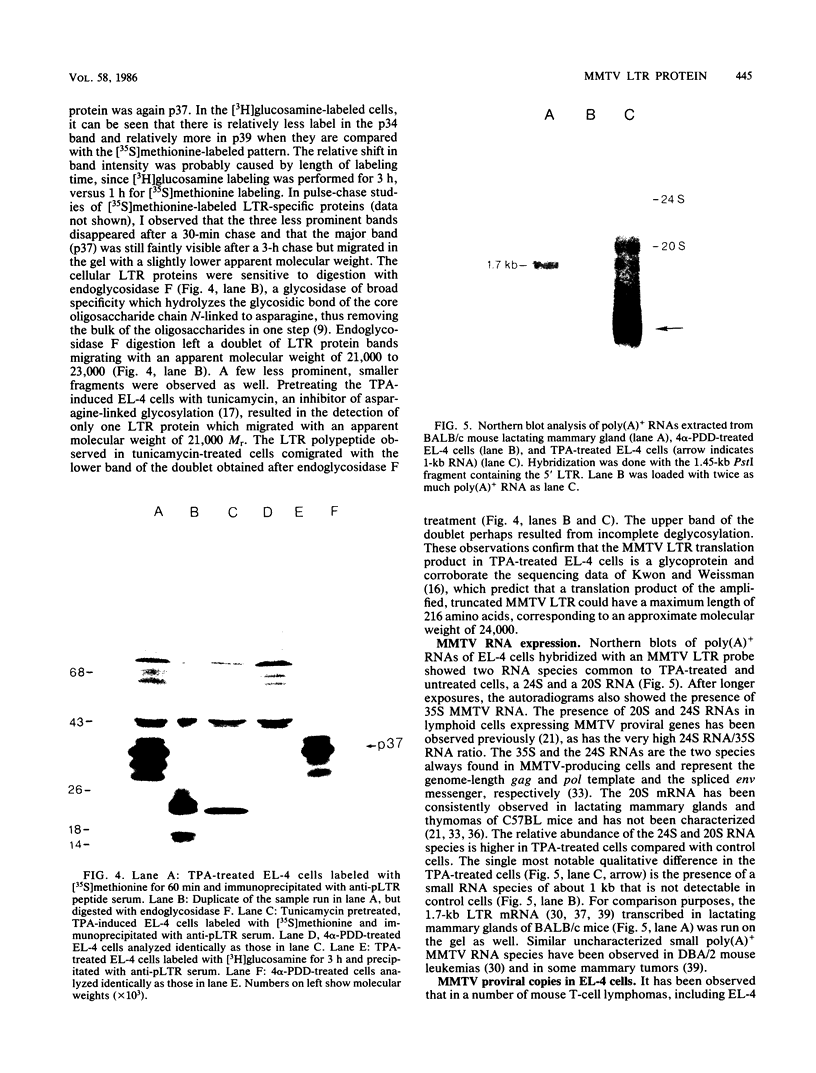
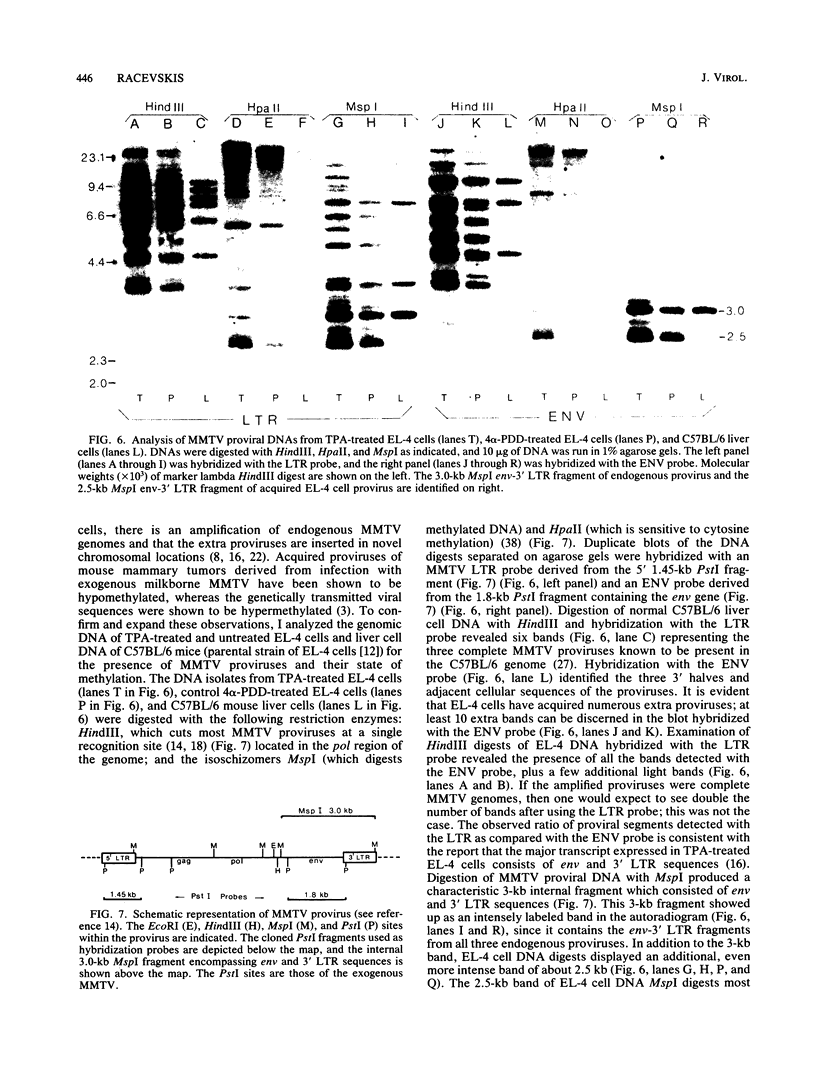
Exposure of C57BL/6 mouse EL-4 T-cell leukemia cells to phorbol ester (12-O-tetradecanoylphorbol-13-acetate) (TPA) induced the synthesis of protein products encoded by the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) region. Analysis of TPA-treated EL-4 cells with antiserum raised against a synthetic peptide predicted by the MMTV LTR open reading frame sequence detected a polypeptide migrating in gels with an apparent molecular weight of 37,000 Mr, as well as three less prominent proteins with apparent molecular weights of 31,000, 34,000, and 39,000. Tryptic peptide analysis established the identity of the immunoprecipitated cellular proteins with the LTR proteins obtained from in vitro translation of MMTV genomic RNA. All four proteins were glycosylated and were derived from one initial nonglycosylated translation product of 21,000 Mr. The 21,000-Mr apoprotein could be detected after digestion with endoglycosidase F or pretreatment of cells with tunicamycin. Untreated EL-4 cells synthesized three species of MMTV mRNA: 35S, 24S, and 20S. TPA treatment resulted in an increased level of transcription of the three mRNAs and the appearance of a new 1-kilobase mRNA. At least 10 acquired MMTV proviruses are present in the EL-4 genome, and examination of the degree of proviral methylation revealed extensive demethylation. However, no qualitative differences in the state of proviral methylation were apparent between TPA-treated and untreated cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arya S. K. Phorbol ester-mediated stimulation of the synthesis of mouse mammary tumour virus. Nature. 1980 Mar 6;284(5751):71–72. doi: 10.1038/284071a0. [DOI] [PubMed] [Google Scholar]
- Carr J. K., Traina-Dorge V. L., Cohen J. C. Mouse mammary tumor virus gene expression regulated in trans by Lps locus. Virology. 1985 Nov;147(1):210–213. doi: 10.1016/0042-6822(85)90241-7. [DOI] [PubMed] [Google Scholar]
- Cohen J. C. Methylation of milk-borne and genetically transmitted mouse mammary tumor virus proviral DNA. Cell. 1980 Mar;19(3):653–662. doi: 10.1016/s0092-8674(80)80042-0. [DOI] [PubMed] [Google Scholar]
- Dickson C., Peters G. Protein-coding potential of mouse mammary tumor virus genome RNA as examined by in vitro translation. J Virol. 1981 Jan;37(1):36–47. doi: 10.1128/jvi.37.1.36-47.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Smith R., Peters G. In vitro synthesis of polypeptides encoded by the long terminal repeat region of mouse mammary tumour virus DNA. Nature. 1981 Jun 11;291(5815):511–513. doi: 10.1038/291511a0. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Fleurdelys B., Hager G. L. Further evidence for the protein coding potential of the mouse mammary tumor virus long terminal repeat: nucleotide sequence of an endogenous proviral long terminal repeat. J Virol. 1983 Mar;45(3):941–949. doi: 10.1128/jvi.45.3.941-949.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Huang A. L., Hager G. L. Regulatory and coding potential of the mouse mammary tumor virus long terminal redundancy. J Virol. 1981 Jan;37(1):226–238. doi: 10.1128/jvi.37.1.226-238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J., Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984 Jan;49(1):92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. H., Alexander S. endo-beta-N-acetylglucosaminidase F: endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4540–4544. doi: 10.1073/pnas.79.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkind P. R., Sarkar N. H. Integration of new endogenous mouse mammary tumor virus proviral DNA at common sites in the DNA of mammary tumors of C3Hf mice and hypomethylation of the endogenous mouse mammary tumor virus proviral DNA in C3Hf mammary tumors and spleens. J Virol. 1983 Jan;45(1):114–123. doi: 10.1128/jvi.45.1.114-123.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Pearson K., Buetti E., Diggelmann H. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1982;1(1):3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORER P. A. Studies in antibody response of mice to tumour inoculation. Br J Cancer. 1950 Dec;4(4):372–379. doi: 10.1038/bjc.1950.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzburg W. H., Groner B. The chromosomal integration site determines the tissue-specific methylation of mouse mammary tumour virus proviral genes. EMBO J. 1984 May;3(5):1129–1135. doi: 10.1002/j.1460-2075.1984.tb01941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N. E., Kennedy N., Rahmsdorf U., Groner B. Hormone-responsive expression of an endogenous proviral gene of mouse mammary tumor virus after molecular cloning and gene transfer into cultured cells. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2038–2042. doi: 10.1073/pnas.78.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N., Knedlitschek G., Groner B., Hynes N. E., Herrlich P., Michalides R., van Ooyen A. J. Long terminal repeats of endogenous mouse mammary tumour virus contain a long open reading frame which extends into adjacent sequences. Nature. 1982 Feb 18;295(5850):622–624. doi: 10.1038/295622a0. [DOI] [PubMed] [Google Scholar]
- Kwon B. S., Weissman S. M. Mouse mammary tumor virus-related sequences in mouse lymphocytes are inducible by 12-O-tetradecanoyl phorbol-13-acetate. J Virol. 1984 Dec;52(3):1000–1004. doi: 10.1128/jvi.52.3.1000-1004.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt R., Schlesinger S., Kornfeld S. Tunicamycin inhibits glycosylation and multiplication of Sindbis and vesicular stomatitis viruses. J Virol. 1977 Jan;21(1):375–385. doi: 10.1128/jvi.21.1.375-385.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod J. J., Bourgeois S., Defer N., Crépin M. Demethylation and expression of murine mammary tumor proviruses in mouse thymoma cell lines. Proc Natl Acad Sci U S A. 1983 Jan;80(1):110–114. doi: 10.1073/pnas.80.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Hilkens J., Hilgers J., Groner B., Hynes N. E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982 Sep;43(3):819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Weijers P. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol Cell Biol. 1985 Apr;5(4):823–830. doi: 10.1128/mcb.5.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984 Apr 19;308(5961):693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Peters G., Smith R., Brookes S., Dickson C. Conservation of protein coding potential in the long terminal repeats of exogenous and endogenous mouse mammary tumor viruses. J Virol. 1982 Jun;42(3):880–888. doi: 10.1128/jvi.42.3.880-888.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D. O., Kriz K. G., Marich J. E., Toohey M. G. Sequence organization and molecular cloning of mouse mammary tumor virus DNA endogenous to C57BL/6 mice. J Virol. 1985 May;54(2):525–531. doi: 10.1128/jvi.54.2.525-531.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A., Honnen W. J., Li J. S. Studies with inhibitors of oligosaccharide processing indicate a functional role for complex sugars in the transport and proteolysis of Friend mink cell focus-inducing murine leukemia virus envelope proteins. Virology. 1984 Jul 15;136(1):196–210. doi: 10.1016/0042-6822(84)90259-9. [DOI] [PubMed] [Google Scholar]
- Pless D. D., Lennarz W. J. Enzymatic conversion of proteins to glycoproteins. Proc Natl Acad Sci U S A. 1977 Jan;74(1):134–138. doi: 10.1073/pnas.74.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis J., Prakash O. Proteins encoded by the long terminal repeat region of mouse mammary tumor virus: identification by hybrid-selected translation. J Virol. 1984 Sep;51(3):604–610. doi: 10.1128/jvi.51.3.604-610.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis J., Sarkar N. H. ML antigen of DBA/2 mouse leukemias: expression of an endogenous murine mammary tumor virus. J Virol. 1982 Jun;42(3):804–813. doi: 10.1128/jvi.42.3.804-813.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Robertson D. L., Varmus H. E. Dexamethasone induction of the intracellular RNAs of mouse mammary tumor virus. J Virol. 1981 Dec;40(3):673–682. doi: 10.1128/jvi.40.3.673-682.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUECK B., BOYSE E. A., OLD L. J., CARSWELL E. A. ML: A NEW ANTIGEN FOUND IN LEUKAEMIAS AND MAMMARY TUMOURS OF THE MOUSE. Nature. 1964 Sep 5;203:1033–1034. doi: 10.1038/2031033a0. [DOI] [PubMed] [Google Scholar]
- Sen G. C., Racevskis J., Sarkar N. H. Synthesis of murine mammary tumor viral proteins in vitro. J Virol. 1981 Mar;37(3):963–975. doi: 10.1128/jvi.37.3.963-975.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya A. B., Taraschi N. E., Tancin S. L., Long C. A. Regulation of endogenous murine mammary tumor virus expression in C57BL mouse lactating mammary glands: transcription of functional mRNA with a block at the translational level. J Virol. 1983 Jun;46(3):818–828. doi: 10.1128/jvi.46.3.818-828.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. A., Butel J. S., Medina D., Cardiff R. D., Hager G. L. Transcription of mouse mammary tumor virus: identification of a candidate mRNA for the long terminal repeat gene product. J Virol. 1983 Apr;46(1):42–49. doi: 10.1128/jvi.46.1.42-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooyen A. J., Michalides R. J., Nusse R. Structural analysis of a 1.7-kilobase mouse mammary tumor virus-specific RNA. J Virol. 1983 May;46(2):362–370. doi: 10.1128/jvi.46.2.362-370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]