Abstract
Gene targeting provides a powerful tool for dissecting gene function. However, repeated targeting of a single locus remains a practice mostly limited to unicellular organisms that afford simple targeting methodologies. We developed an efficient method to repeatedly target a single locus in Drosophila. In this method, which we term “site-specific integrase mediated repeated targeting” (SIRT), an attP attachment site for the phage phiC31 integrase is first targeted to the vicinity of the gene of interest by homologous recombination. All subsequent modifications of that gene are introduced by phiC31-mediated integration of plasmids carrying an attB attachment site and the desired mutation. This highly efficient integration results in a tandem duplication of the target locus, which is then reduced into a single copy carrying the mutation, likely by the efficient “single strand annealing” mechanism, induced with a DNA double-strand break (DSB). We used SIRT to generate a series of six mutations in the Drosophila nbs gene, ranging from single amino acid replacements and small in-frame deletions to complete deletion of the gene. Because all of the components of SIRT are functional in many different organisms, it is readily adaptable to other multicellular organisms.
Keywords: gene targeting, position effect, site-specific integration, rare-cutting endonuclease, single strand annealing
A comprehensive understanding of gene function may require, in addition to null mutations, other mutations that disrupt specific domains or residues. Ideally, these mutations should be generated at the endogenous location of the gene to ensure their proper expression. This type of mutational analysis has been limited mostly to simpler organisms, in which targeted mutagenesis is efficient and relatively easy. However, the importance of such studies in higher eukaryotes is unequivocal. In organisms for which efficient transgenic techniques exist, such as Drosophila, an alternative is to introduce transgenes carrying different mutations into a null background. The drawback of this approach has been that each transgene is inserted at different positions and is subject to different degrees of chromosomal position effect. In addition, many location specific effects on transcription, such as transvection, are difficult to reproduce by using transgenes. Moreover, the effect of long-range regulatory elements may not be included in the transgene constructs. Over the years, the Drosophila research community has developed several methods to combat position effect. The most effective ones use site-specific integration to compare the effect of different transgenes placed at a single preselected genomic location (1–3). To this end, the bacterial phage phiC31 integrase was recently introduced into Drosophila (4). The integrase catalyzes unidirectional recombination between its attP and attB DNA targets that is irreversible under normal conditions (5). phiC31-mediated integration experiments have been carried out by direct embryo injection. This makes it the simplest and most efficient integration method currently for use in Drosophila. More recently, insulator elements were used in combination with the phiC31 system to further reduce the influence of position effects (6).
Although ectopic expression from transgenes can be useful in certain cases such as overproducing dominant negative mutants or knocking down tissue specific expression by RNAi, the endogenous locus is undoubtedly the best position in which to compare the effect of different mutations of a particular gene. Therefore, Drosophila researchers are in need of an efficient method that would allow repeated modifications of a gene at its endogenous locus. Traditional schemes in higher eukaryotes are inherently labor intensive, requiring several months for a single targeting experiment in Drosophila (7). In this study, we describe the SIRT (site-specific integrase-mediated repeated targeting) method, which combines the phiC31-mediated site-specific integration and the endogenous homologous recombination machinery. Here we demonstrate the power of the SIRT method by generating a series of six precise mutations in the nbs gene, which encodes the Drosophila homolog for the human NBS1 protein that is defective in Nijmegen breakage syndrome patients (8). Mutations range from single amino acid changes to a complete deletion of the gene. The SIRT method is also adaptable for other organisms. To facilitate the placement of att sites and mutations in DNA vectors, we devised a two-step bacterial recombineering method.
Results
Method Overview.
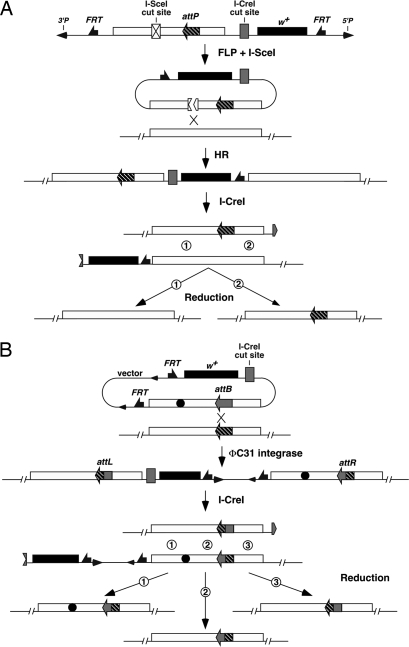
The first step of SIRT is the placement of an attP site close to the locus of interest, which is accomplished by the current ends-in targeting scheme (7, 9). A P element is constructed, which carries the white+ (w+) marker gene and a genomic fragment as targeting homology (Fig. 1A). These two fragments are flanked by FRT targets for the FLP recombinase. An attP and a cut site for the I-SceI endonuclease are placed within the target homologous fragment. Flies carrying this donor element are crossed with flies expressing FLP and I-SceI under the control of the heat shock promoter. Their progeny are heat shocked early in their development. The combined action of FLP and I-SceI generates an extrachromosomal linear DNA. Recombination between this donor and its chromosomal target generates a tandem duplication of the target locus.
Fig. 1.
A schematic overview of SIRT. (A) Current ends-in targeting scheme followed by reduction for attP placement. At the top is the donor P element. FLP and I-SceI generate a linear extrachromosomal donor molecule. Homologous recombination (HR) integrates the donor into the target locus, creating a tandem duplication. I-CreI cuts between the target copies. Recombination at region 1 leads to an unchanged wild-type copy, whereas recombination at region 2 leads to a copy with the attP site. (B) phiC31-mediated site-specific integration followed by I-CreI-induced reduction. Note that the integrated donor is the entire plasmid carrying vector sequences that include FRTs and P ends. The donor plasmid also carries the desired modification (filled circle). Recombination between attP and attB creates attR and attL sites that are no longer the targets for phiC31. I-CreI-induced recombination that occurs at region 1 leads to a target gene carrying the modification and attR. Recombination at region 2 leads to a normal target gene with attR, which can serve as a control locus in subsequent phenotypic analyses. Recombination at region 3 leads to a normal target gene with attL.
Flies with the duplication, which have red eyes (w+), are then crossed to flies expressing the I-CreI rare-cutting endonuclease under the control of the heat shock promoter, and their progeny are heat shocked. The original donor P element carries a strategically placed cut site for I-CreI, so that a future double-strand break (DSB) can be induced between the tandem target copies. This DSB stimulates recombination between the target repeats, converting the duplication to a single copy. The reduction events can be recovered as white-eyed flies, some of which carry attP (Fig. 1A).
Homozygous lines with the properly placed attP are established and used in injection experiments in which donor plasmids are introduced along with a source for phiC31. The donor plasmids are very similar to the one used to create the original attP-containing P element, with important modifications. First, they lack the cut site for I-SceI. Second, an attB replaces attP at the identical position. Third, they carry various modifications of the target gene. The phiC31 integrase inserts the donor plasmid into the chromosome, again creating a tandem duplication of the target gene. These flies are crossed to I-CreI-expressing flies for a second round of reduction, giving rise to the final product (Fig. 1B).
An Allelic Series for Drosophila nbs.
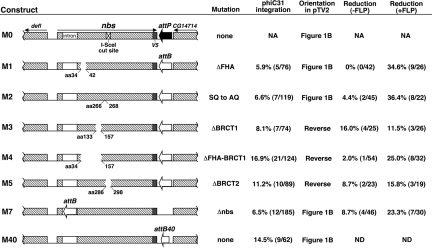
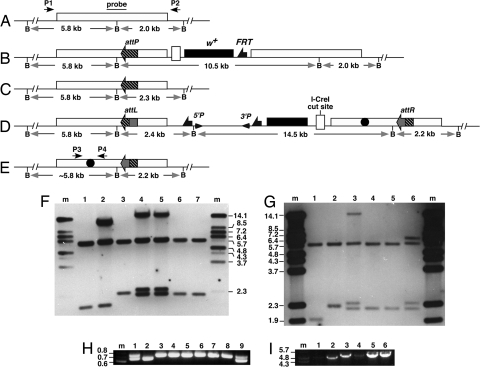
We are interested in the role of Drosophila NBS in genome maintenance (10), and set out to generate an nbs allelic series (Fig. 2). To facilitate the placement of att sites and mutations in DNA vectors, we devised a two-step bacterial recombineering method [see supporting information (SI) Results]. We decided to place an attP site in the intergenic region between nbs and the CG14174 gene. The M0 vector was constructed for the purpose of attP targeting to the genome. This vector is based on the generic gene-targeting vector pTV2 (9) and carries 5 kb of nbs homologous sequences with an I-SceI cut site placed in the middle region. This fragment also carries DNA sequences from the two genes that flank nbs. We placed a short sequence encoding the V5 epitope tag to the C terminus of nbs to facilitate future biochemical studies of NBS. We conducted gene targeting by using published protocols (9) and recovered several nbs-targeted events. The genomic structure in these events was verified by Southern blot analyses (Fig. 3). The presence of attP and V5 were verified by PCR and sequencing (data not shown). We subjected several independent lines to the I-CreI-induced reduction reaction by using published protocols (9). Homozygous viable, white-eyed, flies were subjected to molecular analyses to verify the presence of V5 and attP, and the overall structural integrity of the nbs locus was verified by PCR, sequencing, and Southern blot analyses (Fig. 3, and data not shown). We recovered multiple independent reduction events and selected line nbs434 for use in the rest of the study. Flies with nbs434 have normal viability and fertility, suggesting that the attP insertion did not have an adverse effect on fitness.
Fig. 2.
nbs constructs for SIRT. The genomic regions used to construct the allelic series are shown in the diagram. These regions were cloned into the pTV2 targeting vector giving rise the different M constructs. Coding regions are shown as hatched boxes, with the direction of transcription marked with arrows. Only the first intron of nbs is shown. M0 was used for attP targeting. In M2, serine residues at amino acid (aa) 266 (aa266) and aa268 were mutated to alanine. In M1, M3–5, protein regions deleted were marked with the starting and the ending residues. M40 carried a 40-bp attB site (attB40). The orientation of the nbs inserts are either identical to what is depicted in Fig. 1B or reversed. Frequencies for vasa-phiC31-mediated integration by microinjection are calculated as the percentage of fertile crosses with at least one pigmented progeny. The reduction frequencies were calculated as the number of independent reduction lines that retained the desired mutation divided by the total number of independent white-eyed lines established. −FLP, reduction frequencies without an FLP-mediated vector excision step; +FLP, with the FLP step; NA, not applicable; ND, not done.
Fig. 3.
Molecular analyses of products from various SIRT steps. (A–E) Genomic structures for DNA recovered at various steps of SIRT. The distance between the BamHI (B) sites is labeled. The nbs region used as targeting homology is shown as unfilled rectangular boxes. (A) The starting unmodified nbs region. Probe for Southern blotting used in F and G and primer pair (P1 and P2) for long-range PCR in I are indicated. (B) The nbs duplication after ends-in targeting for attP placement. (C) The nbs locus in line nbs434, a reduction product from B that carries attP. (D) The nbs duplication after phiC31-mediated integration. The 5′ and 3′ ends of P element are shown as arrowheads. An nbs mutation is shown as a filled circle. (E) The final SIRT product carrying the mutation and attR. Primer pair (P3 and P4) used in H are shown. (F) Southern blot analysis of M5. Lane m, markers in kb; lane 1, wild-type as in A; lane 2, nbs duplication as in B; lane 3, nbs434 as in C; lanes 4 and 5, two independent M5 integration lines as in D; lanes 6 and 7, two independent M5 reduction lines as in E. (G) Southern blot analysis of M3. Lane m, markers; lane 1, wild-type; lane 2, nbs434; lane 3, an M3 integration line as in D; lanes 4 and 5, two independent M3 reduction lines (the original 5.8-kb band is slightly smaller due to the deletion); lane 6, a white-eyed line resulted from NHEJ repair. The 14.5-kb mini-white band harbors a deletion about 8 kb in size (compare lanes 3 and 6). (H) PCR characterization of potential M3 reduction events without the FLP-mediated excision step. The top band was from the wild-type nbs copy, the lower one from the M3 mutant copy. Sample 1 is an M3 integration line as in D. Sample 9 retains both nbs copies. All of the other samples have a precisely reduced nbs locus. (I) Long PCR amplification of reduction events using P1 and P2. Lanes 1 and 2, wild-type control; lane 3, nbs434 (the product is slightly bigger than wild type due to the presence of V5 and attP); lanes 4–5, two independent final reduction events with the ΔBRCT1 mutation (M3); lane 6, a final reduction event with the ΔBRCT2 mutation (M5).
We constructed five individual point mutations of nbs contained in constructs M1–5, which were derived from the original M0 construct. In these constructs, an attB replaced attP at the identical position. These plasmids were then individually injected into preblastoderm nbs434 embryos along with a source of phiC31 integrase. First, we injected in vitro-transcribed phiC31 mRNA along with the donor plasmids as previously described (4), and recovered several integration events. For later experiments, we used a transgenic line expressing phiC31 endogenously (vasa-phiC31, 11), by making lines homozygous for both nbs434 and an X-linked vasa-phiC31 transgene. We recovered multiple integration events as flies with red eyes for all of the M constructs using vasa-phiC31. Integration frequencies are reported in Fig. 2. The frequencies that we obtained (between 6% and 17%) are within the range of ones reported for phiC31-mediated integration in Drosophila (4, 11, 12). We verified the structure of at least one integrant for every M construct and six of the seven independent integrants of the M2 construct by Southern blot and sequence analyses (Fig. 3 and data not shown). All of the events created attR and attL sites as a consequence of phiC31-mediated recombination (data not shown). Importantly, independent integration events of a particular construct had identical structures (Fig. 3 and data not shown). Therefore, most if not all white+ events were canonical phiC31-mediated integration of the plasmid at nbs. We also integrated the M40 construct that have a 40-bp attB site in place of the 250-bp attB that we used in other M constructs, and recovered a comparable integration frequency (14.5%, Fig. 2). We did not observe great differences in integration frequencies when different mutation-bearing constructs were compared, suggesting that the structure of the integrating construct did not affect integration frequency greatly.
We subjected at least one integrant line per mutation to I-CreI-induced reduction by crossing them to flies expressing I-CreI. The progeny experienced I-CreI induced white loss in the soma, leading to mosaic eyes. Mosaic males were individually mated to white females to recover germ-line white loss events. With the exception of the ΔFHA mutation (M1), we were able to recover precisely reduced events that had the engineered mutations, although at frequencies that were generally lower than we expected (Fig. 2, the “-FLP” column). The reduced efficiency of white loss was evident in the soma, so that we observed a smaller degree of eye mosaicism than that found with reduction of similar nbs duplications during the gene targeting step (Fig. 1A). We used data obtained from M3 reduction to document this reduced efficiency in the male germ line. Previously, we observed that essentially all mosaic males gave rise to white-eyed progeny after germ-line induction of I-CreI, with an average of 36% of progeny showing white eyes and harboring precisely reduced events (9). For M3 reduction, only 77% (23/30) of the mosaic males gave rise to white-eyed progeny, and most male parents gave rise to less than 5% of the progeny with white eyes. In addition, we recovered many events in which part of the white gene was lost due to deletions possibly generated during nonhomologous end joining (NHEJ) repair of the I-CreI-induced DSB. In these “imprecise” events, both nbs copies were still present, as evidenced by Southern blot and PCR analyses (Fig. 3). These NHEJ events were recovered in previous experiments, but very rarely. However, for M3 reduction, 8/25 independent white-eyed events were NHEJ events. The other 17 events were precise, and 4 of them retained the ΔBRCT1 mutation.
Integration events mediated by phiC31 result in the entire plasmid being integrated, creating a tandem duplication of nbs. The structure of this duplication is slightly different from the one resulting from the integration of the attP-containing donor molecule during gene targeting (compare Fig. 1A and Fig. 1B). We hypothesized that the “extra” DNA (FRTs, P ends and vector backbone) had an adverse effect on the reduction efficiency, due either to the extra length that it added between the nbs copies or to a particular feature of the “extra” sequences. To directly test the effect of the “extra” sequences on reduction efficiency, we excised these sequences by FLP-mediated recombination between FRTs. This excision improved reduction efficiency in the soma, as evidenced by eyes with much greater degrees of mosaicism, and in the germ line, as (i) all males gave rise to white-eyed progeny (n > 100), and (ii) each male generally had more white-eyed progeny. In addition, most if not all white-eyed flies were the result of precise reduction events. For M3 reduction after FLP-mediated vector excision, all 26 events were precise reduction, of which 3 retained ΔBRCT1. We repeated the reduction experiments for all mutations after having used FLP to excise the vector sequences, and recovered generally much improved frequencies for obtaining the desired mutations (Fig. 2, the “+FLP” column). We also attempted to induce reduction using the P transposase, but did not obtain higher reduction efficiency over the “-FLP” control (see SI Results).
All nbs point mutations support viability. However, all except the “SQ to AQ” mutation (in M2) impair fertility.
Deleting the nbs Locus.
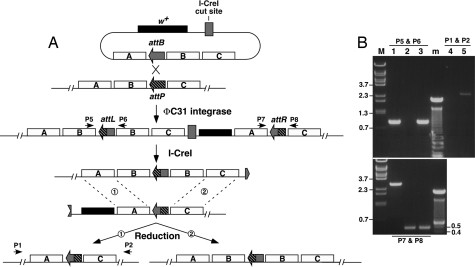
Deleting the entire coding region ensures a null allele of a gene. We designed a scheme to generate a gene deletion using SIRT, which is depicted in Fig. 4. We placed an attB site in the first intron of nbs, less than 50 bp away from the start codon (M7 in Fig. 2). The region to the left of attB corresponds to fragment A in Fig. 4A; the nbs coding region corresponds to fragment B; and the region to the right of attP in M0 corresponds to fragment C. We recovered integration events at a rate (7%) similar to that of other M constructs. We recovered four precise deletion events out of 46 reduction events without implementing an FLP-mediated vector excision step, and 7/30 with such a step. In addition, we also recovered events with a duplicated nbs locus (Fig. 4B). Therefore, SIRT can be used to make gene deletions and duplications. Animals homozygous for the nbs deletion die as pupae, similar to those with the previously identified nbs1 mutation (10).
Fig. 4.
Using SIRT to precisely delete a gene. (A) Fragment B denotes the gene to be deleted. Fragments A and C are flanking homology used to facilitate recombination. An attP placed between fragments B and C is first targeted to the chromosome. On the injected plasmid donor, an attB is placed between fragments A and B instead. After phiC31-mediated integration, a duplication of fragment B is generated to the left side of the marker gene while a deletion of fragment B is made to the right. During I-CreI-induced reduction, recombination between the two fragment A regions (“region 1”) gives rise to a deletion (left), whereas recombination between the fragment C regions (“region 2”) results in a duplication (right). The positions for primers P1, P2, P5–8 are labeled. (B) PCR analyses on nbs deletion and duplication events. Lane M: markers in kb; lane m, 100-bp ladder. Samples in lanes 1–3 were PCR amplified with P5, P6 (Upper) and P7, P8 (Lower) primer pairs. Samples in lanes 4 and 5 were PCR amplified with P1 and P2. Lane 1 was from a white-eyed fly heterozygous for an nbs duplication. The 2.9-kb product in Lower was from the wild-type nbs locus on the balancer chromosome. Lane 2 was from an nbs deletion heterozygote. Lane 3 was from a homozygous M7 integrant (red-eyed, before reduction). Lane 4 was from a duplication heterozygote, same sample as that in lane 1. No PCR product was expected due to the large size of the fragment. Lane 5 was a deletion heterozygote, same sample as that in lane 2.
Discussion
Systematic mutational analysis is a powerful tool for dissecting gene function. In this study, we developed the SIRT method to make such analysis more efficient in the multicellular organism Drosophila melanogaster. A summary flow chart for carrying out SIRT mutagenesis is shown in Fig. S2.
The Advantages of SIRT over Traditional Gene Targeting.
SIRT greatly extends the utilities for the current gene targeting method in Drosophila. Under the current scheme (7), an allelic series for a gene has to be generated, with each modification carried out as independent targeting experiments, each of which could take several months to complete. For loci that are less susceptible to targeted mutagenesis, such an allelic series would be a daunting task. In SIRT, the initial rate-limiting step is placing an att attachment site close to the gene of interest by using standard targeting methods. All subsequent modifications are introduced by means of two highly efficient processes: phiC31-mediated site-specific integration and I-CreI-induced recombination between direct repeats, each of which are many orders of magnitude more efficient than integration mediated by the endogenous recombination machinery. We recovered integrants at frequencies in line with phiC31-mediated integration at other genomic sites (4, 11, 12), although at the lower end. This could be due to the particular attP position, or simply our inexperience in embryo injection (G.G. and C.M. had not performed embryo injection before this study.).
During homologous recombination mediated targeting, imprecise integration events can be quite frequent for certain loci, generating unanticipated mutations at the target locus (7). Multiple independent events are generally needed to ensure the recovery of a precisely modified gene. On the other hand, phiC31-mediated integration does not involve new DNA synthesis, thus preserving the integrity of both the donor plasmid and the target locus. For each of the six nbs mutations (M1–M5, M7) that we generated, we recovered multiple independent integration events that were structurally identical to each other (Fig. 3 and data not shown). Therefore, even a single integration event may be sufficient for subsequent reduction experiments.
In SIRT, the tandem duplication as the result of a phiC31-mediated integration is reduced by recombination induced by I-CreI cutting. Such recombination likely occurs via the single-strand annealing (SSA) mechanism (9, 13, 14). SSA involves extensive processing of DSBs into 3′ overhangs, which is followed by annealing of single-stranded regions from both sides of the DSB. Reduction by SSA can be very efficient in the germ line. For a typical reduction reaction induced by the I-CreI endonuclease, >30% of the progeny would inherit a correctly reduced target locus (9). We observed lower reduction efficiency for the second reduction step in SIRT (Fig. 1B). This was due to the vector sequences present between the target copies as the result of phiC31 inserting the entire plasmid. Presumably, the “extra” sequence required more extensive processing, thus reducing the efficiency of SSA repair. Nevertheless, we were able to recover precise reduction events, although at reduced frequencies. Under unfavorable conditions, we recommend first eliminating the “extra” sequence by FLP-mediated excision.
SIRT is a multistep process that requires strategic placement of several DNA elements in the plasmid donor, i.e., the cut site for I-SceI, att sites, and point mutations. We developed a two-step recombineering scheme so that different combinations of these elements could be used to modify a single donor vector, creating a series of very similar constructs for both the initial gene targeting and the subsequent SIRT manipulations (see Fig. S1 and SI Materials and Methods). The relatively large size of att sites currently in use (≈250 bp) could interfere with normal gene expression of the region. It is encouraging that 39-bp att sites can support efficient recombination in human cells (15). Bischof et al. (11) showed that a 54-bp attP supported recombination with a larger attB. Results from using our M40 construct suggest that a 40-bp attB can efficiently recombine with a larger attP. Therefore, a 40-bp attB and a 54-bp attP might be sufficient for efficient integration in Drosophila.
Additional Utilities of SIRT.
Besides small modifications, e.g., point mutations and epitope tags, SIRT is also suitable for making gene deletions. This is most desirable for making a null allele of a gene. We generated a precise deletion of the nbs locus using a scheme (Fig. 4), which is similar to a previous study using ends-in targeting to make deletions (16). By changing the distance between the two att sites, one could recover deletions with different sizes.
The initial investment of targeting an att site renders the entire genomic region, not only the initial gene of interest, susceptible to SIRT-mediated manipulations. For example, one can use the nbs-proximal attP site to mutate the nearby CG14174 gene. The size limit for the region that can be modified using SIRT is determined by the clonable size of the genomic fragment under investigation. With the P[acman] system recently developed for Drosophila that facilitates cloning and integration of large genomic fragments (17), it is reasonable to speculate that a region 100 kb or more next to a previously targeted att site could be susceptible to SIRT-mediated mutagenesis. If one integrates such a large genomic fragment by phiC31-mediated integration, the duplicated target copies would share very extensive sequence homology that is likely to enhance subsequent reduction reactions.
All of the components of SIRT are likely to be universally functional. phiC31 integrase and several rare-cutting endonucleases have been introduced into several organisms, from bacteria to plants and mammals (18, 19). SSA repair seems to be operating in every system studied, a process that can be harnessed to reduce target duplications. Therefore, SIRT can be readily adapted for multicellular systems with an existing gene-targeting methodology, such as mouse ES cells. In addition, due to the versatility of the SIRT method, a collection of targeted att insertions genome-wide may be a valuable complement to the genome-wide knock-out lines currently under construction.
Materials and Methods
Plasmid construction.
Primer sequences are listed in Table S1. Primers used for sequencing are not listed, and their positions and sequences are available upon request.
nbs targeting construct.
A 4.8-kb nbs genomic fragment was PCR amplified by using primers nbs2057d and nbs6819u, which added flanking KpnI sites, and cloned by using the TOPO-TA cloning kit from Invitrogen. An I-SceI cut site was cloned into a HindIII site within this fragment by using annealed oligos of I-site-H3plus and I-site-H3minus, with the HindIII site eliminated in the process. A V5 epitope tag was added to the AflII site at the C terminus of nbs by using annealed oligos: nbs-V5+ and nbs-V5−, with the AflII site eliminated in the process. This KpnI fragment was cloned into the pTV2 targeting vector (9).
nbs mutant constructs.
Point mutations of nbs were introduced into the KpnI fragment in TOPO vector by using the Phusion site-directed mutagensis kit from NEB. Primers nbs5447d and nbs5398u were used for ΔFHA (as in M1); nbs4726d and nbs4725u for SQ to AQ (in M2); nbs5148d and nbs5147u for ΔBRCT1 (in M3); nbs5447d and nbs5147u for ΔFHA-BRCT1 (in M4); and nbs4630d and nbs4629u for ΔBRCT2 (in M5). The entire nbs coding region was sequenced to verify that no other mutation was introduced. These constructs were cloned into pTV2 using the KpnI sites. The attP and attB sites were introduced by recombineering (see SI Materials and Methods).
Stocks and Genetics.
Ends-in targeting of attP using the M0 vector was performed according to published protocols (9), and I-CreI-induced reduction crosses were also performed according to published protocols (9). I-CreI-induced reduction occurred in premeiotic germ cells so that multiple events from the same male parent could be identical events that had been amplified by germ cell mitoses. We followed a common practice of retaining only one event per male parent to ensure having independent events for subsequent experiments. Because these males were mated individually to three females, and individual males did not show systematic differences in fertility (we routinely recovered 100–150 progeny per male), the percentages of males with at least one of the desired product (Fig. 2) are likely a reasonable measure of the frequency at which different events of interest occurred. phiC31-mediated site-specific integration by microinjection was performed according to published protocols (4, 11). An X-linked vasa-phiC31 line was kindly provided by Drs. J. Bischof and K. Basler (11). The constitutively active 70FLP10 line was used to excise plasmid vector before I-CreI-induced reduction (9). Flies with both an integrated M construct and 70FLP10 were heat shocked during early development at 38°C for 1 h.
Molecular Analyses.
Southern blot analyses were performed by using the nonradioactive kit from Amersham Bioscience. DNA was cut with BamHI, AflII, and PstI enzymes individually. A 1-kb nbs probe (Fig. 3A) was PCR amplified with primers nbs2970d and nbs3942u. Another probe was amplified with primers nbs4496d and nbs5467u. The presence of the ΔFHA mutation in M1 was verified by PCR with primers nbs5467u and nbs5275d to produce a 200-bp product for wild-type samples and a 180-bp product for mutant samples. The presence of the SQ to AQ mutation in M2 was verified by allelic PCR with primers FR7 and nbs6968u for mutant samples, and FR6 and nbs6968u for wild-type samples. Each pair gave rise to a 2.2-kb product. The presence of the ΔBRCT1 mutation in M3 was verified by PCR with primers nbs4933d and nbs5663u to produce a 730-bp product for wild-type samples and 650-bp for mutant samples. The presence of the ΔFHA-BRCT1 mutation in M4 was verified by PCR with primers nbs4933d and nbs5663u to produce a 730-bp product for wild-type samples and 356-bp for mutant samples. The presence of the ΔBRCT2 mutation in M5 was verified by PCR with primers nbs4535d and nbs4766u to produce a 230-bp product for wild-type and 200-bp for mutant samples. The primer pair nbs6998d and nbs1991u (P1 and P2, respectively, in Figs. 3A and 4A) lies outside of the nbs homology used for gene targeting. They were used to verify the 5-kb fragment from reduced lines. These primers were also used to verify the nbs deletion generated using the M7 construct. The deletion resulted in a 2.3-kb fragment. This product was sequenced to verify the deletion junction. Primers nbs2776d and nbs5663u were used to amplify the 500-bp deletion junction in M7 (P7 and P8 in Fig. 4A). Primers 3169u and nbs5275d were used to amplify the 900-bp duplication junction in M7 (P5 and P6 in Fig. 4A).
Supplementary Material
Acknowledgments.
We thank Germana Collazzo and Cassie Rauser for assistance in constructing the nbs targeting clones; Natalia Wesolowska for assistance in determining the conditions for recombineering; Drs. Michael Lichten, Bruce Paterson, and Michelle Beaucher at the National Cancer Institute (NCI) for comments on the manuscript; and Drs. Michele Calos (Stanford University), Konrad Basler (University of Zurich), and Don Court (NCI) for sending reagents. Our research is supported by the intramural research program of NCI.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805843105/DCSupplemental.
References
- 1.Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn C, Handler AM. Site-specific genomic targeting in Drosophila. Proc Natl Acad Sci USA. 2005;102:12483–12488. doi: 10.1073/pnas.0504305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberstein A, Pare A, Kaplan L, Small S. Site-specific transgenesis by Cre-mediated recombination in Drosophila. Nat Methods. 2005;2:583–585. doi: 10.1038/nmeth775. [DOI] [PubMed] [Google Scholar]
- 4.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groth AC, Calos MP. Phage integrases: Biology and applications. J Mol Biol. 2004;335:667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 6.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 8.Kondratenko I, Paschenko O, Polyakov A, Bologov A. Nijmegen breakage syndrome. Adv Exp Med Biol. 2007;601:61–67. doi: 10.1007/978-0-387-72005-0_6. [DOI] [PubMed] [Google Scholar]
- 9.Rong YS, et al. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi X, et al. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci USA. 2005;102:15167–15172. doi: 10.1073/pnas.0504981102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preston CR, Engels W, Flores C. Efficient repair of DNA breaks in Drosophila: Evidence for single-strand annealing and competition with other repair pathways. Genetics. 2002;161:711–720. doi: 10.1093/genetics/161.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rong YS, Golic KG. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 2003;165:1831–1842. doi: 10.1093/genetics/165.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie HB, Golic KG. Gene deletions by ends-in targeting in Drosophila melanogaster. Genetics. 2004;168:1477–1489. doi: 10.1534/genetics.104.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 18.Keravala A, Calos MP. Site-specific chromosomal integration mediated by phiC31 integrase. Methods Mol Biol. 2008;435:165–173. doi: 10.1007/978-1-59745-232-8_12. [DOI] [PubMed] [Google Scholar]
- 19.Belfort M, Roberts RJ. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.