Abstract
A primary step in activating the alternative nuclear factor-κB (NF-κB) pathway requires NF-κB2/p100 processing to generate p52. In most cases, stimuli-induced p100 processing is dependent on NF-κB-inducing kinase/IκB kinase α-mediated phosphorylation and ubiquitination. Here, we report that post-translational modification of p100 at specific sites by the small ubiquitin-like modifier (SUMO) is another determining factor for stimuli-induced p100 processing. The results show that basal SUMO modification is required for stimuli-induced p100 phosphorylation and that blocking SUMOylation of p100, either by site-directed mutation or by short interfering RNA-targeted diminution of E2 SUMO-conjugating enzyme Ubc9, inhibits various physiological stimuli-induced p100 processing and ultimate activation of the alternative NF-κB pathway. Together, these findings show the crucial role of SUMO1 modification in p100 processing and provide mechanistic insights into the participation of SUMO1 modification in the regulation of signal transduction.
Keywords: NF-κB2, p100, phosphorylation, processing, SUMOylation
Introduction
Processing of the nuclear factor-κB2 (NF-κB2) precursor protein p100 to p52 is a main step in the activation of the alternative NF-κB pathway. A crucial molecular event in stimuli-induced p100 processing is activation of NF-κB-inducing kinase (NIK)- and IκB kinase α (IKKα)-mediated phosphorylation of p100 at Ser 866 and Ser 870 (Senftleben et al, 2001; Xiao et al, 2001). This is required for stimuli-induced recruitment of E3 ligase β-transducin repeat containing protein (β-TrCP) to the p100 complex, and subsequent ubiquitination and processing of p100 to generate p52 (Xiao et al, 2001; Liang et al, 2006).
In addition to phosphorylation, SUMOylation also activates or suppresses NF-κB activation. For example, NEMO/IKKγ, the regulatory subunit of IKK, is SUMOylated in response to genotoxic stress, which is required for NEMO phosphorylation and ultimately IKK/NF-κB activation (Huang et al, 2003; Wu et al, 2006). In stimuli-induced polyubiquitination and degradation of IκBα, the small ubiquitin-like modifier (SUMO) competes with ubiquitin for IκBα conjugation, therefore blocking IκBα degradation (Desterro et al, 1998). Nevertheless, whether and how SUMOylation is involved in the regulation of the alternative NF-κB pathway remains to be determined.
Similar to ubiquitination, SUMOylation is a dynamic process that is mediated by activating (E1), conjugating (E2) and ligating (E3) enzymes (Di Bacco & Gill, 2006). The SUMO modification of substrates is reversible and SUMO conjugation can be removed by a family of SUMO-specific proteases. SUMOylation has emerged as an important regulatory mechanism for protein activity, stability and localization (Hay, 2005). Here, we have examined the role of SUMOylation in the stimuli-induced processing of NF-κB2/p100. The results indicate that a threshold of basal p100 SUMOylation creates a priority pool of p100 that are competent for stimuli-induced phosphorylation and subsequent β-TrCP recruitment, polyubiquitination and ultimate p52 generation and NF-κB transcriptional activation. Together, these findings reveal a new regulatory mechanism of activation of the alternative NF-κB pathway.
Results And Discussion
Modification of p100 by SUMOylation
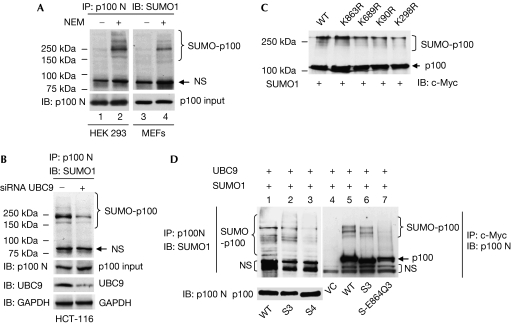
To test whether p100 is a SUMO substrate, we compared p100 modification patterns in the presence or absence of SUMO proteases inhibitor N-ethylmaleimide (NEM). In the presence of NEM, multiple bands above 100 kDa were detected in both human embryonic kidney (HEK) 293 cells and murine embryonic fibroblasts (MEFs; Fig 1A, compare lanes 1 and 2, 3 and 4). Similar SUMO patterns were detected under denaturing conditions in human colon tumour (HCT)-116 cells (supplementary Fig 1A online). The fact that anti-SUMO1 did not crossreact with ubiquitin protein (supplementary Fig 1B online) indicated that most of the SUMOylated bands detected (Fig 1A; supplementary Fig 1A online) represented SUMOylated p100. Furthermore, knockdown of UBC9, the only known SUMO E2-conjugating enzyme for the SUMOylation pathway (Gong et al, 1997; Johnson & Blobel, 1997), greatly reduced SUMO1 conjugation to p100 (Fig 1B), confirming that p100 is a SUMO substrate.
Figure 1.
In vitro and in vivo SUMO1 modification of p100. (A) SUMO1-conjugated p100 was detected by immunoprecipitation (IP) with anti-p100 N (recognizing both p100 and p52) and subsequent immunoblotting (IB) with anti-SUMO1. (B) HCT-116 cells were transfected with siRNA UBC9 oligonucleotides and control oligonucleotides. At 72 h after transfection, the cells were collected for IP and IB. (C) In vitro SUMOylation assay was performed by using in vitro-translated c-Myc-p100 and p100 mutants with single mutated SUMO site. SUMO1-conjugated p100 and non-conjugated p100 were detected by IB with anti-c-Myc. (D) HCT-116 cells were transfected with p100 or p100 SUMO mutants along with UBC9 and SUMO1 expression plasmids. S3, p100-KKK90/298/689RRR; S3-E864Q, p100-KKK90/298/689RRR-E864Q; S4, p100-KKKK90/298/689/863RRRR. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HA, haemagglutinin; HCT-116, human colon tumour 116 cells; HEK 293, human embryonic kidney 293 cells; MEFs, murine embryonic fibroblast cells; NEM, N-ethylmaleimide; NIK, NF-κB-inducing kinase; NS, non-specific; siRNA, short interfering RNA; SUMO1, small ubiquitin-like modifier 1; UBC9, ubiquitin-conjugating enzyme 9; VC, vector control; WT, wild type.
Next, we tried to identify potential SUMOylation sites. By using the online programme SUMOplot™ (Abgent, San Diego, CA, USA), we identified four putative SUMOylation sites of p100: Lys 90, Lys 298, Lys 689 and Lys 863. We mutated each of these lysines (K) to arginine (R) in an attempt to disrupt p100 SUMOylation. The results showed that single K to R substitution only slightly inhibited in vitro p100 SUMOylation (Fig 1C), indicating the involvement of several SUMO sites. We then generated the double, triple and quadruple lysine mutants. It has been reported that of the amino-acid residues in the SUMO motif, glutamic acid (E) is the most highly conserved residue apart from lysine (Rodriguez et al, 2001; Sapetschnig et al, 2002). Therefore, to confirm p100 SUMOylation further, we also generated a p100 SUMO mutant with three lysine residues replaced with arginine residues, along with a Glu 864 to glutamine (Q) mutation to test whether E to Q mutation in SUMO motif (VKED, 862–865) exerts similar effects as lysine mutation. By comparing the in vivo SUMOylation patterns of the mutants with those of wild-type p100, we found that various combinations of double lysine mutations had no effect on p100 SUMOylation (data not shown), and that SUMO1 conjugation was only slightly inhibited in the triple SUMO mutant (S3; Fig 1D, compare lanes 1 and 2, lanes 5 and 6). However, disruption of all four potential SUMOylation motifs (S4 and S3-E864Q) markedly reduced SUMO1 modification of p100 (Fig 1D, lanes 3 and 7). Thus, Lys 90, Lys 298, Lys 689 and Lys 863 were identified as SUMOylation sites of p100.
SUMOylation is essential for induced p100 processing
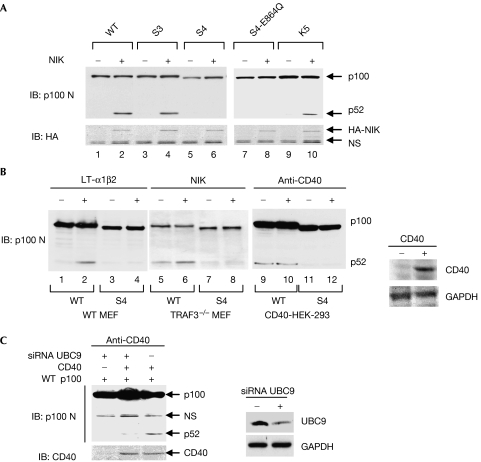
To explore the functional role of p100 SUMOylation, we compared the abilities of individual p100 SUMOylation mutants in NIK-induced p100 processing (Xiao et al, 2001). Consistent with their ability in SUMOylation, all the single, double and triple p100 K/R mutants were processed as efficiently as wild-type p100 (Fig 2A; supplementary Fig 2 online; data not shown). Markedly, NIK failed to induce processing of SUMO-deficient mutants (S4 and S3-E864Q; Fig 2A, lanes 6 and 8), suggesting that a threshold of basal SUMO modification, involving at least four SUMO sites, is required for NIK-induced p100 processing. To distinguish further the SUMO motifs from random lysine sites in NIK-induced p100 processing, we tested whether a combination of random mutation of the lysine sites of p100 would cause a similar effect on p100 processing. As expected, a combined substitution of carboxy- and amino-terminal lysines with arginines (K5) did not affect NIK-induced p100 processing (Fig 2A, compare lanes 8 and 10), confirming that it is the lack of SUMO modification that is responsible for the inhibition of NIK-induced p100 processing.
Figure 2.
SUMO1 modification is required for the induced p100 processing. (A) HCT-116 cells were transfected with wild-type (WT) p100 or p100 SUMO mutants along with empty vector or NIK. The K5 mutant stands for p100-KKKKK153/179/182/856/863RRRRR. (B) Left panel: the WT MEFs were transfected with WT p100 and S4 mutant, followed by treatment with recombinant LT-α1β2 proteins (1 μg/ml) for 16 h. Middle panel: TRAF-deficient MEFs were transfected with WT p100 and S4 mutant, with or without NIK. Right panel: HEK 293 cells were transfected with WT p100 and S4 mutant, followed by transfection with human CD40 expression vector. At 24 h after second transfection, the transfected cells were treated with anti-CD40 (10 μg/ml) for 5 h. Expression of CD40 was confirmed with immunoblotting with anti-CD40. (C) HEK 293 cells were transfected first with siRNA UBC9 and control oligonucleotides followed by transfection with p100 (WT and S4), and CD40 expression vector 24 h later. At 72 h after first transfection, the cells were stimulated with anti-CD40 (10 μg/ml) for a further 5 h. GAPDH, glyceraldehyd-3-phosphate dehydrogenase; HA, haemagglutinin; HCT-116 cells, human colon tumour 116 cells; HEK 293 cells, human embryonic kidney cells; IB, immunoblotting; MEF, murine embryonic fibroblast; NIK, NF-κB-inducing kinase; NS, non-specific; siRNA, short interfering RNA; SUMO1, small ubiquitin-like modifier 1; TRAF, tumour necrosis factor receptor-associated factor.
Next, we examined whether the observation with NIK could be extended to other cell systems with different stimuli. We found that in wild-type MEFs, lymphotoxin β-receptor (LTβR) ligation failed to induce processing of the p100 S4 mutant (Fig 2B, compare lanes 2 and 4). In tumour necrosis factor receptor-associated factor 3 (TRAF3)-deficient MEFs, the noncanonical NF-κB pathway is constitutively activated (He et al, 2006, 2007). Consistent with the previous report, we also observed that the constitutive processing of p100 and NIK expression could further enhance p100 processing in TRAF3−/− cells (Fig 2B, lanes 5 and 6). Again, regardless of the presence or absence of NIK stimulation, processing of the p100 S4 mutant to p52 was completely blocked (Fig 2B, lanes 7 and 8). Processing of the S4 mutant was also blocked in CD40-transfected HEK 293 cells (Fig 2B, lanes 9–12). Furthermore, depletion of UBC9 by short interfering RNA (siRNA) significantly inhibited CD40-induced p100 processing (Fig 2C). Collectively, these results established that SUMO1 modification is a general requirement for stimuli-induced p100 processing.
SUMOylation is required for p100 phosphorylation
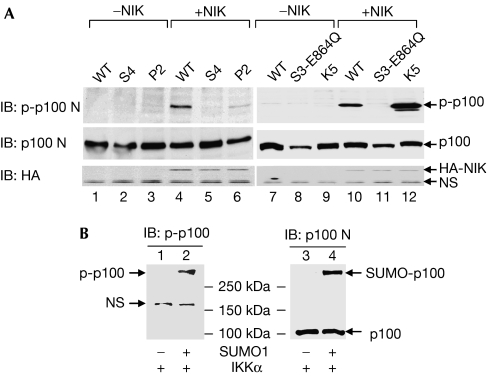
Induction of p100 phosphorylation at S866/870 is required for stimuli-induced p100 processing (Xiao et al, 2001; Liang et al, 2006). Using the p100 SUMO-deficient mutant and the p100 phosphorylation mutant (p100-SS866/870AA, labelled as P2) as tools, we tested next whether SUMOylation controls p100 processing by regulating p100 phosphorylation. Consistent with their ability in SUMOylation, all the tested single, double and triple SUMO mutants, as well as the K5 mutant, were phosphorylated after NIK stimulation as efficiently as wild-type p100 (Fig 3A; supplementary Fig 3A online; data not shown). Remarkably, NIK failed to induce phosphorylation of the SUMO-deficient mutants (S4 and S3-E864Q; Fig 3A, lanes 5 and 11). Thus these results indicate that SUMO modification is required for stimuli-induced p100 phosphorylation. Consistent with this suggestion, in murine B cells stably transfected with CD40 (M12-CD40; Liang et al, 2006), stimulation with anti-mouse CD40 rapidly induced p100 phosphorylation at S866/870 (supplementary Fig 3B online, middle panel). Interestingly, all the bands recognized as phosphorylated p100 were around and above 150 kDa, and those bands also represented SUMOylated p100 (supplementary Fig 3B online, upper and lower panels), indicating that only SUMOylated p100 can be phosphorylated. Collectively, these results strongly suggest that basal SUMOylation is required for NIK-induced p100 phosphorylation.
Figure 3.
Basal SUMO1 conjugation is required for inducible phosphorylation of p100. (A) HCT-116 cells were transfected with wild-type (WT) p100 and S4 and P2 mutants, with or without NIK. Phosphorylation of p100 was detected by immunoblotting (IB) with anti-phospho-p100-S866/870. Expression of HA-NIK was verified by immunoblotting with anti-HA. (B) In vitro translation and subsequent in vitro SUMOylation assay were performed as described in the Methods. HA, haemagglutinin; HCT-116 cells, human colon tumour 116 cells; NIK, NF-κB-inducing kinase; NS, non-specific; SUMO1, small ubiquitin-like modifier 1.
To gain direct evidence that basal SUMO modification is required for induced p100 phosphorylation, we performed an in vitro phosphorylation assay. Using in vitro-translated p100 as the substrate and immunoprecipitated IKKα as the enzyme source, we found that only SUMOylated p100 was efficiently phosphorylated (Fig 3B, compare lanes 1 and 2, 3 and 4). This finding provides direct evidence that basal SUMOylation is a prerequisite for p100 phosphorylation.
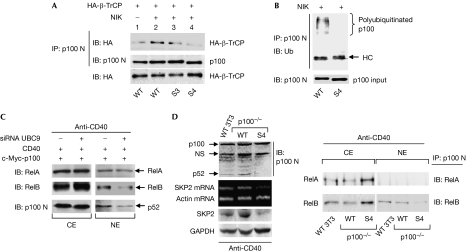
SUMOylation controls polyubiquitination of p100
To understand how SUMOylation is involved in stimuli-induced p100 phosphorylation and subsequent processing, we examined whether SUMOylation affects the cellular distribution of p100 and its interactions with IKKα; the results excluded these possibilities (supplementary Fig 4A,B online). Recent studies suggest that SUMOylation-dependent ubiquitination is a new regulatory principle that is important in cellular processes (Sun et al, 2007). Therefore, we examined whether blocking p100 SUMOylation prevents stimuli-induced recruitment of ubiquitin E3 ligase β-TrCP to the p100 complex and subsequent p100 polyubiquitination, a process essential for induced p100 processing (Fong & Sun, 2002; Xiao et al, 2004). We found that under NIK stimulation, although the recruitment of β-TrCP to wild-type p100 and S3 was markedly increased (Fig 4A, compare lanes 1 and 2, 3), the interaction between β-TrCP and the p100 S4 mutant was disrupted (Fig 4A, compare lanes 2, 3 and 4), and that the subsequent ubiquitination of S4 p100 was also blocked (Fig 4B). These findings provide further evidence that SUMO conjugation controls ubiquitin-dependent action (Prudden et al, 2007; Uzunova et al, 2007). Inhibition of p100 processing ultimately restrains the nuclear translocation of p52 and the nuclear association of p52 and its binding partners RelB and/or RelA (Coope et al, 2002). Consistent with this, we found that inhibition of p100 SUMOylation, either by siRNA downregulation of UBC9 or by structural mutation (S4 mutant), suppressed CD40- or NIK-induced p52 generation and nuclear translocation of the p52/RelB dimer, respectively (Fig 4C; supplementary Fig 5 online). It is noteworthy that the nuclear translocation of RelA was not affected in the above-mentioned studies. Similar results were obtained in NIH 3T3 p100−/− cells transiently transfected with S4 mutant after NIK stimulation (supplementary Fig 6A online) or in p100−/− stable cell lines with reconstituted S4 mutant treated with physiological stimuli, such as CD40 (Fig 4D, right panel), confirming that activation of the alternative NF-κB pathway preferably involves RelB. Activation of the alternative NF-κB pathway ultimately activates p52/RelB-mediated transcriptional activation of the S-phase kinase-associated protein 2 (SKP2; Schneider et al, 2006; Barre & Perkins, 2007). As anticipated, reconstitution of NIH 3T3 p100−/− cells with stably transfected wild-type p100 but not the S4 mutant restored CD40-mediated SKP2 transcriptional activation (Fig 4D, left panel). Similar results were obtained with p100−/− cells transiently transfected with p100 and stimulated by NIK (supplementary Fig 6B online). These findings thus validated the functional consequence of SUMO modification of p100 in the regulation of the alternative NF-κB pathway.
Figure 4.
SUMO modification of p100 is required for activation of the alternative NF-κB pathway. (A) The HCT-116 cells were transfected with c-Myc-tagged wild-type (WT) p100 and S3 and S4 mutants along with HA-β-TrCP, with or without NIK. The interaction of p100 and β-TrCP was tested by immunoprecipitation (IP) with anti-p100 N and immunoblotting (IB) with anti-HA. (B) The HCT-116 cells were transfected with WT p100 and the S4 mutant along with NIK. Polyubiquitination of the p100 complex was examined by IP with anti-p100 N and IB with an antibody against ubiquitin (Ub). (C) Downregulation of UBC9 inhibits CD40-induced nuclear translocation of p52/RelB. HEK 293 cells were treated as described in Fig 2C. The CE and NE fractions were used for the IP with anti-c-Myc and subsequent IB with RelA, RelB and p100N. (D) The p100−/− NIH 3T3 stable cell lines reconstituted with WT p100 or the S4 mutant were transfected with CD40 expression plasmid. At 48 h after transfection, the cells were subjected to anti-CD40 stimulation. The total messenger RNAs (mRNAs) and the whole-cell lysate were used for evaluating SKP2 mRNA and SKP2 protein level, respectively. To minimize the interference resulting from unequal mRNA input, the probes for β-actin and the probes for SKP2 were added to the same reverse transcription–PCR reaction tube. CE, cytoplasmic extract; HA, haemagglutinin; HC, heavy chain; HCT-116 cells, human colon tumour 116 cells; HEK 293 cells, human embryonic kidney cells; NE, nuclear extract; NF-κB, nuclear factor-κB; NIK, NF-κB-inducing kinase; NS, non-specific; siRNA, short interfering RNA; SKP2, S-phase kinase-associated protein 2; SUMO, small ubiquitin-like modifier; TrCP, transducin repeat containing protein; UBC9, ubiquitin-conjugating enzyme 9.
In summary, our findings provide the first demonstration, to our knowledge, that basal SUMOylation of p100 is required for stimuli-induced p100 phosphorylation, processing and activation of the alternative NF-κB pathway. SUMOylation of p100 could be of physiological relevance in the processes that involve deregulated activation of the alternative NF-κB pathway.
Methods
Reagents and antibodies. Recombinant mouse LT-α1β2 proteins were obtained from R&D Systems (Minneapolis, MN, USA); NE-PER Nuclear and Cytoplasmic Extraction Reagents were obtained from Pierce (Rockford, IL, USA). Anti-p100 N-terminal and anti-c-Myc tag (9E10) were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA); anti-haemagglutinin (HA) and anti-ubiquitin were obtained from Abgent; and anti-mouse CD40 was obtained from Biolegend (San Diego, CA, USA). Anti-SUMO1 was obtained from Zymed Laboratories (San Francisco, CA, USA). All other antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA).
Cell culture. All cell culture reagents were purchased from Mediatech Inc. (Herndon, VA, USA). HEK 293 cells, NIH 3T3 fibroblasts and MEFs were grown in Dulbecco's modified Eagle's medium with 10% FBS. TRAF3−/− MEFs were kindly provided by Dr G. Cheng and were cultured as described previously (He et al, 2006, 2007). HCT-116 cells were grown in McCoy's A medium containing 10% FBS. Murine B cells were cultured in RPMI 1640 supplemented with 10% FCS, 10 μM 2-mercaptoethanol and antibiotics (BCM-10).
Plasmids and transfection. The N-terminal c-Myc-tagged p100 was generated by inserting a c-Myc tag upstream from the p100 coding sequence. All the mutants generated by PCR were verified by automated DNA sequencing.
The expression vectors of HA-NIK, p100 phosphorylation mutant (SS866/870AA) and human HA-β-TrCP have been described previously (Xiao et al, 2001). The UBC9 and SUMO1 expression plasmids were kind gifts from Dr M. Kuehn. The oligonucleotide for UBC9 siRNA was GCAGAGGCCTACACGATTTAC (Integrated DNA Technologies Inc., Coralville, IA, USA). Negative control siRNA was obtained from Ambion (Austin, TX, USA). Transient transfections were performed by using Fugene™ (Roche Applied Science, Indianapolis, IN, USA), according to the protocol provided. The stable cell lines were generated by G418 selection. The human CD40 expression vector was purchased from Open Biosystems Inc. (Huntsville, AL, USA).
Immunoblotting and co-immunoprecipitation. Whole-cell lysates were prepared in immunoprecipitation lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40 and 10% glycerol. Unless stated in the figure legend, 20 mM NEM (Sigma, St Louis, MO, USA) was added to the lysis buffer for immunoprecipitation but not for immunoblotting. Immunoblotting and immunoprecipitation were performed as described previously (Hu et al, 2007).
In vitro translation and SUMO1 conjugation assays. In vitro translations were carried out by using the rabbit reticulocyte Lysate TNT Coupled Transcription/Translation System (Promega, Madison, WI, USA) according to the protocol provided. A 7 μl portion of the translated protein mixture was used for in vitro SUMOylation assay by using the in vitro SUMOylation kit (LAE Biotech International, Rockville, MD, USA) according to the provided protocol. After in vitro SUMOylation reaction, the reaction mixtures were subjected to immunoblotting. Non-SUMOylated p100 and SUMOylated p100 were detected by using an antibody recognizing the c-Myc tag.
In vitro p100 phosphorylation (S866/870) assay. The in vitro phosphorylation assay was carried out as described previously (Hu et al, 2004). Briefly, after in vitro translation of p100, 10 μl of the reaction mixture containing non-purified p100 was used for an in vitro SUMOylation assay. All the reagents except the SUMO1 substrate were added to the control reaction. A 10 μl portion of the reaction mixture was used for the subsequent phosphorylation reaction. In vitro phosphorylation of p100 was carried out by mixing the precipitated IKKα complex with the above-mentioned reaction mixture in kinase buffer containing 200 μM ATP (Cell Signaling Technology). The total volume for the reaction was 50 μl and the reaction was carried out at 30°C for 30 min. The entire reaction mixture was used for immunoblotting. Phosphorylated p100 was detected by using an antibody against phosphorylated p100 at S866/870.
RNA isolation and reverse transcription–PCR. Total RNA was isolated by using TRIzol reagent (Invitrogen, Cincinnati, OH, USA) according to the manufacturer's protocol. The primers used to amplify 210 bases of the SKP2 gene were 5′-CTGCTATCAGGCATGGGTGTCTCG-3′ and 5′-CTGGAAGGGAGTCCCAGGAGACAC-3′. The 146 bases of the housekeeping gene β-actin were amplified by using primers 5′-CATGGAGTCCTGTGGCATCCACGAAACT-3′ and 5′-ATCTCCTTCTGCATCCTGTCGGCAAT-3′.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Data
Acknowledgments
We thank C. Bakkenist for critical reading of the paper and valuable suggestions. TRAF3−/− MEFs were kindly provided by G. Cheng. Murine B-cell line M12.4.1 stably transfected with human CD40 (M12–CD40) was kindly provided by G.A. Bishop. This investigation was supported by the United States Public Health Service grant K22 CA111394 and Hillman Foundation fellowship to J.H. and National Institutes of Health/National Cancer Institute R01 CA116616 to G.X.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barre B, Perkins ND (2007) A cell cycle regulatory network controlling NF-κB subunit activity and function. EMBO J 26: 4841–4855 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC (2002) CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J 21: 5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT (1998) SUMO1 modification of IκBα inhibits NF-κB activation. Mol Cell 2: 233–239 [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Gill G (2006) SUMO-specific proteases and the cell cycle. An essential role for SENP5 in cell proliferation. Cell Cycle 5: 2310–2313 [DOI] [PubMed] [Google Scholar]
- Fong A, Sun SC (2002) Genetic evidence for the essential role of β-transducin repeat-containing protein in the inducible processing of NF-κB2/p100. J Biol Chem 277: 22111–22114 [DOI] [PubMed] [Google Scholar]
- Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET (1997) Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J Biol Chem 272: 28198–28201 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, Dempsey PW, Cheng G (2006) Rescue of TRAF3-null mice by p100 NF-κB deficiency. J Exp Med 203: 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Saha SK, Kang JR, Zarnegar B, Cheng G (2007) Specificity of TRAF3 in its negative regulation of the noncanonical NF-κB pathway. J Biol Chem 282: 3688–3694 [DOI] [PubMed] [Google Scholar]
- Hu J, Nakano H, Sakurai H, Colburn NH (2004) Insufficient p65 phosphorylation at S536 specifically contributes to the lack of NF-κB activation and transformation in resistant JB6 cells. Carcinogenesis 25: 1991–2003 [DOI] [PubMed] [Google Scholar]
- Hu J, Straub J, Xiao D, Singh SV, Yang HS, Sonenberg N, Vatsyayan J (2007) Phenethyl isothiocyanate, a cancer chemopreventive constituent of cruciferous vegetables, inhibits cap-dependent translation by regulating the level and phosphorylation of 4E-BP1. Cancer Res 67: 3569–3573 [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S (2003) Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell 115: 565–576 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802 [DOI] [PubMed] [Google Scholar]
- Liang C, Zhang M, Sun SC (2006) β-TrCP binding and processing of NF-κB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal 18: 1309–1317 [DOI] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654–12659 [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G (2002) Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J 21: 5206–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM (2006) IKKα controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J 25: 3801–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U et al. (2001) Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 293: 1495–1499 [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to sumoylated proteins. EMBO J 26: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K et al. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science 311: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC (2001) NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell 7: 401–409 [DOI] [PubMed] [Google Scholar]
- Xiao G, Fong A, Sun SC (2004) Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J Biol Chem 279: 30099–30105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Data