Abstract
Nonlinear chromatin configurations can juxtapose widely separated elements within a genomic locus; however, it remains unclear how these structures are established and contribute to transcriptional control. A 5′-remote locus control region (LCR) regulates the human growth hormone (hGH-N) gene. HSI, a pituitary-specific component of the hGH LCR, establishes a domain of polymerase II (PolII) transcription 5′ to hGH-N. Repression of this transcriptional domain by HSI deletion or PolII blockade decreases hGH-N expression. Here, we show that hGH-N activation is accompanied by positioning of the hGH-N promoter to this LCR transcriptional domain. Selectively blocking LCR transcription inhibits the formation of this active ‘looped' conformation. Thus, HSI is crucial for establishing a domain of noncoding PolII transcription, and this domain is intimately linked with chromatin organization of the active hGH-N locus. This integration of LCR transcription with chromatin reconfiguration constitutes a robust pathway for long-range gene activation.
Keywords: epigenetics, locus control region, looping, transcription
Introduction
Long-range controls have a crucial and often predominant role in metazoan gene activation (Ho et al, 2004; Dean, 2006). Mechanisms of long-range gene activation have been grouped into several structural models (Forsberg & Bresnick, 2001; Ho et al, 2004; Dean, 2006). However, the degree to which these models accurately reflect the full array of locus control region (LCR) functions remains to be determined and might vary among loci. The human growth hormone (hGH) cluster contains one pituitary-specific hGH-N gene and four placenta-specific paralogues (Fig 1A; Chen et al, 1989). These five genes are activated by a set of LCR determinants located 15–32 kb upstream of the cluster (Jones et al, 1995; Su et al, 2000; Ho et al, 2002). The positioning of a B-lymphocyte-specific gene, CD79b, between the pituitary-specific HSI and the hGH-N promoter increases the complexity of the hGH gene cluster (Bennani-Baiti et al, 1998), as does the location of the more distal HSIII-HSV elements within introns of the striated muscle sodium channel gene, SCN4A (Fig 1A). Defining how this complex locus achieves robust and tissue-specific expression of its corresponding genes should determine crucial relationships between chromatin structure and gene regulatory pathways.
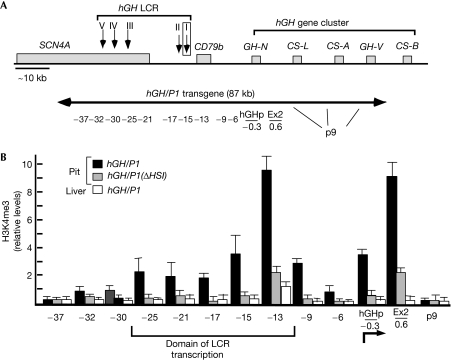
Figure 1.
H3K4me3 modifications that are both pituitary specific and HSI dependent encompass the hGH LCR and hGH-N. (A) Diagram of 100 kb of the human genome on chromosome 17q22 encompassing the hGH locus and the hGH/P1 transgene. The region shown contains the five genes of the hGH cluster, the B-lymphocyte-expressed CD79b gene and the striated muscle-specific gene SCN4A (grey rectangles). All genes are transcribed in the same orientation (from left to right). The five DNase I HS that constitute the hGH LCR are indicated by downward arrows. The extent of the 87 kb hGH/P1 transgene is shown below by the double-headed arrow. The positions of the 13 PCR amplimers used to survey the locus for H3K4me3 modification enrichment are indicated below the arrow as coordinates (kb) relative to the start site of hGH-N transcription. The 99 bp deletion in the hGH/P1(ΔHSI) transgene that removes two of the three pituitary Pit-1 binding sites is denoted by the boxed HSI. Note that the ‘HSI' amplimer (−15) used in these studies detects sequences that are adjacent to this deleted segment in the hGH/P1(ΔHSI) transgene. The p9 amplimer corresponds to a conserved sequence located 5′ to each of the four placentally expressed genes in the cluster. (B) H3K4me3 modifications in mouse transgenic pituitary (Pit) and liver chromatin and in the absence of HSI. The chromatin immunoprecipitation antibody was specific to the H3K4me3 modification. Each bar represents the mean±s.d. of four independent assays of the indicated chromatin samples: hGH/P1 pituitary chromatin (black bars), hGH/P1(ΔHSI) pituitary chromatin (grey bars) or hGH/P1 liver chromatin (white bars). Each histogram bar represents the ratio of the DNA amplification signal from Southern blot detected in the immunoprecipitated sample to that in the corresponding input sample. Each ratio was normalized to the corresponding ratio for a DNA segment within the endogenous skeletal muscle-specific mMyoD gene (Weintraub et al, 1991; Sawado et al, 2003). Each ratio, after normalization to mMyoD, is expressed on the Y axis in arbitrary units. The amplimer set corresponding to each set of bars is noted below the X axis. The site of hGH-N transcription initiation is noted below the X axis (angled arrow) and the previously defined ‘LCR domain of transcription' is indicated by the labeled bracket below the histogram. All comparisons between hGH/P1 modifications in the pituitary compared with liver are significant (P<0.001) except at −37 and p9. Comparisons between hGH/P1 and hGH/P1ΔHSI between −25 and Ex2 inclusive are significant (P<0.001). Comparisons between hGHΔHSI pituitary and hGH/P1 liver are significant only at Ex2 (P<0.01). H3K4me3, histone H3 lysine 4 trimethylation; hGH, human growth hormone; LCR, locus control region.
Previous studies have shown that the hGH LCR and the hGH-N promoter are encompassed within a 32 kb domain of acetylated histones H3 and H4 in pituitary chromatin (Elefant et al, 2000); this domain is centered at HSI,II. In mouse transgenic models, selective deletion of the pituitary-specific HSI results in the loss of histone acetylation throughout the LCR and a marked decrease in hGH-N transcription (Ho et al, 2006). HSI is also crucial for establishing a pituitary-specific ‘domain of transcription' that encompasses the LCR and the adjacent CD79b gene. Repression of transcription in the CD79b subregion of this domain, by insertion of a polymerase II (PolII) termination element between HSI and CD79b, results in a marked reduction in hGH-N expression (Ho et al, 2006). The mechanistic link between the HSI-dependent domain of noncoding transcription and enhancement of hGH-N expression is investigated further in this paper. A comparison of the higher order chromatin configurations of the active and silent hGH loci supports a model in which the promoter activity of hGH-N is enhanced by its positioning into the PolII-enriched environment of the LCR transcriptional domain.
Results And Discussion
Formation of an HSI-dependent chromatin conformation
In contrast to the domain of histone acetylation that directly links the hGH LCR with the hGH-N promoter, the domain of PolII transcription at the active hGH locus, which encompasses the LCR and adjacent CD79b region, is separated from the active hGH-N gene by a nontranscribed gap (Ho et al, 2006). To define further the structure of the active hGH locus, we carried out chromatin immunoprecipitation (ChIP) analysis of histone H3 lysine 4 trimethylation (H3K4-me3) at the hGH/P1 transgene locus. H3K4-me3 modifications are associated with active chromatin structure and often tightly linked to PolII elongation (Gerber & Shilatifard, 2003; Berger, 2007). The hGH/P1 transgene extends from −40 to +45 kb relative to the transcriptional start site of hGH-N (Fig 1A). The H3K4-me3 modifications at the active hGH/P1 transgene locus in the mouse pituitary paralleled PolII distribution; they extended through the LCR and adjacent CD79b region, and were separated from the hGH-N promoter by an intervening gap of unmodified chromatin. The modifications were greatly diminished when HSI was deleted (Fig 1B). The concordant gaps in PolII occupancy and H3K4-me3 modification between the LCR and the hGH-N promoter suggested that activation of the hGH-N promoter might be mediated by nonlinear interactions with its LCR.
The higher order configuration of the active hGH-N locus was explored by using chromosome conformation capture (3C; Cullen et al, 1993; Dekker et al, 2002; Tolhuis et al, 2002). Conditions were established in which the BglII restriction enzyme effectively and reproducibly digests primary chromatin preparations (Fig 2A; supplementary Fig 1 online). A single nucleotide divergence between the 5′-flanking regions of the hGH-N and hGH-V/hCS genes distinguishes BglII fragments that encompass the hGH-N promoter from those encompassing the hGH-V/hCS promoters (supplementary Fig 2 online). The initial set of PCR analyses of 3C ligation products used a ‘GH/CS' anchor primer that recognizes all of the related hGH/hCS genes. BglII ligation products containing the hGH-N promoter region were distinguished from those containing the placenta-expressed paralogue by size (supplementary Fig 2 online), thereby allowing a direct comparison of the proximity of the hGH-N promoter or the promoters of the placenta-expressed paralogue to other defined sites within the locus. The 3C studies represent a minimum of four independent experiments performed on each of two separate hGH/P1 transgenic lines. Previous studies have shown that the pattern of histone acetylation and the level of hGH-N expression are similar in these two lines (Su et al, 2000; Ho et al, 2006). The analysis of liver chromatin (nonexpressing cells) from these mice (Fig 2B, white bars) showed that the efficiency of ligation of the hGH-N promoter fragment to each of the 5′ restriction fragments decreased as the distance between the GH/CS anchor and upstream primers increased. This relationship is consistent with a linear conformation of the cluster in liver chromatin (Dekker et al, 2002). By contrast, the analysis of pituitary chromatin from the same two hGH/P1 transgenic lines (Fig 2B, black bars) showed that the ratio of ligation products shifted in favour of the fragment containing the hGH-N promoter when the anchor GH/CS primer was paired with the primer associated with HSI,II or adjacent CD79b restriction fragments. This pattern of ligation frequencies indicated a close positioning between the HSI,II region and the hGH-N promoter despite the intervening linear 14.5 kb. The PCR products generated by amplification between the GH/CS primer and the HSII primer were purified and sequenced to confirm their identity. These data support a selective, long-range interaction between the HSI,II region and the hGH-N promoter.
Figure 2.
Interaction between HSI,II and the activated hGH-N promoter is HSI dependent and pituitary specific. (A) The BglII map of the hGH/P1 transgene. Each BglII site is indicted below the locus map by a tick mark. The position of each of the PCR primers is also shown (arrowheads); the GH/CS primer, common to all four GH-related genes (large arrowheads), functioned as the anchor primer in these studies. The four upstream primers detect individual BglII fragments that contain HSV, p4, HSII, CD79b or p1 regions. (B) Summary of the 3C analyses in hGH/P1(TerF) pituitary (Pit) chromatin. 3C assays were performed on pituitaries (black bars) or livers (white bars) of mice representing two hGH/P1 transgenic lines and on pituitaries of mice representing two hGH/P1(ΔHSI) lines (grey bars). GH/CS, the anchor primer for these studies, was paired with each of the four 5′ primers in the PCR analysis of the ligation products. The panels below the histogram show representative ethidium bromide-stained agarose gels containing the indicated PCR amplification products; the distinct bands representing the hGH-N- and hGH-V/hCS-specific ligation products are indicated by arrowheads at the left of the top panel. The intensities of the bands were quantified. Lane C (control) contains the PCR amplifications of the ligation products of a BglII-digested hGH/P1 plasmid DNA. Lane marked − contains the products of the assay performed using the hGH/P1 pituitary chromatin preparation in the absence of T4 DNA ligase. The band in the CD79b (−) sample represents a background amplification product that is insensitive to BglII. The histogram values represent ratios of the signal of the hGH-N-specific ligation product to the combined placental gene-specific ligation products (supplementary Figs 1,2 online). Each histogram bar represents the mean±s.d. of four or more independent studies on each of the two transgenic lines. In comparison of the value obtained from hGH/P1 pituitary chromatin to the indicated value, a single asterisk indicates P<0.01 and a double asterisk indicates P<0.001. 3C, chromosome conformation capture; hGH, human growth hormone; LCR, locus control region.
To validate the above results, a reciprocal 3C assay was performed using HSII as the anchor primer (Fig 3A). In these assays, the combination of the HSII anchor and GH/CS primer generated the expected doublet. Ligations to each of the other primers generated a single, specific PCR product (Fig 3B). For each set of amplifications, the PCR signal was normalized to the random ligation control (Fig 3B, lane C, ligation of hGH/P1 plasmid DNA) and to ligation fragments generated from the endogenous, universally expressed ERCC3 locus (Palstra et al, 2003; Fig 3C). The high frequency of ligation between the HSII anchor and CD79b in both liver and pituitary chromatin samples is consistent with their proximity to the locus in the linear structure (Liu & Garrard, 2005). By contrast, there was a significantly higher ligation frequency between the HSII anchor and the hGH-N promoter fragment in pituitary compared with liver chromatin from the same mouse (Fig 3B). The region between CD79b and hGH-N, represented by the p1 fragment, had a lower ligation frequency to HSII (Fig 3B). The low-level association between the HSII anchor and the p1 fragment is consistent with their linear proximity. The similar ligation frequency between p1 and the HSII anchor fragment in liver chromatin from the same mouse (Figs 2, 3) further suggests that these low-level interactions do not contribute significantly to the activating function of the hGH LCR. These 3C results confirmed a specific close contact between the HSI,II region and the hGH-N promoter in pituitary chromatin.
Figure 3.
Reversal of amplification primers in the 3C assay confirms the specific association between hGH-N and the HSI,II region. (A) Map of primer positions for 3C using the HSII region primer as the anchor. The symbols are as in Fig 2A. (B) HSI-dependent association between HSI,II and the hGH-N promoter, and evidence for a pituitary (Pit)-specific interaction between HSII and HSIII-HSV regions. The semiquantitative PCR analyses of 3C ligation products were normalized to parallel analyses of the endogenous ERCC3 locus (C). Each bar represents the mean±s.d. of four independent assays of the indicated chromatin samples: hGH/P1 pituitary chromatin (black bars), hGH/P1(ΔHSI) pituitary chromatin (grey bars) and control liver chromatin (white bars). Lane C contains a PCR analysis of ligation products of BglII-digested hGH/P1 plasmid DNA and represents random ligation of equimolar quantities of each of the BglII fragments in the locus under the conditions of the assay. The ligation efficiency shown on the Y axis was calculated using the equation (signaltissue/signalrandom control)/(signalERCC3 ligation/signalERCC3 loading). (C) 3C analysis of the control ERCC3 locus. The indicated head-to-head ligation frequency of two adjacent BglII fragments released from the ubiquitously expressed ERCC3 locus during the 3C analysis was determined and used as an internal control for each 3C assay. PCR within a unique BglII fragment was used as the loading control for each analysis. (D) A looping model of pituitary-specific hGH-N activation. The 3C studies of the hGH/P1 and hGH/P1(ΔHSI) transgene loci in the pituitary and the hGH/P1 transgene locus in the liver are summarized in a diagrammatic format. 3C, chromosome conformation capture; hGH, human growth hormone.
Next, structural alterations at the active hGH locus were assessed for their dependence on HSI. The 3C assay was carried out using pituitary chromatin from two hGH/P1(ΔHSI) transgenic mouse lines (Fig 2B, grey bars). PCR analyses showed a pattern of ligation products in the pituitary chromatin of the hGH/P1(ΔHS)I mice that was essentially identical to those at the inactive hGH/P1 locus in liver chromatin (Fig 2B, compare grey with white bars). Thus, looping of HSI,II to the active hGH-N promoter in pituitary chromatin is dependent on HSI. The HSI dependency of HSI,II juxtaposition to the hGH-N promoter was confirmed by using the HSII anchor primer (Fig 3B, grey bars). This HSI dependency links the specific chromatin conformation at the hGH locus in the pituitary to enhancement of hGH-N transgene expression (Ho et al, 2006).
In addition to the contact between the HSI,II region and the hGH-N promoter, 3C analysis also showed that the HSI,II region comes into close proximity with HSIII-HSV; the ligation frequency between HSII and HSIII-HSV fragments is significantly greater in the pituitary than in the liver of the hGH/P1 mouse (Fig 3B). Of note, this interaction is maintained in the HSI-deleted transgene. Limited functional tests of HSIII-HSV have suggested that this region might have boundary functions (Jones et al, 1995), and acetylation of histones H3/H4 at HSV is also unique in its HSI independence (Ho et al, 2006). Surprisingly, although the HSI,II fragment crosslinks to both the HSIII-HSV region and the hGH-N promoter, there is no evidence for crosslinking between HSIII-HSV and the hGH-N promoter. This relationship might reflect dynamic switching of the HSI,II interactions between HSV and the hGH-N promoter (Liu & Garrard, 2005). High-resolution 3C analyses of the HSIII-HSV region will be useful in exploring further the configuration of this set of long-range interactions.
The chromatin conformations at the hGH transgene locus in pituitary and liver, and at the pituitary locus lacking an active HSI are schematically compared (Fig 3D). The HSI-dependent interaction between the hGH-N promoter and the LCR/CD79b domain of transcription is specific to the active locus in the pituitary. In the absence of HSI, this interaction is lost, whereas the interaction between HSII and the more 5′ end of the LCR is retained.
Looping is dependent on the LCR domain of transcription
To define further the basis for the interaction between the hGH-N promoter and the HSI,II regions, we determined whether looping was dependent on the presence of the LCR noncoding transcriptional domain or, alternatively, whether HSI might be sufficient for this interaction. We have previously shown that insertion of a PolII termination element (TerF) 3′ to HSI (Fig 4A, hGH/P1(TerF) transgene) selectively interrupts the downstream CD79b transcriptional subdomain and results in a six- to eightfold decrease in hGH-N transgene expression (Ho et al, 2006; Fig 4A, inset). This insertion has no significant impact on HSI formation or on HSI-dependent histone acetylation (Ho et al, 2006). 3C assays were performed on pituitary chromatin from two hGH/P1(TerF) transgenic mouse lines. The analyses, using both GH/CS and HSII primers as anchors in separate studies (Fig 4B,C), showed that TerF insertion effectively blocked looping between HSI,II and the hGH-N promoter. The frequency of the crosslinking between HSI,II and the hGH-N promoter was reduced to the level seen in the absence of HSI (compare Figs 2, 3 and 4). Interactions between HSI,II and HSIII-HSV regions were retained in the presence of TerF (Fig 4C; data summarized in Fig 4D). We conclude that the full chromatin conformation at the active hGH locus is dependent on a functional LCR/CD79b domain of transcription. HSI, in the absence of this function, is not sufficient for chromatin reconfiguration and gene activation.
Figure 4.
Interruption of the locus control region domain of transcription by insertion of a polymerase II termination element (TerF) destabilizes the active chromatin conformation at the hGH locus. (A) A map of the hGH cluster with the position of the TerF PolII termination element indicated. The angled ‘TerF' arrow indicates the position and orientation of the inserted TerF PolII termination element. The diagram in the box shows the impact of the TerF mutation in these lines on the levels of noncoding transcripts 5′ and 3′ to the insertion site (Ho et al, 2006). The TerF insertion selectively represses the transcriptional activity of PolII in the CD79b subdomain of the LCR/CD79b domain of transcription (stippled ovals). (B) Summary of the 3C analysis of the hGH/P1(TerF) pituitary (Pit) chromatin using the GH/CS primer as the anchor. The 3C was carried out on pituitaries of mice representing two hGH/P1(TerF) transgenic lines. Representative semiquantitative PCR assays are shown below the map. Each histogram bar represents the mean±s.d. of three separate analyses from each of two lines (n=6). The previous results of the 3C analysis of the hGH/P1 pituitary chromatin (black bars) are shown to facilitate direct comparison with the hGH/P1(TerF) results (stippled bars); **P<0.001. (C) Summary of the 3C analysis of the hGH/P1(TerF) pituitary chromatin using the HSII primer as an anchor. The 3C was performed as in (A); **P<0.001. (D) The spatial conformation of hGH/P1(TerF) locus in the pituitary in the absence of CD79b noncoding transcription. 3C, chromosome conformation capture; hGH, human growth hormone; LCR, locus control region.
Activation and enhancement of hGH-N transcription by its LCR reflects a complex and multifaceted process. The crucial role of HSI in this process is evident from its essential functions in establishing linear domains of histone modification within the locus and in establishing the domain of noncoding PolII enrichment 5′ to the hGH cluster. The formation of the HSI-dependent LCR/CD79b domain of transcription seems to have a crucial function in the gene activation process; interruption of this domain by insertion of a PolII terminator results in the loss of the chromatin conformation specific to the active locus and in a corresponding decrease in hGH-N expression. Remarkably, disruption of the LCR transcriptional domain does not alter histone acetylation throughout the locus (Ho et al, 2006). This suggests that the HSI-dependent histone acetylation at the hGH locus is independent of LCR transcription and is, by itself, insufficient for hGH-N activation. These relationships support a model in which histone acetylation constitutes an early step in locus activation. This initial step is followed by the establishment of the LCR/CD79b domain of transcription with subsequent reorganization of the chromatin locus and enhancement of hGH-N expression.
These studies showed that an active LCR/CD79b domain of transcription is essential and a probable prerequisite to looping between HSI,II and the hGH-N promoter. These data, along with our previous studies of PIT-1 trans-acting factor binding in HSI and the promoter of hGH-N (Shewchuk et al, 1999; Ho et al, 2002), support a model in which occupancy of PIT-1 at both HSI and the hGH-N promoter triggers interactions between the two regions that are subsequently stabilized by the noncoding transcriptional activity (Fig 3D). The final chromatin conformation at the active locus places the hGH-N promoter in close proximity to the PolII-enriched LCR/CD79b transcriptional domain. This relocation of a promoter into an environment rich in elongating PolII is reminiscent of reports of close physical relationships between certain activated genes and PolII-enriched subnuclear ‘transcriptional factories' (Iborra et al, 1996; Grande et al, 1997; Osborne et al, 2004; Ragoczy et al, 2006). Whether hGH-N gene activation is linked to an association with subnuclear transcription factories, or whether the juxtaposition of the promoter with the local accumulation of PolII within its LCR is sufficient will be of interest in future studies. Irrespective of this relationship, the observation that loop formation and expression at the hGH locus are both lost when LCR transcription is interrupted suggests that formation of this domain and looping are interdependent and mutually sustaining processes in the pathway of long-range transcriptional control.
Methods
Transgenic mouse lines. All transgenic mouse lines used in this study were established as described previously; the hGH/P1 mouse lines were 809F and 811D (copy numbers of transgene are 4 and 5, respectively; Su et al, 2000), hGH/P1(ΔHSI) lines were 960G and 969E (copy numbers 3 and 4; Ho et al, 2002), and hGH/P1(TerF) lines were 1301C and 1301G (copy numbers are 3 and 5; Ho et al, 2002). ChIP and 3C assays were performed using 3- to 4-month-old compound transgenic mouse lines carrying hGH/P1, hGH/P1(ΔHSI) or hGH/P1(TerF) transgene along with a growth hormone-releasing factor (hGRF) transgene under the control of the metallothionine promoter. The hGRF transgene stimulates selective expansion of the somatotrope population with consequent pituitary hypertrophy (Mayo et al, 1988).
Chromatin immunoprecipitation assays. ChIP was performed as described previously (Ho et al, 2006). The sequences of the primers have also been described previously (Ho et al, 2006). Each ratio of bound fraction to input was normalized to the corresponding ratio for a DNA segment within the endogenous skeletal muscle-specific mMyoD gene (Weintraub et al, 1991; Sawado et al, 2003).
Chromatin conformation capture assay. The 3C procedure was performed as described previously (Tolhuis et al, 2002), but with modifications as described in the supplementary information online.
PCR analysis of chromatin conformation capture ligation products. The semiquantitative PCR analysis is described in the supplementary information online. The ligation efficiency was determined as a ratio of products ligated to the hGH-N promoter relative to the hCS/hGH-V promoter fragments when the GH/CS primer functioned as the anchor primer (Figs 2B, 4B), or relative to an internal control at an endogenous ercc3 locus when the HSII primer functioned as the anchor primer (Figs 3B, 4C). Data were plotted as mean+s.d. and t-tests were performed to determine the level of significance in binary comparisons.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank G. Blobel for a critical review of the manuscript. This work was funded by National Institutes of Health grants R01HD25147 and R01HD046737 (N.E.C. and S.A.L.) and K01DK064011 (Y.H.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bennani-Baiti IM, Cooke NE, Liebhaber SA (1998) Physical linkage of the human growth hormone gene cluster and the CD79b (Ig beta/B29) gene. Genomics 48: 258–264 [DOI] [PubMed] [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Chen EY, Liao YC, Smith DH, Barrera-Saldana HA, Gelinas RE, Seeburg PH (1989) The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics 4: 479–497 [DOI] [PubMed] [Google Scholar]
- Cullen KE, Kladde MP, Seyfred MA (1993) Interaction between transcription regulatory regions of prolactin chromatin. Science 261: 203–206 [DOI] [PubMed] [Google Scholar]
- Dean A (2006) On a chromosome far, far away: LCRs and gene expression. Trends Genet 22: 38–45 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Elefant F, Cooke NE, Liebhaber SA (2000) Targeted recruitment of histone acetyltransferase activity to a locus control region. J Biol Chem 275: 13827–13834 [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Bresnick EH (2001) Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23: 820–830 [DOI] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A (2003) Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem 278: 26303–26306 [DOI] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, de Jong L, van Driel R (1997) Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci 110: 1781–1791 [DOI] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Cooke N, Liebhaber S (2002) A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol Cell 9: 291–302 [DOI] [PubMed] [Google Scholar]
- Ho Y, Liebhaber SA, Cooke NE (2004) Activation of the human GH gene cluster: roles for targeted chromatin modification. Trends Endocrinol Metab 15: 40–45 [DOI] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Liebhaber SA, Cooke NE (2006) Locus control region transcription plays an active role in long-range gene activation. Mol Cell 23: 365–375 [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR (1996) Active RNA polymerases are localized within discrete transcription ‘factories' in human nuclei. J Cell Sci 109: 1427–1436 [DOI] [PubMed] [Google Scholar]
- Jones BK, Monks BR, Liebhaber SA, Cooke NE (1995) The human growth hormone gene is regulated by a multicomponent locus control region. Mol Cell Biol 15: 7010–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Garrard WT (2005) Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol 25: 3220–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo KE, Hammer RE, Swanson LW, Brinster RL, Rosenfeld MG, Evans RM (1988) Dramatic pituitary hyperplasia in transgenic mice expressing a human growth hormone-releasing factor gene. Mol Endocrinol 2: 606–612 [DOI] [PubMed] [Google Scholar]
- Osborne CS et al. (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M (2006) The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20: 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M (2003) The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewchuk BM, Asa SL, Cooke NE, Liebhaber SA (1999) Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J Biol Chem 274: 35725–35733 [DOI] [PubMed] [Google Scholar]
- Su Y, Liebhaber SA, Cooke NE (2000) The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J Biol Chem 275: 7902–7909 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Weintraub H et al. (1991) The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251: 761–766 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information