Abstract
Microcephalin (MCPH1) has a crucial role in the DNA damage response by promoting the expression of Checkpoint kinase 1 (CHK1) and Breast cancer susceptibility gene 1 (BRCA1); however, the mechanism of this regulation remains unclear. Here, we show that MCPH1 regulates CHK1 and BRCA1 through the interaction with E2F1 on the promoters of both genes. MCPH1 also regulates other E2F target genes involved in DNA repair and apoptosis such as RAD51, DDB2, TOPBP1, p73 and caspases. MCPH1 interacts with E2F1 on the p73 promoter, and regulates p73 induction and E2F1-induced apoptosis as a result of DNA damage. MCPH1 forms oligomers through the second and third BRCT domains. An MCPH1 mutant containing only its oligomerization domain has a dominant-negative role by blocking MCPH1 binding to E2F1. It also inhibits p73 induction in DNA damage and E2F1-dependent apoptosis. Taken together, MCPH1 cooperates with E2F1 to regulate genes involved in DNA repair, checkpoint and apoptosis, and might participate in the maintenance of genomic integrity.
Keywords: MCPH1/BRIT1, E2F1, p73, Chk1, oligomerization
Introduction
Primary microcephaly is an autosomal recessive disorder characterized by reduced brain size. The first causative gene identified encodes microcephalin (MCPH1; Jackson et al, 2002). Microcephalin, also known as BRIT1 (BRCT-repeat inhibitor of human telomerase reverse transcriptase (TERT) expression), was independently identified in a genetic screen for transcriptional repressors of human TERT (Lin & Elledge, 2003). Mutation of MCPH1 results in premature onset of condensin II-mediated chromosome condensation (Trimborn et al, 2006). MCPH1 contains three Breast cancer susceptibility gene 1 (BRCA1)-carboxy-terminal (BRCT) domains. It is induced by DNA damage, and regulates both Checkpoint kinase 1 (CHK1) and BRCA1 (Xu et al, 2004; Lin et al, 2005). It has a role in the maintenance of chromosomal integrity and infrared- or ultraviolet-induced foci formation (Rai et al, 2006). MCPH1 also acts downstream from CHK1 in regulating Cell division cycle 25A (Cdc25A) stability and consequently preventing premature entry into mitosis (Alderton et al, 2006). A function of MCPH1 as a tumour suppressor has been suggested by an association of genomic instability and metastasis with decreased levels of MCPH1 in human cancer (Rai et al, 2006). So far, the mechanism by which MCPH1 regulates CHK1 and BRCA1 remains unclear.
The E2F transcription factor family has a crucial role in the regulation of cell-cycle progression. In addition to genes directly involved in cellular proliferation, E2F also regulates genes responsible for checkpoint activation, DNA repair, apoptosis, chromatin assembly, condensation and chromosome segregation (Dimova & Dyson, 2005), supporting the role of E2F in the maintenance of genomic integrity. The function of E2F in chromosome stability is supported further by the observation that the combined loss of both E2F1 and E2F2 leads to polyploidy in the liver, salivary gland and exocrine pancreas (Li et al, 2003), and loss of E2F3 results in aneuploidy (Saavedra et al, 2003). Although the role of E2F1 in the control of cell-cycle-related genes has been well established, how E2F participates in genomic surveillance remains to be investigated.
Results And Discussion
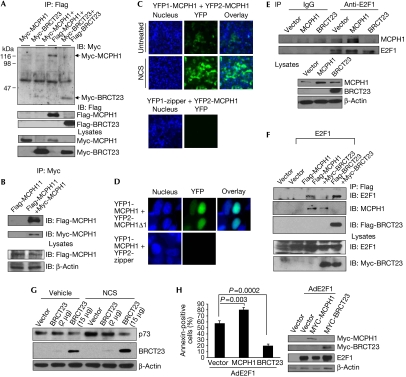
Our previous studies have shown the crucial role of a BRCT domain-containing protein DNA topoisomerase IIβ binding protein 1 (TOPBP1) in controlling the transcriptional activities of E2F1 (Liu et al, 2003, 2004, 2006). We compared the sequences of TOPBP1 and MCPH1 and found that they share 24% sequence identity and 32% sequence similarity. The sequence homology spans throughout the entire 835 amino-acid (aa)-coding sequence of MCPH1 and the fifth to eighth BRCT domains of TOPBP1. This homology region includes the E2F1-binding domain (sixth to eighth BRCT domains) of TOPBP1; therefore, we speculated that MCPH1 could interact with and regulate E2F1. Indeed, an interaction between endogenous MCPH1 and E2F1 was detected by reciprocal co-immunoprecipitation in human embryonic kidney 293 (HEK293) cells either during normal growing conditions or on treatment of a radiomimetic agent neocarzinostatin (NCS) or adriamycin (Fig 1A,B). The complex formation increased after NCS (1.7-fold) or adriamycin (1.5-fold) treatment, corresponding with a modest induction of E2F1 and MCPH1. An interaction between endogenous MCPH1 and E2F1 was also observed in the T98G glioblastoma cell line (data not shown). An in vitro glutathione S-transferase pulldown assay showed a further direct binding between MCPH1 and E2F1 (supplementary Fig S1 online). Next, we mapped the E2F1-binding domain to the second and third BRCT domains of MCPH1 (Fig 1C,D). MCPH1 could also associate with E2F2; however, their interaction seemed to be much weaker (Fig 1E). No interaction between MCPH1 and E2F3 was detected (Fig 1F). To detect the interaction between MCPH1 and E2F1 in cultured cells with minimal perturbation of the normal cellular environment, we carried out a bimolecular fluorescence complementation assay. When both YFP1-E2F1 (YFP for yellow fluorescent protein) and YFP2-MCPH1 proteins were expressed in either 293T or NIH3T3 cells, the interaction of MCPH1 and E2F1 led to reconstitution of functional YFP (Fig 1G). The variation of YFP intensity is probably due to various levels of protein expression in transient transfection. This interaction depends on the amino terminus of E2F1 (aa 1–150; Fig 1H,I). By contrast, a control zipper domain fusion protein YFP1-zipper could not interact with YFP2-MCPH1.
Figure 1.
MCPH1 interacts with E2F1. (A,B) HEK293 cells were left untreated (No tx) or treated with 300 ng/ml NCS for 4 h or 2 μM adriamycin (Adr) for 24 h. The cells were collected for immunoprecipitation (IP) using a rabbit MCPH1 (A) or mouse E2F1 antibody (B) or a control IgG followed by immunoblotting (IB) as indicated. Input represents 10% of lysates added to each IP reaction. (C) Schematic structure of MCPH1 protein. (D–F) 293T cells were co-transfected with Myc-tagged wild-type or mutant MCPH1 along with E2F1, E2F2 or E2F3. The next day, cells were collected for IP with anti-Myc beads followed by immunoblotting. (G,H) 293T or NIH3T3 cells were co-transfected with YFP1-E2F1, YFP1-E2F1(1–150) or YFP1-E2F1(151–437) and YFP2-MCPH1, or a control YFP1-zipper (containing a zipper domain derived from GCN4) and YFP2-MCPH1. The next day, cells were fixed and nuclei were stained with Hoechst 33258. (I) Cellular lysates of 293T cells in (H) were analysed for expression of YFP1-E2F1 or its deletional mutants and YFP2-MCPH1 using a GFP antibody for immunoblotting. BRCT, BRCA1 carboxy-terminal; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; HEK, human embryonic kidney cells; MCPH1, microcephalin; NCS, neocarzinostatin; YFP, yellow fluorescent protein.
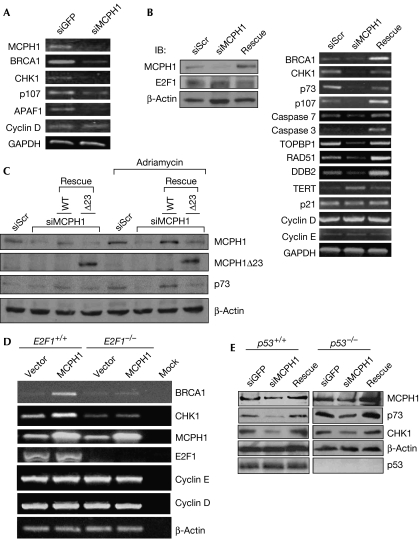
To test whether MCPH1 is involved in E2F1 function, we knocked down the expression of MCPH1 in HEK293 cells. Consistent with previous reports, depletion of MCPH1 inhibited the expression of BRCA1 and CHK1 (Fig 2A). The expression of two E2F1 target genes p107 and Apoptotic peptidase activating factor 1 (APAF1), but not cyclin D, was also decreased by MCPH1 short interfering RNA (siRNA), suggesting that MCPH1 controls some of the E2F1 functions. We also established a U2OS cell line stably expressing MCPH1 siRNA. Knockdown of MCPH1 inhibited the expression of CHK1 and BRCA1 in these cells, and also inhibited the expression of several E2F target genes, including p73, p107, and caspases 3 and 7 (Fig 2B). As MCPH1 is required for the maintenance of genomic integrity, we speculated that it might regulate other E2F target genes involved in DNA repair such as TOPBP1, RAD51 and DDB2. The expression of these genes was also dependent on MCPH1. These effects were derived from the depletion of MCPH1, as they could be rescued by reconstitution of MCPH1 expression with an MCPH1 complementary DNA (cDNA) carrying several silent mutations within the MCPH1 siRNA target sequence. Interestingly, MCPH1 siRNA did not alter the expression of cyclin E, cyclin D or p21. Depletion of MCPH1 induced the expression of TERT (Fig 2B), supporting an inhibitory function on TERT expression (Lin & Elledge, 2003). Next, we examined the role of MCPH1 in p73 and CHK1 expression during DNA damage and found that adriamycin-mediated induction of p73 was inhibited by MCPH1 depletion in HEK293 cells (Fig 2C). The effect was rescued only by reconstitution of MCPH1 expression with wild-type MCPH1, but not by a truncated mutant MCPH1Δ23, which failed to bind to E2F1. Taken together, these results strongly support the crucial role of MCPH1 in the regulation of E2F target genes involved in DNA repair, checkpoint activation and apoptosis under physiological conditions as well as during DNA damage.
Figure 2.
The physiological role of MCPH1 in the control of E2F1 target genes and induction of p73 by adriamycin. (A) HEK293 cells were transfected with a control siGFP or siMCPH1. The next day, RNA was extracted and RT–PCR was performed. (B) The scrambled siRNA (siScr) or siMCPH1 in stable U2OS-TREX cells was induced by doxycycline for 2 days and some cells were then transfected with siRNA-resistant MCPH1 cDNA (Rescue) or a control vector to rescue MCPH1 expression. The transfected cells continued to be cultured in a medium containing doxycycline. RNA was extracted 24 h later and RT–PCR was performed. Left panels: Western blot analysis from cell lysates. (C) HEK293 cells were transfected with pSUPER-siScr or pSUPER-siMCPH1, with or without an siRNA-resistant MCPH1 expression vector expressing either wild-type (WT) MCPH1 or a truncated mutant MCPH1Δ23 (Δ23) as indicated. After 24 h, cells were treated with 5 μM adriamycin for 24 h and then collected for immunoblotting. (D) E2F1+/+ and E2F1−/− MEFs were transfected with an MCPH1 expression vector or an empty vector. After 48 h, cells were collected and total RNA was extracted. Gene expression was analysed by RT–PCR. (E) p53+/+ HCT116 and p53−/− HCT116 cells were infected with Ad-siGFP, Ad-siMCPH1 or Ad-siMCPH1 along with Ad-MCPH1 expressing siRNA-resistant MCPH1 (rescue) at an m.o.i. of 50 each. After 48 h, cells were collected for immunoblotting. APAF1, Apoptotic peptidase activating factor 1; BRCA1, Breast cancer susceptibility gene 1; cDNA, complementary DNA; CHK1, Checkpoint kinase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HCT, human colon carcinoma; HEK, human embryonic kidney cells; IB, immunoblotting; MCPH1, microcephalin; m.o.i., multiplicity of infection; RT–PCR, reverse transcription–PCR; siGFP, short interfering green fluorescent protein; TERT, telomerase reverse transcriptase; TOPBP1, DNA topoisomerase IIβ binding protein 1.
To investigate whether the regulation of CHK1 and BRCA1 by MCPH1 depends on E2F1, we examined the effect of MCPH1 overexpression on CHK1 and BRCA1 in E2F1−/− mouse embryonic fibroblasts (MEFs). As shown in Fig 2D, MCPH1 induced the expression of CHK1 and BRCA1 in E2F1+/+ MEFs. The induction was largely abrogated in E2F1−/− MEFs, indicating the requirement of E2F1 in MCPH1-mediated induction of CHK1 and BRCA1. There was a marginal induction of both CHK1 and BRCA1 by MCPH1 overexpression in E2F1−/− MEFs. This might be mediated by other E2Fs such as E2F2, as MCPH1 might interact weakly with E2F2 (Fig 1E). Consistent with the results in Fig 2B, MCPH1 failed to induce the expression of cyclin D and E, suggesting that MCPH1 specifically regulates a subset of E2F1 target genes such as CHK1 and BRCA1. Conversely, p53 was not required for the promoting effect of MCPH1 on CHK1 or p73 expression (Fig 2E). Therefore, we conclude that MCPH1 regulates CHK1 and BRCA1 through E2F1 action.
As E2F1 can directly activate CHK1 and BRCA1 gene promoters (supplementary information online), we speculated that the interaction between MCPH1 and E2F1 might upregulate the transcriptional activity of E2F1 on CHK1 and BRCA1 promoters. Indeed, MCPH1 enhanced the transcriptional activities of E2F1 and E2F2, but not E2F3 or E2F4, in an E2F activity reporter assay (Fig 3A,B; supplementary Fig S2 online). This is consistent with its binding to E2F1 and E2F2, but not to E2F3. To test the occupancy of MCPH1 on CHK1, BRCA1 and p73 promoters, we performed chromatin immunoprecipitation (ChIP) in U2OS, HEK293, MDA-MB453, p53+/+ HCT116 and p53−/− HCT116 cell lines. The results showed that MCPH1 and E2F1 co-occupied on these promoters in U2OS (Fig 3C,D) and HEK293 cells (Fig 3E), and that this co-occupancy could be induced by adriamycin or NCS treatment. A ChIP–re-ChIP assay further confirmed the presence of MCPH1–E2F1 complex formation on CHK1, p73 and caspase 7 promoters in HEK293, U2OS and MDA-MB453 cells (Fig 3F). In line with the role of E2F1 rather than p53 in the regulation, we found co-occupancy of MCPH1 and E2F1, but not p53, on CHK1 promoter in both p53+/+ and p53−/− HCT116 cells, indicating that the co-occupancy of MCPH1 and E2F1 does not require p53 (Fig 3G). Both E2F1 and MCPH1 are also found on the caspase 7 promoter. Interestingly, although p53 does not regulate CHK1, we observed occupancy of p53 on caspase 7 promoter (Fig 3G), which is consistent with the role of p53 in caspase 7 regulation (Joshi et al, 2007). The data provide the first evidence for a promoter-binding activity of MCPH1 in the regulation of BRCA1, CHK1, p73 and caspase 7. Taken together, these results establish a role for MCPH1 in the regulation of E2F1 functions.
Figure 3.
MCPH1 enhances E2F1-mediated transactivation and occupies on CHK1, BRCA1, caspase-7 and p73 promoters. (A,B) E2F activity was measured with a p14ARF promoter-luciferase activity assay in the absence or presence of MCPH1 in HEK293 cells. Each sample was tested in triplicate and the results were reproduced in another cell line T98G (supplementary Fig S2 online). *P=0.006 (t-test, two-tailed) compared with the E2F1-alone group; **P<0.0001 (t-test, two-tailed) compared with the E2F-alone group. (C–G) ChIP assay performed in U2OS (C,D), HEK293 (E) or p53+/+ and p53−/− HCT116 cells (G) using antibodies against E2F1, MCPH1 or p53, respectively. Some cells were treated with 300 ng/ml NCS for 4 h or 2 μM adriamycin for 24 h before collection. The IgG lane represents a control reaction using normal rabbit or mouse IgG for ChIP. Input represents 0.5% of the total amount of chromatin added to each ChIP reaction. For re-ChIP (F), the NCS-treated HEK293, U2OS or MDA-MB453 were subjected to ChIP assay using a mouse E2F1 antibody, then re-ChIP with a rabbit MCPH1 antibody or control normal rabbit IgG. Input DNA and protein-bound DNA fragments were amplified with primer pairs derived from each promoter as indicated. BRCA1, Breast cancer susceptibility gene 1; ChIP, chromatin immunoprecipitation; CHK1, Checkpoint kinase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HCT, human colon carcinoma cells; HEK, human embryonic kidney cells; MCPH1, microcephalin; NCS, neocarzinostatin.
Depletion of MCPH1 did not affect the expression of cyclins E and D, which are responsible for G1- to S-phase transition. Instead, MCPH1 is required for the expression of p73, APAF1, and caspases 3 and 7, which have been shown to be important mediators for E2F1-induced apoptosis. These results indicate that MCPH1 might not be involved in the proliferative activity of E2F1, but that it regulates its pro-apoptotic function. Consistent with this is the result that depletion of MCPH1 does not alter cell-cycle profiles under normal growing conditions (supplementary Fig S3 online; Xu et al, 2004; Lin et al, 2005). Instead, MCPH1 depletion significantly inhibited E2F1-induced apoptosis in U2OS cells, and reconstitution of the expression of MCPH1 restored the apoptosis induced by E2F1 (Fig 4A; supplementary Fig S4 online). The apoptosis was specific for E2F1 and could not be induced by AdE2F2, AdE2F3 and AdE2F4 (Fig 4B). To test further the role of MCPH1 in E2F1-mediated apoptosis during DNA damage, we assayed its effect on the apoptosis induced by adriamycin in HEK293 cells, in which adriamycin-induced apoptosis has been shown to be specifically dependent on E2F1 (Wang et al, 2004). Overexpression of MCPH1 enhanced adriamycin-induced apoptosis (Fig 4C), whereas depletion of MCPH1 by two different MCPH1 RNAis inhibited both adriamycin-induced apoptosis and p73 expression (Fig 4D). Taken together, we conclude that MCPH1 cooperates with E2F1 in the activation of checkpoint/DNA repair proteins, as well as pro-apoptotic proteins, and that it is required for the pro-apoptotic function of E2F1.
Figure 4.
Role of MCPH1 in E2F1- and adriamycin-induced apoptosis. (A,B) MCPH1 was knocked down and then rescued in doxycycline-inducible U2OS-TREX cells as in Fig 2B. Cells were then infected with AdCMV, AdE2F1, AdE2F2, AdE2F3 or AdE2F4 at an m.o.i. of 200. The cells were then cultured in a serum-free medium with 2 μg/ml doxycycline for another 48 h before collecting for annexin V staining. The data shown are the mean±s.e. of three independent experiments. **P=0.005 (t-test, two-tailed) compared with the siScr group; *P=0.02 (t-test, two-tailed) compared with the siMCPH1 group. Upper panels: Western blot analysis of the cells as described. (C) HEK293 cells were transfected with MCPH1 or a control vector. The next day, cells were treated with 5 μM adriamycin for 48 h before collection for propidium iodide staining. Apoptotic cells (sub-G1 fractions) were analysed by flow cytometry. The data shown are the mean of triplicate experiments and the P-values (two-tailed t-test). Upper panels: Western blot analysis from the cell lysates as described. (D) HEK293 cells were transfected with siScr or two different MCPH1 siRNAs (siM#1 and siM#2). The next day, cells were treated with adriamycin (5 μM) for 24 h and then measured for caspase-3/7 activity. The cell lysates were collected for immunoblottings (upper panels). HEK, human embryonic kidney cells; MCPH1, microcephalin; m.o.i., multiplicity of infection; PARP, poly(ADP-ribose) polymerase; siScr, scrambled siRNA (Liu et al, 2006); siM, MCPH1 siRNA.
Several BRCT domain proteins such as TOPBP1 (Liu et al, 2006) and Crumbs homologue 2 (CRB2; Du et al, 2004) can form oligomers and this structural change regulates their functions. The fact that the sequence homologous region between MCPH1 and TOPBP1 contains an oligomerization domain (the seventh and eighth BRCT domains of TopBP1) prompted us to investigate whether MCPH1 also forms oligomers. There is an intermolecular interaction between Flag- and Myc-tagged MCPH1 as shown by reciprocal immunoprecipitation (Fig 5A,B). Flag-MCPH1 interacted only with the second and third BRCT domains (referred to as BRCT23 hereafter), but not with the first BRCT domain or with the portion between the first and second BRCT domains (supplementary Fig S5 online). An MCPH1 deletional mutant (Δ1) lacking the first BRCT domain still bound to MCPH1, although less efficiently; however, an MCPH1 deletional mutant (Δ23) lacking the BRCT23 domain completely lost the interaction (supplementary Fig S6 online). Finally, BRCT23 is sufficient to self-associate (Fig 5A). Therefore, we conclude that BRCT23 is responsible for the oligomerization of MCPH1; however, an optimal interaction might need the first BRCT domain. A bimolecular fluorescence complementation assay further confirmed the self-association of MCPH1 in living cells during normal growth and on NCS treatment (Fig 5C). MCPH1 also interacted with MCPH1Δ1 in this assay (Fig 5D). Taken together, these data support a BRCT23-dependent oligomerization of MCPH1.
Figure 5.
MCPH1 forms oligomers through its second and third BRCT domains. An MCPH1 dominant-negative mutant containing only the oligomerization domain inhibits p73 induction by DNA damage and E2F1-dependent apoptosis. (A,B) 293T cells were co-transfected with Myc-tagged or/and Flag-tagged wild-type MCPH1 or their truncated mutants. The next day, cells were collected for immunoprecipitation (IP) using anti-Flag (A) or anti-Myc beads (B). The immunoprecipitates were subjected to immunoblotting (IB) using rabbit Myc and mouse Flag antibodies. (C) 293T cells were transfected with YFP1-MCPH1 and YFP2-MCPH1, or with YFP1-zipper and YFP2-MCPH1 for 24 h, and then either left untreated or treated with NCS at 300 ng/ml for 4 h. Nuclei were stained with Hoechst 33258. (D) 293T cells were co-transfected with YFP1-MCPH1 and YFP2-MCPH1Δ1 or YFP2-zipper. The next day, nuclei were stained and fluorescent images were captured. (E) HEK293 cells were transfected with MCPH1 or Myc-BRCT23. The next day, the cells were collected for immunoprecipitation using a mouse E2F1 antibody or a control mouse IgG followed by immunoblotting. (F) HEK293 cells were transfected with Flag-MCPH1, Myc-BRCT23 and HA-E2F1 expression vectors as indicated. After 48 h, cells were collected for immunoprecipitation using anti-Flag beads followed by immunoblotting. (G) U2OS cells were transfected with various concentrations of Myc-BRCT23. The next day, cells were exposed to 300 ng/ml NCS for 4 h and then collected for immunoblotting. (H) U2OS cells were transfected with MCPH1 or Myc-BRCT23. The next day, cells were infected with AdCMV or AdE2F1 at an m.o.i. of 400 and then cultured under serum-free medium for 48 h before annexin V staining. The data shown are the mean of three independent experiments and the P-values (two-tailed t-test). Right panels: Western blot analysis for the cells as described. BRCT, BRCA1 carboxy-terminal; HEK, human embryonic kidney cells; MCPH1, microcephalin; m.o.i., multiplicity of infection; NCS, neocarzinostatin; YFP, yellow fluorescent protein.
Next, we tested whether BRCT23 could function as a dominant-negative inhibitor to block MCPH1 function in E2F1 binding and activation. Indeed, overexpression of BRCT23 inhibited the interaction of endogenous MCPH1 and E2F1 (Fig 5E) and on co-overexpression with BRCT23 (Fig 5F). We investigated further the dominant-negative effect of BRCT23 on the induction of p73 after DNA damage. BRCT23 significantly blocked the induction of p73 by NCS in a dose-dependent manner (Fig 5G). BRCT23 also inhibited adriamycin-mediated induction of p73 in HEK293 cells (supplementary Fig S7 online). Finally, we examined the effect of MCPHI and BRCT23 on the pro-apoptotic activity of E2F1. MCPH1 enhanced E2F1-dependent apoptosis, whereas BRCT23 inhibited E2F1-induced apoptosis (Fig 5H). These results show a dominant-negative effect of BRCT23 in inhibiting E2F1-mediated apoptosis and the crucial role of MCPH1 in p73 induction during DNA damage. These data also indicate that in addition to BRCT23 (Figs 1D, 5F), other domains of MCPH1 are required for the cooperation with E2F1 in transcriptional activation. The dominant-negative activity of BRCT23 could be due to competition with endogenous MCPH1 for E2F1 binding and/or forming a non-functional complex with endogenous MCPH1 through oligomerization.
Compared with normal tissues, several types of human cancer such as prostate, ovarian and breast cancer express much lower concentrations of MCPH1, and the concentrations of MCPH1 are correlated with genomic instability and metastasis (Rai et al, 2006). It would be interesting to determine whether cancer cells containing lower concentrations of MCPH1 are defective in the expression of p73, APAF1, caspases, BRCA1, CHK1, RAD51 or DDB2. Any deficiency in the expression of these genes might contribute to genomic instability in cancer. Given the crucial role of E2F1–p73 axis in the chemotherapeutic response, decreased concentrations of MCPH1 in cancer might have dampened the apoptotic response to chemotherapy owing to lesser induction of p73.
The fact that the C-terminal second and third BRCT motifs of MCPH1 are required for both oligomerization and E2F1 binding suggests that E2F1 might bind to MCPH1 oligomers. Although the oligomerization domain is required for E2F1 binding, the N terminus of MCPH1 is also required for cooperation with E2F1 in transcriptional activation. It is unclear at this moment how the interaction between MCPH1 and E2F1 facilitates transcriptional activation. However, it is important to note that activation of E2F1 target genes by MCPH1 is guided towards genes involved in genomic surveillance. MCPH1 binds to chromatin (Lin et al, 2005) and recruits other complexes such as Mediator of DNA damage checkpoint 1 (MDC1), p53 Binding protein 1 (p53BP1) and Nijmegen breakage syndrome 1 (NBS1) to chromatin (Rai et al, 2006); therefore, MCPH1 could alter chromatin structure around promoters of its target genes. It is also possible that the N terminus of MCPH1 (for example, the first BRCT domain) recruits other factors that modify E2F1 or chromatin and activate transcription. Further investigation is needed to determine the mechanism by which MCPH1 regulates E2F1 activity, and the physiological role of E2F2–MCPH1 binding remains to be determined. A minor induction of BRCA1 and CHK1 by MCPH1 overexpression in E2F1−/− MEFs (Fig 2D) suggests an E2F1-independent activity of MCPH1. We propose that E2F2 might, to a lesser degree, compensate for E2F1 loss during normal growth. However, only E2F1 is induced after DNA damage (Lin et al, 2001); therefore, E2F1 is largely responsible for induction of its target genes by DNA-damaging agents.
In summary, we have identified a new function for MCPH1 in transcriptional regulation. By forming a complex with E2F1, MCPH1 regulates a subset of crucial E2F target genes involved in DNA repair, checkpoint activation and apoptosis. We have also shown the crucial role of MCPH1 in the expression and induction of p73 during DNA damage and E2F1-dependent apoptosis. Through regulation of these genes, the MCPH1–E2F1 complex might be important in the maintenance of genomic integrity and correct response to DNA damage. The identification of another BRCT domain-containing protein for E2F1 regulation also has further implications. The pRb family members have long been considered to be the chief regulators of E2F. TOPBP1 and MCPH1 seem to be a new class of regulators for E2F1; they both contain several BRCT motifs and interact with the N terminus of E2F1 through their BRCT motifs. TOPBP1 is a negative regulator, whereas MCPH1 is a positive regulator of E2F1 target genes in checkpoint/repair and apoptosis. Both proteins are also directly involved in DNA damage checkpoint activation; therefore, they might coordinate the DNA damage response signalling with transcriptional regulation.
Methods
Cell culture. HEK293, 293T, T98G and U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS); p53+/+ HCT116 and p53−/− HCT116 cells in McCoy's 5A medium with 10% FBS; and MDA-MB453 cells in DMEM with 2 mM glutamine, non-essential amino acids and 10% FBS. E2F1+/+ or E2F1−/− primary MEFs (Liu et al, 2003) were cultured in DMEM with 10% FBS. Transfection was performed by the standard calcium phosphate method or by using the Gene Pulser Xcell electroporation system (Bio-Rad, Hercules, CA, USA) or FuGENE 6 transfection reagent (Roche Applied Science, Basel, Switzerland) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
Chromatin immunoprecipitation assay. ChIPs were performed as previously described (Liu et al, 2004). For re-ChIP assay, endogenous E2F1 complexes were first immunoprecipitated using a mouse E2F1 antibody. The complexes were eluted by incubation for 0.5 h at 37°C in 10 mM dithiothreitol, diluted 20 times in RIPA buffer and subjected to a second chromatin immunoprecipitation using a rabbit MCPH1 antibody. The detailed procedures and primer sequences for PCR are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Fig S1–S7
Acknowledgments
We acknowledge S.W. Michnick for YFP1 and YFP2 vectors, and B. Vogelstein for p53−/− HCT116 cells. This study was supported by grants from National Institutes of Health (CA 100857) and the Department of Defense Breast Cancer Research Program (W81XWH-04-1-0442). W.-C.L. is a Leukemia & Lymphoma Society Scholar.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, Jeggo PA, O'Driscoll M (2006) Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol 8: 725–733 [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ (2005) The E2F transcriptional network: old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Du LL, Moser BA, Russell P (2004) Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J Biol Chem 279: 38409–38414 [DOI] [PubMed] [Google Scholar]
- Jackson AP et al. (2002) Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet 71: 136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Rastogi S, Morris M, Carastro LM, DeCook C, Seto E, Chellappan SP (2007) Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J 401: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FX, Zhu JW, Tessem JS, Beilke J, Varella-Garcia M, Jensen J, Hogan CJ, DeGregori J (2003) The development of diabetes in E2f1/E2f2 mutant mice reveals important roles for bone marrow-derived cells in preventing islet cell loss. Proc Natl Acad Sci USA 100: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Elledge SJ (2003) Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113: 881–889 [DOI] [PubMed] [Google Scholar]
- Lin SY, Rai R, Li K, Xu ZX, Elledge SJ (2005) BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1–Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci USA 102: 15105–15109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Lin FT, Nevins JR (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev 15: 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lin FT, Ruppert JM, Lin WC (2003) Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol 23: 3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Luo Y, Lin FT, Lin WC (2004) TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev 18: 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Paik JC, Wang B, Lin FT, Lin WC (2006) Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J 25: 4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R et al. (2006) BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell 10: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra HI, Maiti B, Timmers C, Altura R, Tokuyama Y, Fukasawa K, Leone G (2003) Inactivation of E2F3 results in centrosome amplification. Cancer Cell 3: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Schindler D, Neitzel H, Hirano T (2006) Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle 5: 322–326 [DOI] [PubMed] [Google Scholar]
- Wang B, Liu K, Lin FT, Lin WC (2004) A role for 14-3-3 τ in E2F1 stabilization and DNA damage-induced apoptosis. J Biol Chem 279: 54140–54152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lee J, Stern DF (2004) Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem 279: 34091–34094 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Fig S1–S7