Abstract
We have recently identified a new form of post-translational regulation of BACE1 (β-site amyloid precursor protein (APP)-cleaving enzyme 1), a membrane protein that acts as the rate-limiting enzyme in the generation of the Alzheimer disease amyloid β-peptide (Aβ). Specifically, BACE1 is transiently acetylated on seven lysine residues in the lumen of the endoplasmic reticulum/endoplasmic reticulum–Golgi intermediate compartment (ER/ERGIC). The acetylated intermediates of the nascent protein are able to reach the Golgi apparatus, whereas the non-acetylated ones are retained and degraded in a post-ER compartment. Here, we report that the serine protease PCSK9 (proprotein convertase subtilisin kexin type 9) contributes to the disposal of non-acetylated BACE1. Both overexpression and small interfering RNA-mediated downregulation of PCSK9 affected the levels of BACE1. The downregulation of PCSK9 affected the levels of the loss-of-acetylation mutants (BACE1Ala and BACE1Arg) but not those of the gain-of-acetylation mutant (BACE1Gln). In addition, Pcsk9−/− mice showed increased levels of BACE1 and Aβ in the brain. Finally, we found that nascent low-density lipoprotein receptor, a known substrate of PCSK9, is also acetylated.
Keywords: BACE1, PCSK9, lysine acetylation, Alzheimer disease
Introduction
The efficiency of folding of nascent membrane and secretory proteins is enhanced by transient or permanent post-translational modifications that occur in the early secretory pathway. These modifications work in tight association with chaperones and proteolytic enzymes, which degrade misfolded protein intermediates (Trombetta & Parodi, 2003; Meusser et al, 2005). Ultimately, the disposal of misfolded protein intermediates represents a fundamental function of the early secretory pathway.
We have recently identified a new form of post-translational regulation of the membrane protein BACE1 (β-site amyloid precursor protein (APP)-cleaving enzyme 1) that involves transient acetylation of the nascent protein in the early secretory pathway (Costantini et al, 2007). The system works in such a way that nascent BACE1 is acetylated on seven lysine residues of the amino-terminal domain while in the endoplasmic reticulum/endoplasmic reticulum–Golgi intermediate compartment (ER/ERGIC) and then deacetylated in the lumen of the Golgi apparatus when the protein has achieved conformational maturation. Non-acetylated intermediates of the nascent protein are not allowed to reach the Golgi apparatus and are degraded in a post-ER compartment through a mechanism that does not seem to involve the proteasome (Costantini et al, 2007).
PCSK9 (proprotein convertase subtilisin kexin type 9), also known as neural-apoptosis-regulated convertase 1, is a prohormone–proprotein convertase of the subtilisin (S8) family of serine proteases (Attie, 2004; Maxwell & Breslow, 2005). Specifically, PCSK9 is part of the S8A subfamily, which includes only PCSK9 and S1P (site-1 protease), a protease that is involved in the post-translational processing and activation of many ER-based transcription factors (SREBP, ATF6, CREBH, OASIS, Tisp40 and LZIP/CREB3) in the ERGIC/cis-Golgi system (Yoshida, 2007).
PCSK9 is synthesized as an approximately 75 kDa proprotein (proPCSK9, P-form); the N-terminal prodomain is then removed by autocatalysis (Seidah et al, 2003; Benjannet et al, 2004) generating an approximately 65 kDa form (C-form) that is secreted into the extracellular milieu (Grozdanov et al, 2006). Studies performed with the low-density lipoprotein receptor (LDLr) indicate that PCSK9 can dispose of LDLr both in the ‘post-ER compartment' of the secretory pathway (Maxwell & Breslow, 2004; Maxwell et al, 2005) and in the ‘endosomal–lysosomal compartment' (Benjannet et al, 2006). The post-ER mechanism requires the proteolytic activity of PCSK9, whereas the endosomal–lysosomal mechanism does not (Park et al, 2004; McNutt et al, 2007). The endosomal–lysosomal mechanism is dependent on autosomal recessive hypercholesterolaemia (ARH) adaptor protein-mediated internalization of the PCSK9 (C-form)–LDLr complex into the endocytic pathway (Park et al, 2004; Qian et al, 2007). Overexpression of full-length PCSK9 in the absence of ARH results in degradation of LDLr through an endocytosis-independent mechanism (Park et al, 2004; Qian et al, 2007) that resembles the post-ER mechanism initially described by Maxwell & Breslow (2004) and Maxwell et al (2005).
Here, we used biochemical and genetic approaches to investigate whether PCSK9 is involved in the disposal of non-acetylated intermediates of BACE1 in the early secretory pathway.
Results And Discussion
Overexpression of PCSK9 decreases the levels of BACE1
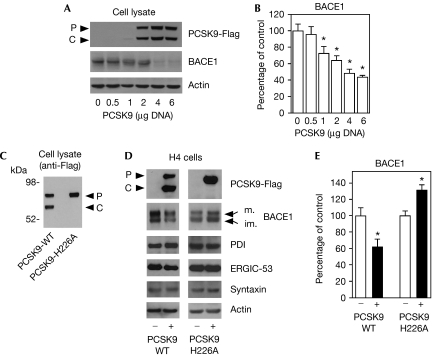
To assess whether PCSK9 is involved in the disposal of BACE1, we transfected Chinese hamster ovary (CHO) cells with human PCSK9-Flag. Fig 1A,B shows that the expression of PCSK9 decreased the steady-state levels of BACE1 in a gene-dose-dependent manner. Next, we transfected human neuroglioma (H4) cells and assessed whether the above events could be observed in a different cellular system. H4 cells also have the advantage of allowing better visualization of the mature and immature forms of BACE1 on SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Fig 1D,E (left panels) shows that overexpression of wild-type PCSK9 successfully reduced the levels of endogenous BACE1 in H4 cells. The fact that both forms of BACE1 decreased (Fig 1D, left panel) seems to suggest that PCSK9 is primarily acting in the early secretory pathway. In fact, a preferential degradation of the mature form in the endosomal–lysosomal pathway would only result in decreased levels of mature BACE1. By contrast, a preferential degradation of the immature form in the early secretory pathway would also result in decreased levels of the mature form.
Figure 1.
Overexpression of PCSK9 decreases the steady-state levels of BACE1. (A,B) CHO cells were transiently transfected with PCSK9-Flag cDNA. Cell lysates were analysed by SDS–PAGE and immunoblotting 48 h after transfection. A representative western blot is shown in (A), whereas the average (n=3)±s.d. is shown in (B); *P<0.05. (C) Western blot showing the migration of the wild-type (PCSK9-WT) and the H226A catalytically inactive (PCSK9-H226A) forms of PCSK9. (D,E) human neuroglioma (H4) cells were transfected with 5 μg DNA of the wild-type or the H226A mutant form of PCSK9. Total cell lysates were analysed with the indicated antibodies. A representative western blot is shown in (D), whereas the average (n=3)±s.d. is shown in (E); *P<0.05. Mature (m) and immature (im) BACE1 are indicated. PDI (protein disulphide isomerase; ER marker), ERGIC-53 (ERGIC marker), syntaxin (Golgi marker) and actin acted as controls. BACE1, β-site amyloid precursor protein (APP)-cleaving enzyme 1; CHO, Chinese hamster ovary; ER, endoplasmic reticulum; ERGIC, ER–Golgi intermediate compartment; PCSK9, proprotein convertase subtilisin kexin type 9; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
The proteolytic activity of PCSK9 requires a triad of residues (Asp 186, His 226 and Ser 386), which constitute the ‘catalytic triad' of the protease. Mutation of one of these residues abolishes the proteolytic activity of PCSK9 and its ability to undergo processing to the C-form (Benjannet et al, 2004, 2006; McNutt et al, 2007). Previous studies with the LDLr seem to indicate that the functional inactivation of PCSK9 can differentiate between the post-ER and the endosomal–lysosomal mechanisms. In fact, the former requires the catalytic activity of PCSK9, whereas the latter does not (Park et al, 2004; McNutt et al, 2007). Therefore, we mutated His 226 to Ala and generated a commonly used H226A mutant version of PCSK9 (Benjannet et al, 2004, 2006). As expected, the H226A mutant was unable to undergo processing and seemed to migrate as the P-form of the protein on SDS–PAGE (Fig 1C). When we analysed specific abilities to degrade BACE1, we found that, in contrast to wild-type PCSK9, the H226A mutant was unable to reduce the steady-state levels of BACE1 but rather caused a small increase (Fig 1D,E, right panels). The opposite behaviour of the wild-type and catalytic mutant forms of PCSK9 suggest that the above events occur in the early secretory pathway, most probably in a ‘post-ER compartment'. As stated above, this pattern is also supported by the ability of wild-type PCSK9 to degrade the immature form of BACE1 (Fig 1D,E). The apparent dominant-negative effect of the H226A mutant form is probably caused by its ability to interfere with the interaction of endogenous PCSK9 (the enzyme) and BACE1 (the substrate). In fact, the H226A mutation abolished only the catalytic activity, but not the ligand/substrate-binding activity, of transgenic PCSK9 (supplementary Fig S1E online). Finally, the effect induced by overexpression of the wild-type and H226A mutant forms of PCSK9 was specific to BACE1 and was not accompanied by generalized changes in the expression levels of control proteins, such as PDI, ERGIC-53 and syntaxin, which are ER-, ERGIC- and Golgi-based proteins, respectively (Fig 1D).
Downregulation of PCSK9 increases the levels of BACE1
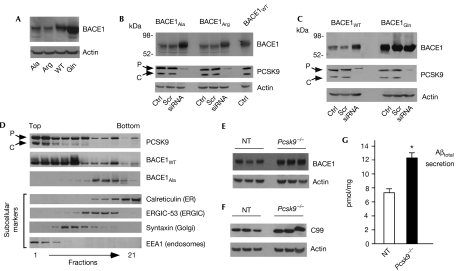
Previously, we showed that BACE1 is acetylated on seven lysine residues (Costantini et al, 2007). We also showed that substitution of these residues with either alanine or arginine generated ‘loss-of-acetylation' mutants that were not able to reach the Golgi apparatus. As a consequence, they were rapidly degraded in a ‘post-ER' compartment without the apparent requirement of the proteasome (Costantini et al, 2007). By contrast, substitution of the same residues with glutamine, which mimics the effects of lysine acetylation, generated a ‘gain-of-acetylation' mutant that showed rapid translocation along the secretory pathway and a longer half-life (Costantini et al, 2007). Finally, we also showed that the various steady-state levels of loss-of-acetylation and gain-of-acetylation mutants (Fig 2A) were not caused by differences in the efficiency of transcription or translation (Costantini et al, 2007).
Figure 2.
PCSK9 is responsible for the disposal of the non-acetylated intermediates of BACE1. (A) Steady-state levels of BACE1Ala, BACE1Arg, BACE1WT and BACE1Gln in stably transfected CHO cells. The mutants used here have already been characterized (Costantini et al, 2007). (B) CHO cells stably expressing BACE1Ala and BACE1Arg were treated with PCSK9 siRNA for 3 days before western blot analysis of total cell lysates. The steady-state levels of wild-type BACE1 (BACE1WT) are shown for comparison. (C) CHO cells stably expressing BACE1WT and BACE1Gln were treated and analysed as above. (D) The subcellular distribution of BACE1WT and the lysine-to-alanine mutant (BACE1Ala) forms of BACE1, and of endogenous PCSK9 was analysed by SDS–PAGE and immunoblotting after separation of intracellular membranes on a 10–24% discontinuous Nycodenz gradient. Membranes were developed in parallel on the same film. The appropriate subcellular markers are indicated. (E,F) Brain homogenate (neocortex) of non-transgenic (NT) and Pcsk9−/− mice was analysed with the indicated antibodies (LiCor Odyssey Infrared images and appropriate quantification are shown in supplementary Fig S3 online). (G) ELISA determination of total amyloid β-peptide (Aβ) in the samples used for (E,F). Values are the average (n=9)±s.d; *P<0.05. BACE1, β-site amyloid precursor protein (APP)-cleaving enzyme 1; CHO, Chinese hamster ovary; Ctrl, control (no treatment); ER, endoplasmic reticulum; ERGIC, ER–Golgi intermediate compartment; PCSK9, proprotein convertase subtilisin kexin type 9; Scr, scrambled (non-silencing) siRNA; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; siRNA, small interfering RNA (targeted against Pcsk9).
To assess whether PCSK9 is responsible for the disposal of non-acetylated intermediates of BACE1, we used small interfering RNA (siRNA) targeted against Pcsk9 in cells stably expressing either the wild-type (BACE1WT) or one of the above mutant forms (BACE1Ala, BACE1Arg and BACE1Gln) of BACE1, which have already been characterized (Costantini et al, 2007). Fig 2B shows that downregulation of PCSK9 completely normalized the steady-state levels of both BACE1Ala and BACE1Arg, indicating that PCSK9 is essential for the removal of non-acetylated intermediates of BACE1. Interestingly, ‘knockdown' of PCSK9 increased the levels of BACE1WT but not those of BACE1Gln (Fig 2C), suggesting that PCSK9 does not affect the acetylated forms of BACE1 (mimicked by the lysine-to-glutamine substitution).
Previously we showed that the subcellular distribution pattern of the loss-of-acetylation mutant forms of BACE1 (BACE1Ala and BACE1Arg) differed from that of the wild-type (BACE1WT) and the gain-of-acetylation mutant (BACE1Gln) forms (Costantini et al, 2007). In fact, BACE1Ala and BACE1Arg were unable to reach the Golgi apparatus and remained confined to the ER/ERGIC system (Fig 2D; see also Costantini et al, 2007). Interestingly, the distribution of BACE1Ala seems to overlap with that of PCSK9 only in the ER/ERGIC (Fig 2D), suggesting that the interaction between PCSK9 and the non-acetylated intermediates of nascent BACE1 occurs early in the secretory pathway. It is also worth remembering that nascent BACE1 contains a propeptide sequence at the N terminus that is removed in the Golgi apparatus. However, non-acetylated BACE1 mutants (BACE1Ala and BACE1Arg) are unable to reach the Golgi apparatus. As a consequence, their prodomain is not removed from the N terminus (Costantini et al, 2007). Therefore, it is likely that the disposal of non-acetylated intermediates of nascent BACE1 by PCSK9 occurs in the ER/ERGIC system, which is also the system in which the non-acetylated intermediates of BACE1 accumulate. This pattern would be consistent with the ability of PCSK9 to degrade immature (ER/ERGIC-based) BACE1 and with the differing behaviour of the wild-type and H226A forms of PCSK9 (Fig 1D,E). The fact that the ER/ERGIC fractions predominantly contain full-length (P-form) PCSK9 does not contradict our conclusions. In fact, S127R and D129G mutant forms, two naturally occurring PCSK9 mutants (associated with familial hypercholesterolaemia) that are not processed to the C-form and are retained in the early secretory pathway, cause a marked degradation of LDLr without being secreted into the conditioned media (Homer et al, 2008). It is worth noting that the above mutants retain the Asp 186/His 226/Ser 386 proteolytic triad, suggesting that the processing of PCSK9 to the C-form is not required for the degradation of its substrates in the ‘post-ER compartment', provided that both the proteolytic and ligand/substrate-binding activities are preserved.
Finally, we decided to analyse the steady-state levels of endogenous BACE1 in the neocortex of Pcsk9−/− mice. Fig 2E–G and supplementary Fig S3 online show that PCSK9 ‘knockout' animals have higher levels of BACE1 (Fig 2E; supplementary Fig S3 online), C99 (Fig 2F; supplementary Fig S3 online) and Aβ (Fig 2G). C99 is the immediate product of BACE1-mediated cleavage of APP, whereas Aβ represents one of the final products of the sequential processing of APP by BACE1 and γ-secretase (Puglielli, 2008). Parallel changes in C99 and Aβ directly reflect the steady-state levels and activity of BACE1. Taken together, the above results indicate that PCSK9 levels in the brain affect the metabolism of BACE1 and the rate of Aβ generation.
Secreted PCSK9 stimulates the degradation of BACE1
Studies performed with the LDLr indicate that PCSK9 can also dispose of LDLr in the ‘endosomal–lysosomal compartment' (Benjannet et al, 2006). This event does not require the catalytic activity of the protease because similar levels of LDLr degradation can be observed also with the catalytically inactive forms of PCSK9 (Park et al, 2004; McNutt et al, 2007).
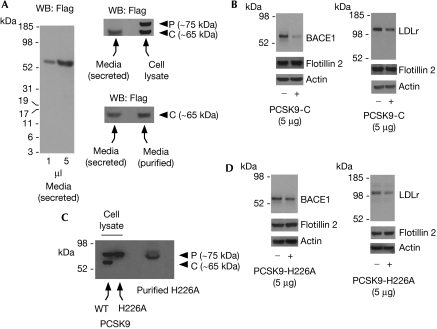
To assess whether a similar mechanism was involved also in the degradation of BACE1, we purified secreted PCSK9 from the conditioned media of stably expressing cells by using Flag antibodies covalently attached to immobilized agarose beads. As expected, when we analysed the conditioned media, secreted PCSK9 seemed to migrate with the molecular mass of the C-form on SDS–PAGE, indicating successful processing (Fig 3A). Fig 3B shows that the addition of affinity-purified secreted (C-form) PCSK9 to the media of BACE1- or LDLr-expressing cells reduced the steady-state levels of both BACE1 and LDLr. However, this effect did not require the catalytic activity of the protease. In fact, a reduction in both BACE1 and LDLr was observed also with affinity-purified PCSK9-H226A (Fig 3C,D).
Figure 3.
The disposal of BACE1 by secreted PCSK9 (C-form) does not require catalytic activity. (A) Conditioned medium from CHO cells stably expressing wild-type PCSK9 was analysed by western blot. The migration of secreted PCSK9 (C-form) in the conditioned media (left panel) can be compared with the migration of the P- and C-form of the protein resolved from a total cell lysate (upper right panel). The migration of secreted PCSK9-C before and after purification is shown in the lower right panel. (B) BACE1- and LDLr-expressing cells were incubated with secreted (C-form) PCSK9 for 8 h before western blot analysis of total cell lysates. Secreted PCSK9 was purified by affinity chromatography from the conditioned media of stably expressing cells (A) before the incubation. (C,D) The experiment described in (B) was repeated with the catalytically inactive PCSK9-H226A. PCSK9-H226A was purified by affinity chromatography from cell extracts of stably expressing cells (C) before the incubation (D). Flotillin 2 (cell-surface protein) and actin acted as controls. BACE1, β-site amyloid precursor protein (APP)-cleaving enzyme 1; CHO, Chinese hamster ovary; LDLr, low-density lipoprotein receptor; PCSK9, proprotein convertase subtilisin kexin type 9.
This result is consistent with the dominant-negative behaviour described in Fig 1D. In fact, although catalytically inactive, the H226A mutant can still bind to its substrates (BACE1 and LDLr) at the cell surface, and can induce internalization and lysosomal-mediated degradation (Park et al, 2004; McNutt et al, 2007; see also supplementary Fig S1E online). In essence, when taken together, the above results indicate that PCSK9 can stimulate the degradation of BACE1 through two different mechanisms: the first requires the catalytic activity of the protease and can be observed after transfection of full-length PCSK9 (Fig 1), whereas the second does not require the catalytic activity of the protease and is observed following the addition of secreted PCSK9 to the media (Fig 3). The above results are reminiscent of the ‘post-ER' and ‘endosomal–lysosomal' mechanisms already described and characterized for the LDLr (Maxwell & Breslow, 2004; Park et al, 2004; Maxwell et al, 2005; Benjannet et al, 2006; McNutt et al, 2007) and indicate that BACE1 is a new substrate for PCSK9. This conclusion is further supported by the fact that PCSK9 can interact with BACE1 in vitro (see supplementary Fig S1 online). Interestingly, the effect of the wild-type and H226A mutant forms of PCSK9 was similar overall in the case of LDLr but not in the case of BACE1 (Fig 3B, D). Whether this is caused by different binding affinities to the substrates remains to be determined.
The immature form of LDLr is acetylated
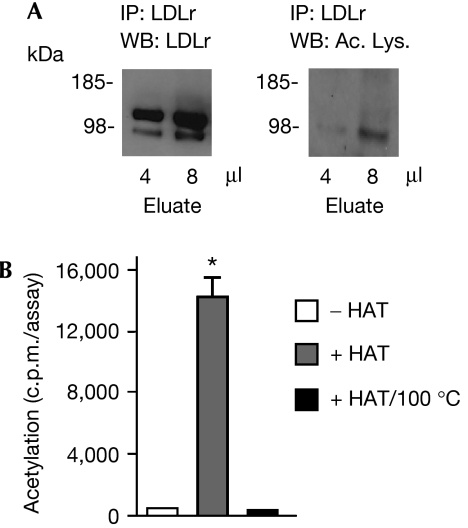
The similarities between the degradation of BACE1 (reported here) and LDLr seem to suggest that PCSK9 might act as, or contribute to, the machinery that disposes of non-acetylated intermediates of nascent membrane proteins. In fact, the results obtained with siRNA (Fig 2) indicate that the non-acetylated intermediates of BACE1 constitute the preferential substrate of PCSK9, at least in the case of BACE1. Therefore, we decided to assess whether LDLr is also acetylated. The analysis of the acetylation pattern of LDLr shows that the immature form––but not the mature form––of the receptor can be recognized with acetylated lysine antibodies (Fig 4A). The in vitro acetylation of lysine residues seems to be highly specific, in that only those lysine residues that are normally acetylated in vivo can also be acetylated in vitro following incubation with an acetyl-CoA:lysine acetyltransferase (Masumoto et al, 2005; Yuan et al, 2005; Costantini et al, 2007). Therefore, we decided to use an in vitro acetylation system previously used for BACE1 (Costantini et al, 2007) and assess whether LDLr could act as a substrate for lysine acetylation. A Myc-tagged (at the carboxyl terminus) version of full-length LDLr was purified by affinity chromatography from stably transfected CHO cells and then incubated with radiolabelled acetyl-CoA, a donor of the acetyl group, and the histone acetyltransferase domain of recombinant CREB-binding protein (CBP/p300). Fig 4B shows that LDLr can be acetylated in vitro, provided that it is incubated in the presence of an enzymatically active acetyltransferase.
Figure 4.
Low-density lipoprotein receptor, a known substrate of PCSK9, is acetylated. (A) LDLr was immunoprecipitated from stably transfected CHO cells and analysed by immunoblotting with the indicated antibodies. (B) LDLr was purified by affinity chromatography (ProFound system) and incubated in the presence of HAT and [3H]acetyl-CoA for 1 h at 30°C (Costantini et al, 2007). LDLr was then immunoprecipitated and analysed on a scintillation liquid counter. The results are the average (n=4)±s.d.; *P<0.0005. Ac. Lys., acetylated lysine antibody; CHO, Chinese hamster ovary; HAT, histone acetyltransferase; LDLr, low-density lipoprotein receptor; PCSK9, proprotein convertase subtilisin kexin type 9.
Taken together, the above results seem to indicate that LDLr is the second known membrane protein to undergo acetylation in the secretory pathway; they also indicate that PCSK9 might act as (or contribute to) the common machinery that disposes of non-acetylated intermediates of nascent membrane proteins in a post-ER compartment. Whether the disposal of non-acetylated intermediates is a direct function of PCSK9 or other unidentified proteases that require PCSK9 is still unknown.
Methods
Antibodies and western blot analysis. Western blotting was performed on a 4–12% Bis–Tris SDS–PAGE system (NuPAGE; Invitrogen, Carlsbad, CA, USA) as described previously (Costantini et al, 2006, 2007; Ko & Puglielli, 2007). The following antibodies were used in this study: anti-acetylated lysine (monoclonal; Abcam, Cambridge, MA, USA); anti-BACE1 (polyclonal; Abcam); anti-calreticulin (polyclonal; Abcam); anti-ERGIC 53 (polyclonal; Sigma, St Louis, MO, USA); anti-syntaxin (monoclonal; Abcam); anti-EEA1 (monoclonal; BD Transduction Laboratories, San José, CA, USA); anti-c-Myc (polyclonal; Sigma); anti-actin (polyclonal; Cell Signaling, Danvers, MA, USA); anti-PDI (protein disulphide isomerase-ER marker; monoclonal; Abcam); anti-Flag (monoclonal; Sigma; anti-PCSK9 (polyclonal; Cayman Chemical, Ann Arbor, MI, USA); anti-LDLr (a generous gift from Dr Alan Attie, University of Wisconsin-Madison); anti-NGF (polyclonal; Chemicon Millipore, Billerica, MA, USA); anti-flotillin 2 (polyclonal; Cell Signaling); anti-C99 (monoclonal; MBL, Woburn, MA, USA), anti-APP C-terminal (polyclonal; Chemicon) and anti-BSA (monoclonal; Abcam). Catalogue numbers are reported in the supplementary information online.
Pixel densities (for signal area) of scanned images were calculated with Adobe Photoshop; densitometry (for signal density) was analysed with the EpiChemi3 DarkroomTM (UVP Bioimaging Systems, Upland, CA, USA) using Labworks Image Acquisition and Analysis Software 4.5. In addition, samples were also imaged and quantified using the LiCor Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA). For infrared imaging, goat anti-rabbit Alexa Fluor 680-conjugated secondary antibodies were used instead of horseradish-peroxidase-conjugated secondary antibodies. For quantification, values were normalized to actin (as loading control).
cDNA. The complementary DNA for human PCSK9-Flag was a generous gift from Dr Jan L. Breslow (The Rockefeller University). The PCSK9-H226A mutation was created with the Quick-Change Site Directed Mutagenesis kit (Stratagene, Cedar Creek, TX, USA), as described previously (Elagoz et al, 2002). PCSK9 and PCSK9-H226A cDNA were inserted into pCMV-Tag4A (Stratagene; for stable and transient transfection) and pTNT® (Promega, Madison, WI, USA; for in vitro translation) vectors. The in vitro translation was performed with the TNT® T7 Quick Coupled Transcription/Translation System (Promega), as suggested by the manufacturer. The cDNA for human LDLr was a generous gift from Dr Alan D. Attie (University of Wisconsin-Madison). LDLr was subcloned into a pcDNA3.1A vector (Invitrogen) to produce a C-terminal Myc-tagged fusion protein.
siRNA treatment. The pools of siRNA duplexes designed against Pcsk9 were from Dharmacon RNA Technologies (Lafayette, CO, USA; cat. no. MQ054699). Scrambled/non-silencing siRNA was used as the control siRNA. siRNAs were transfected into cells by using the siIMPORTER Transfection Reagent (Upstate,cat. no. 64-101), as suggested by the manufacturer.
Incubation with purified PCSK9. The day before the experiment, CHO cells stably expressing BACE1 or LDLr were plated onto six-well plates at a density of 3 × 105 cells per well and incubated in the presence of serum-free media. On the day of the experiment, fresh serum-free media were added together with 5 μg of purified PCSK9-C or PCSK9-H226A. Cells were incubated for 8 h, rinsed in PBS and collected by scraping. Cell lysates were analysed by SDS–PAGE and western blot.
Statistical analysis. Results are always expressed as mean±s.d. of the indicated number of determinations. The data were analysed by ANOVA and Student's t-test comparison, using GraphPad InStat3 software. P<0.05 was considered to be statistically significant.
Additional methods are detailed in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank A. Attie for critical reading of an early version of this manuscript. We thank J. Breslow for PCSK9 cDNA and A. Attie for LDLr cDNA and antibody. This research was supported by a grant from the National Institutes of Health (to L.P.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Attie AD (2004) The mystery of PCSK9. Arterioscler Thromb Vasc Biol 24: 1337–1339 [DOI] [PubMed] [Google Scholar]
- Benjannet S et al. (2004) NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem 279: 48865–48875 [DOI] [PubMed] [Google Scholar]
- Benjannet S, Rhainds D, Hamelin J, Nassoury N, Seidah NG (2006) The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J Biol Chem 281: 30561–30572 [DOI] [PubMed] [Google Scholar]
- Costantini C, Scrable H, Puglielli L (2006) An aging pathway controls the TrkA to p75(NTR) receptor switch and amyloid β-peptide generation. EMBO J 25: 1997–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini C, Ko MH, Jonas MC, Puglielli L (2007) A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochem J 407: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG (2002) Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J Biol Chem 277: 11265–11275 [DOI] [PubMed] [Google Scholar]
- Grozdanov PN, Petkov PM, Karagyozov LK, Dabeva MD (2006) Expression and localization of PCSK9 in rat hepatic cells. Biochem Cell Biol 84: 80–92 [DOI] [PubMed] [Google Scholar]
- Homer VM et al. (2008) Identification and characterization of two non-secreted PCSK9 mutants associated with familial hypercholesterolemia in cohorts from New Zealand and South Africa. Atherosclerosis 196: 659–666 [DOI] [PubMed] [Google Scholar]
- Ko MH, Puglielli L (2007) The sterol carrier protein SCP-x/pro-SCP-2 gene has transcriptional activity and regulates the Alzheimer disease γ-secretase. J Biol Chem 282: 19742–19752 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Maxwell KN, Breslow JL (2004) Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA 101: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Breslow JL (2005) Proprotein convertase subtilisin kexin 9: the third locus implicated in autosomal dominant hypercholesterolemia. Curr Opin Lipidol 16: 167–172 [DOI] [PubMed] [Google Scholar]
- Maxwell KN, Fisher EA, Breslow JL (2005) Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc Natl Acad Sci USA 102: 2069–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt MC, Lagace TA, Horton JD (2007) Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J Biol Chem 282: 20799–20803 [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T (2005) ERAD: the long road to destruction. Nat Cell Biol 7: 766–772 [DOI] [PubMed] [Google Scholar]
- Park SW, Moon YA, Horton JD (2004) Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J Biol Chem 279: 50630–50638 [DOI] [PubMed] [Google Scholar]
- Puglielli L (2008) Aging of the brain, neurotrophin signaling, and Alzheimer's disease: is IGF1-R the common culprit? Neurobiol Aging 29: 795–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian YW et al. (2007) Secreted PCSK9 downregulates low density lipoprotein receptor through receptor-mediated endocytosis. J Lipid Res 48: 1488–1498 [DOI] [PubMed] [Google Scholar]
- Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M (2003) The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci USA 100: 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Parodi AJ (2003) Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol 19: 649–676 [DOI] [PubMed] [Google Scholar]
- Yoshida H (2007) ER stress and diseases. FEBS J 274: 630–658 [DOI] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307: 269–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information