Abstract
The EKC/KEOPS yeast complex is involved in telomere maintenance and transcription. The Bud32p and kinase-associated endopeptidase 1 (Kae1p) components of the complex are totally conserved in eukarya and archaea. Their genes are fused in several archaeal genomes, suggesting that they physically interact. We report here the structure of the Methanocaldococcus jannaschii Kae1/Bud32 fusion protein MJ1130. Kae1 is an iron protein with an ASKHA fold and Bud32 is an atypical small RIO-type kinase. The structure MJ1130 suggests that association with Kae1 maintains the Bud32 kinase in an inactive state. We indeed show that yeast Kae1p represses the kinase activity of yeast Bud32p. Extensive conserved interactions between MjKae1 and MjBud32 suggest that Kae1p and Bud32p directly interact in both yeast and archaea. Mutations that disrupt the Kae1p/Bud32p interaction in the context of the yeast complex have dramatic effects in vivo and in vitro, similar to those observed with deletion mutations of the respective components. Direct interaction between Kae1p and Bud32p in yeast is required both for the transcription and the telomere homeostasis function of EKC/KEOPS.
Keywords: fusion protein, telomere, transciption
Introduction
Kinase, endopeptidase and other peptides of small size (KEOPS), also called endopeptidase-like kinase chromatin-associated (EKC), is a complex of proteins that has been implicated in transcription and telomere maintenance. The yeast complex consists of five subunits: Bud32p, kinase-associated endopeptidase 1 (Kae1p), Pcc1p, Gon7p and Cgi121p (Downey et al, 2006; Gavin et al, 2006; Kisseleva-Romanova et al, 2006). These proteins have been identified during a genome-wide screen in Saccharomyces cerevisiae for suppressors of cdc13-1, an allele of the telomere-capping protein Cdc13p. This has suggested that KEOPS somehow functions either to promote uncapping or telomere resection when capping is defective. Deletion of individual KEOPS subunits such as Bud32p, Cgi121p and Gon7p leads to decreased telomere sizes, indicating that, in addition to promote telomere uncapping, the KEOPS complex is also required to maintain the appropriate telomeric repeats length.
The same complex was isolated in a parallel study and was dubbed as EKC. Subunits of EKC were shown to be required for the expression of several yeast genes, some of which are involved in the response to the mating pheromone and polarized growth (Kisseleva-Romanova et al, 2006). It was shown that the EKC impacts transcription of galactose-inducible genes by affecting recruitment of the SAGA and Mediator co-activators. Chromatin immunoprecipitation experiments and multiple genetic interactions of PCC1 mutants with mutants of the transcription apparatus and chromatin-modifying enzymes underscore the direct function of the complex in transcription.
Sequence analyses and functional complementation experiments (Lopreiato et al, 2004; Kisseleva-Romanova et al, 2006) have shown that subunits of the EKC/KEOPS are conserved among species and suggested that the function of the complex in transcription and/or telomere maintenance (and possibly other processes) is also conserved in other species. Kae1p is the most highly conserved member of the complex, with sequence identity of roughly 60% between yeast and humans. On the basis of homology to a bacterial protein, it was originally suggested that this protein might be endowed with endopeptidase activity (Mellors and Lo, 1995; Lopreiato et al, 2004). Bud32p is the yeast orthologue of human p53-related protein kinase (PRPK; 32% sequence identity) that was shown to phosphorylate p53 and promote p53-dependent transcriptional activation (Abe et al, 2001; Facchin et al, 2003). The presence of Bud32p and Cgi121p in the same complex in yeast is paralleled by the documented interaction of human PRPK and CGI121. Finally, yeast Pcc1p is the likely orthologue of small conserved human proteins (ESO3 and CTAG1/2), some of which belong to the family of cancer-testis antigens (Kisseleva-Romanova et al, 2006).
Genome analysis has revealed that homologues of yeast Kae1p and Bud32p are present in all eukaryotes and archaea. The genes encoding the archaeal orthologues of Kae1p and Bud32p are juxtaposed in nearly all archaeal genomes, and they are even fused in several archaea, encoding a single bifunctional fusion protein, strongly suggesting that Kae1p and Bud32p are physically and functionally linked (Marcotte et al, 1999). Recently, we have determined the crystal structure and analysed the biochemical function of PAB1159, the orthologue of Kae1p in the archaeon Pyrococcus abyssi (hereafter named PaKae1; Hecker et al, 2007). As predicted by Aravind and Koonin (1999), PaKae1 belongs to the ASKHA protein family containing acetate and sugar kinase, Hsp70 chaperone proteins and actin. All these proteins have ATPase activity. PaKae1 was shown to bind the ATP analogue AMPPNP in a canonical manner, occupying a groove situated between the two domains of the protein. Surprisingly, the nucleotide was found to be directly bound through its phosphate groups to a non-heme iron. Mutation of residues coordinating this iron in the yeast Kae1p causes lethality, demonstrating the importance of the metal binding for function (Kisseleva-Romanova et al, 2006). Although yeast Kae1p was proposed to possess endopeptidase activity (Mellors and Lo, 1995), the crystal structure of PaKae1 as well as biochemical activity tests do not support this hypothesis. Rather, it was shown that PaKae1 binds cooperatively to single and double-stranded DNA and induces DNA conformational changes without significant DNA lengthening or shortening. PaKae1 also exhibits a class I apurinic endonuclease activity (AP-lyase), suggesting an important function of this protein in the maintenance of genome integrity (Hecker et al, 2007).
To get further insights on the interactions between Kae1 and Bud32 in the KEOPS/EKC complex, we investigated the archaeal fusion protein. The orthologues of yeast Kae1p (MjKae1) and Bud32p (MjBud32) in M. jannaschii are fused into a single protein MJ1130 (MjKae1/Bud32). We solved the structure of this fusion protein by X-ray crystallography showing that Bud32 is an atypical protein kinase, the smallest for which the structure was solved so far. The structure of MJ1130 suggests that association with Kae1 maintains the Bud32 kinase in an inactive state. We indeed show that yeast Kae1p represses the kinase activity of yeast Bud32p. We further observed the existence of extensive intramolecular interactions between the MjKae1 and MjBud32 moieties of MJ1130, also suggesting that the two yeast proteins interact directly. On the basis of the MJ1130 structure, we constructed a model for the yeast Kae1p/Bud32p interaction surface and further sampled this interaction by in vivo and in vitro experiments. Importantly, mutations that disrupt the interaction between the two proteins strongly affect the function of the yeast EKC/KEOPS and elicit major phenotypic effects both on transcription and telomere maintenance, linking the function of the complex to its structural integrity.
Results
The structure of MJ1130 reveals complex formation between MjKae1 and MjBud32
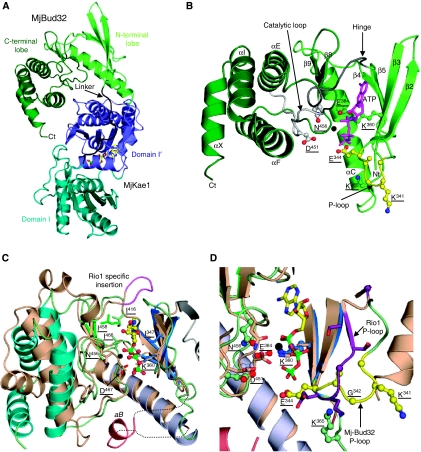
Purified MjKae1/Bud32 exhibits a pink colour in solution resulting from a broad absorbance band centred at 492 nm, suggesting that MjKae1/Bud32 contains an Fe3+ ion (Hecker et al, 2007; see Supplementary Figure S1). The crystal structure of MjKae1/Bud32 was solved to 3.2 Å resolution by single-wavelength anomalous dispersion (SAD) phasing from a selenomethionine-labelled protein crystal and refined against 3.05 Å resolution native data. The N-terminal part (residues 1–323) corresponds to MjKae1 (Hecker et al, 2007), whereas the C-terminal part (residues 341–532) covers the MjBud32 kinase (Figure 1A, Supplementary Figure S2 and S3). Both parts are connected by a linker comprising residues 324–340. The MjKae1/Bud32 fusion protein has an elongated shape with MjBud32 attached to the opposite side of the ATP-binding groove of MjKae1 (Figure 1A). The asymmetric unit of the crystal contains two identical copies of MjKae1/Bud32 (r.m.s.d. of 0.5 and 0.6 Å for MjKae1 and MjBud32, respectively). However, the elbow angle between MjKae1 and MjBud32 differs by 5°.
Figure 1.
MJ1130 structure and comparison with RIO1 kinase. (A) Ribbon representation of MJ1130. Mj-Kae1 is coloured in light (subdomain I) and dark (subdomain I′) blue, MjBud32 is represented in light (N-terminal lobe) and dark (C-terminal) green. The linker is in black and AMPPNP is shown as sticks. (B) Ribbon representation of the MjBud32: N- and C-terminal lobes are in light and dark green, respectively, and the hinge region is in grey. The catalytic and P-loops are in light grey and yellow, respectively. The ATP (purple sticks) and manganese ion (black sphere) have been modelled as indicated in panel C. (C) A model for ATP binding to MjBud32 was obtained by superposing it to RIO1 (salmon) with ATP (ball and sticks) and manganese (black sphere; PDB code: 1ZP9). Conserved residues involved in ATP and Mn2+ binding are shown as sticks. For clarity, only the labels for MjBud32 residues are given. RIO1 helix αB from the loop connecting β3 to αC, which becomes largely disordered upon ATP binding, is shown in pink. The insertion of five residues observed in the loop connecting the N- and C-terminal lobes from RIO1 kinases is coloured in light purple. (D) Comparison of the active sites of MjBud32 and RIO1 kinase. Side chains from active site residues from MjBud32 and RIO1 are shown as ball and sticks. For clarity, only the labels for MjBud32 residues are given. The MjBud32 and RIO1 P-loops are colored yellow and purple, respectively.
The present MjKae1 crystal structure is very similar to that of the P. abyssi orthologue (PaKae1) (r.m.s.d. of 1.1 Å over 308 Cα atoms; 56% sequence identity; Hecker et al, 2007). MjKae1 adopts a fold characteristic of the members of the ASKHA superfamily of phosphotransferases (Hurley, 1996; Buss et al, 2001). This fold is made up of a repeated five-stranded α/β domain (I and I′) (Hecker et al, 2007). AMPPNP (a non-hydrolysable ATP analogue) is bound at the C-terminal edge of the domain I β-sheet. Kae1 was annotated previously as an O-sialoglycoprotease (Mellors and Lo, 1995), but the structure did not confirm this.
MjBud32 (residues 340–530), which has strong sequence identity with the yeast Bud32p kinase (Hanks and Hunter, 1995; Facchin et al, 2002), adopts the protein kinase fold consisting of two lobes (Figure 1A and B). The N-terminal lobe (residues 340–420), corresponding to the ATP-binding domain, comprises a four-stranded antiparallel β-sheet (β2β3β5β4) as well as helix αC (between strands β3 and β4). The C-terminal lobe (residues 421–532) contains four α-helices (αE to αX) and a short β-hairpin (strands β8 and β9) and is much smaller than usually observed for kinases. This domain is responsible for binding the protein/peptide substrate and for initiating phosphotransfer in eukaryotic kinases.
MjBud32 has strong structural similarity with RIO kinases
Despite low sequence identity, MjBud32 is most closely related to the RIO protein kinase family (r.m.s.d. of 1.94 Å with Archaeoglobus fulgidus RIO1, PDB code 1zth, Z-score 7.2, for 244 Cα positions, 18% sequence identity; Figure 1C and D) (LaRonde-LeBlanc and Wlodawer, 2005; Laronde-Leblanc et al, 2005). However, some significant differences are apparent. Generally, protein kinases have a five-stranded sheet in the N-terminal lobe, but in MjBud32 this domain has only four strands (strand β1 is missing). The N-terminal residues that should correspond to this strand (333–339) belong to the linker between MjBud32 and MjKae1. This linker (residues 324–339) is only visible in one copy of the asymmetric unit and adopts an elongated conformation that bypasses the β1 strand. The P-loop of MjBud32 is fully visible in only one copy of MJ1130 where it adopts a conformation distinct from that of RIO1 (Figure 1D). ATP generally binds in the groove between the N- and C-terminal lobes of protein kinases and contacts the metal binding and catalytic loops as well as the P-loop. Although MjKae1/Bud32 was incubated with a 10-fold molar excess of AMPPNP, no nucleotide could be observed in the binding pocket of MjBud32, contrary to MjKae1, which has the nucleotide clearly bound at its active site (Supplementary Figure S4).
MjBud32 is a structural model for yeast Bud32p
We modelled ATP/Mn2+ into the active site of MjBud32 using the structure of the RIO1/AMPPNP complex as template (PDB code: 1zp9; Figure 1C and D). Slight movements of ATP and minor side-chain adjustments allowed construction of a realistic model, which was not further adjusted by calculations. On the other hand, the structure–function relationship in yeast Bud32p has been probed by site-directed mutagenesis on the basis of sequence alignment with the cAMP-dependent protein kinase, PKA (Facchin et al, 2002), and we analysed these mutations against our modelled MjBud32–ATP complex (MjBud32 residues are underlined). In our model, the adenosine moiety points into a hydrophobic pocket (Ile347, Ile416, Ile458, Ile466) that is very well conserved between RIO1 and MjBud32 (Figure 1C). RIO1 contains a five-residue insertion (residues 152–156, purple in Figure 1C) in the linker between the two lobes. This peptide buries partly the adenine ring of ATP. This flap is absent both in RIO2 and MjBud32, resulting in a more open ATP-binding pocket than that for RIO1 kinase.
A Mn2+ ion coordinates the α and β ATP phosphate moieties in RIO1 and thereby helps to orient the γ phosphate for transfer. In the model of the MjBud32–ATP complex, the γ-phosphate group of ATP is facing Asp451 and Asn456. The homologous residue of the latter asparagine contacts a Mn2+ ion in RIO1 kinase. In our MjBud32–ATP model, these interactions with the phosphates can be maintained and the coordination sphere of the metal can be completed by the totally conserved side chains of Asn456 from the catalytic loop and Asp467 from the metal-binding loop (Figure 1C and Supplementary Figure S3). Asp467 is part of the conserved DFG, which adopts a conformation similar to that found in other protein kinases. Replacement of the homologous Asp161 and Asn166 in Bud32p by alanine dramatically affects activity showing that Bud32p has a canonical metal-binding site involved in phosphate transfer. In MjBud32, the α and γ phosphate groups from ATP interact with Lys360, which corresponds to Lys52 from yeast Bud32p (Figure 1D and Supplementary Figure S3). Mutation of Lys52 into Ala in yeast Bud32p resulted in complete loss of catalytic activity (Facchin et al, 2002). In eukaryotic kinases, this lysine is involved in the coordination of two phosphate groups from ATP. The equivalent Lys360 from strand β3 also makes a salt bridge with the totally conserved carboxylate from Glu384, whose mutation into Ala in yeast Bud32p (Glu76) also abolishes catalytic activity (Figure 1D). Our model confirms that MjBud32 possesses an active ATP-binding pocket compatible with the kinase activity as measured in vitro for the yeast and human orthologues (Facchin et al, 2002).
Mutation in yeast Bud32p of the invariant Gly25 (Gly342) of the P-loop into valine completely abolished both kinase activity and binding of the ATP analogue FSBA. Residue Gly342 is in a turn that follows the linker region connecting MjKae1 to MjBud32 (Figure 1D). In the superposed structure, Gly342 is at 6.6 Å from the homologous RIO1 glycine (Gly58) and is directed away from the ATP-binding pocket. We suspect that the present ATP-binding pocket of the MjBud32 is in an inactive conformation, explaining the absence of any bound nucleotide.
The kinase activity of Bud32p is inhibited by Kae1p
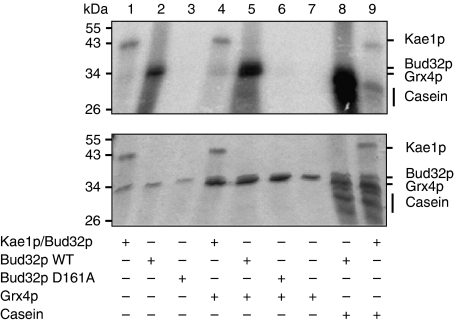
The absence of nucleotide in the structure of the MjBud32 moiety suggested that Kae1 maintains Bud32 in an inactive conformation in the complex. It was previously shown that yeast Bud32p is capable of phosphorylating casein and glutaredoxin Grx4 in vitro (Stocchetto et al, 1997; Lopreiato et al, 2004). We therefore tested the effect of Kae1p on the Bud32p kinase activity. Bud32p phosphorylated Grx4p and casein as expected (Figure 2, lanes 5 and 8) but not the D161A Bud32p mutant, confirming that the phosphorylation was dependent of Bud32 (lanes 5 and 6). The kinase activity was strongly diminished in the presence of Kae1p (lanes 4 and 9). We also noticed a strong autophosphorylation of yeast Bud32p (lane 2), which was also strongly reduced by Kae1p (lane 1). Similar results were obtained for the archaeal MJ1130 protein (see Supplementary Figure S5). Our results indicate that both the yeast and archaeal Bud32 phosphotransferase activities are inhibited by Kae1. This fits well with the fact that we could not detect AMPPNP in the nucleotide-binding site of MjBud32, suggesting that the presence of Kae1 maintains Bud32 in an inactive conformation both in the crystal and in solution.
Figure 2.
Bud32p kinase activity is inhibited by Kae1p. The phosphotransferase activities of Bud32p and the Bud32p/Kae1p complex on Grx4p and dephosphorylated casein were tested. PhosphoImager scan (upper panel) and Coomassie blue staining of SDS–PAGE gels (lower panel) of the reaction mixtures are shown.
MjKae1 and MjBud32 form a tight intramolecular complex
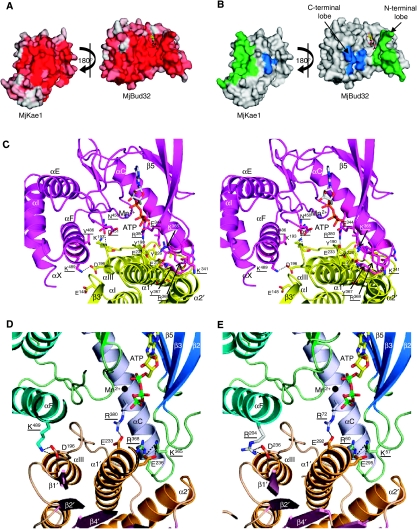
Although yeast Kae1p and Bud32p have been shown to belong to the same complex (Gavin et al, 2002; Downey et al, 2006; Kisseleva-Romanova et al, 2006) and interact in a two-hybrid system (Ho et al, 2002; Lopreiato et al, 2004), a direct interaction between the two proteins has not been proven. Physical and/or functional interaction between the archaeal orthologues of yeast Kae1p was strongly suggested by the very frequent juxtaposition of the archaeal KAE1 and BUD32 genes and the presence of these genes as a fusion in a few archaeal genomes (e.g., MJ1130; Marcotte et al, 1999). The interaction between MjKae1 and MjBud32 occurs between conserved surface patches (Figure 3A and B) and engages roughly 2000 Å2 surface area, suggesting formation of a stable protein–protein complex (Lo Conte et al, 1999). Both the N- and C-terminal lobes of MjBud32 are involved in the interaction, whereas for MjKae1, only the C-terminal domain I′ forms contacts (Figure 3C), confirming previous yeast two-hybrid data (Lopreiato et al, 2004). The C-terminal domain I′ of Kae1 carries an insertion between β3′ and α1′, which consists of three helices (helices αI, αII and αIII), which together with helices α1′ and α2′ form a five-helix bundle. This provides the main platform for interaction with MjBud32 (Figures 1A and 3A–D, Supplementary Figure S2). The residues of MjKae1 that bind to MjBud32 are mainly provided by the 20-amino-acids-long linker connecting helices αII and αIII, and by helices α1′ and α2′ (Supplementary Figure S2). Residues from the helices α1′ and α2′ are facing residues from the MjBud32 N-terminal lobe (i.e., from the linker connecting strand β3 to helix αC and from helix αC). In addition, the C-terminal part of MjKae1 helix α2′ is in contact with the region preceding strand β2. MjBud32 provides 17 interacting amino acids (Figure 3C and Supplementary Figure S3) that are clustered in two distinct patches: 12 residues on the N-terminal and five residues on the C-terminal lobe (Figure 3B). Five hydrogen bonds or salt bridges are stabilizing the interface (Figure 3D). Two hydrogen bonds are realized between the Gly194 amide and Asn450 main-chain carbonyl as well as between the Tyr221 Oη and Asp369 carboxylic groups (MjBud32 residues are underlined) and two salt bridges: Asp196/Lys489 and Glu233/Arg380. The complex is further stabilized by packing of hydrophobic residues from MjKae1 (Leu185, Tyr190, Tyr226, Met262 and Met266) and MjBud32 (Tyr367, Leu372, Ile376; Figure 3A). Interestingly, the insertion in domain I′ of MjKae1 that interacts with MjBud32 is not present in homologous sequences from bacteria that do not possess Bud32 orthologous sequences.
Figure 3.
MjKae1/Bud32 complex. (A) Open leaflet representation of the residue conservation at the interface between MjKae1 and MjBud32. Colouring is from light grey (poorly conserved) to red (highly conserved). (B) Open leaflet representation of the interface between MjKae1 and MjBud32. Residues from the interface are coloured blue (small patch) and green (large patch). (C) Stereo view ribbon representation of the interface between MjKae1 (yellow) and MjBud32 (magenta). The side chains from residues involved at the interface are shown as sticks. Hydrogen bonds and salt bridges are depicted as black dashed lines. The ATP nucleotide (grey sticks) bound in the active site of RIO1 is shown superimposed on the MjBud32. (D) Residues at the interface between MjKae1 and MjBud32 (underlined) that were mutated to study the in vivo and in vitro effects of disruption of the yeast complex. The ATP and Mn2+ ligands have been modelled as described for Figure 1C. Salt bridges are depicted by dashed lines. (E) Model of the Kae1p/Bud32p interface (the same zone and colour code as in panel D).
The 5° difference in the elbow angle between the two copies of MjKae1 and MjBud32 (Supplementary Figure S6A) creates a slightly different interface. Four out of five H-bonds/salt bridges are common to both complexes, but the hydrogen bond between Lys183 carbonyl and Arg371 Nη2 is found only in one complex, whereas a salt bridge between Glu148 and Lys489 is specific to the other.
Direct interaction between Kae1p and Bud32p is required for the biological functions of the EKC/KEOPS complex in yeast
The large interaction surface between MjBud32 and MjKae1, together with the strong sequence conservation between the archaeal and yeast proteins (51 and 43% sequence identity for Kae1 and Bud32, respectively) imply that yeast Bud32p and Kae1p also interact directly. This interaction might be required for yeast viability and for the known functions of EKC/KEOPS in transcription and telomere maintenance. The residues involved in the MjKae1/Bud32 interface are well conserved (Figure 3A, Supplementary Figure S2 and S3). Although the sequence of the yeast Kae1p orthologue contains a few insertions compared with that of MjKae1, these should not interfere with complex formation (Supplementary Figures S6B and S6C). From these observations, we deduce that the structure of MJ1130 might serve as a good model for the EKC/KEOPS Kae1p/Bud32p subcomplex and should allow a deeper characterization of the biological function of this complex in eukaryotes. To validate our hypothesis, we have mutated residues that mediate ionic interactions between the Bud32p and Kae1p in yeast Bud32p and Kae1p so as to disrupt their interface. We tested mutations in two interaction patches of Bud32p and analysed their effect on transcription and telomere maintenance. In all cases, we introduced charge inversions (i.e., Asp/Glu to Arg or Arg/Lys to Glu), as we speculated that these would be the most disruptive for complex formation while maintaining the correct fold of the proteins. We further used the yeast amino-acid numbering for this discussion (the homologous MJ1130 numbering is given in italics). We first targeted the ionic interaction between Kae1p–Asp236 (Asp196) and Bud32p–Arg204 (Lys489). The second targeted patch is made of an intricate network of ionic interactions (Figure 3D and E) involving Lys57, Arg60 and possibly Arg72 (Lys365, Arg368 and Arg380) from Bud32p and both Glu292 and Glu295 from Kae1p (Glu233 and Glu236).
The mutant versions of Kae1p and Bud32p were ectopically expressed in strains deleted for the corresponding genes. The growth rate and some EKC/KEOPS complex-associated phenotypes were assessed in mutants to evaluate the functionality of the complex. The status of the Kae1p/Bud32p interaction was directly assessed by immunoprecipitation, and the expression and stability of the mutant proteins were evaluated by western blot.
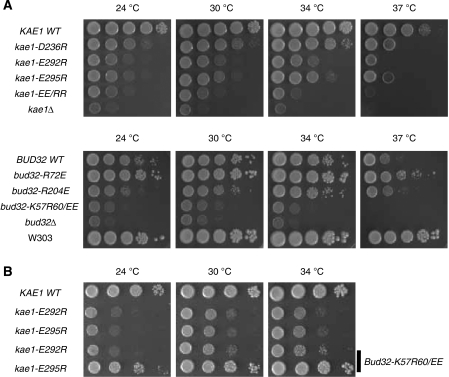
The results of growth complementation assays are shown in Figure 4. The kae1 gene was shown to be essential from a large-scale survey and as a consequence is annotated as essential in some databases; however, we confirm our previous results that this gene is not strictly essential, although kae1-null cells are very sick (Kisseleva-Romanova et al, 2006). As expected, ectopic expression of wild-type Kae1p fully complemented deletion of the kae1 gene. Mutation of either Glu292 or Glu295 (mutants kae1-E292R and kae1-E295R) was deleterious for growth at all temperatures, the kae1-E292R allele being more severe (although not completely inactivating; compare kae1-E292R with kae1Δ in Figure 4A, upper row panels). Introduction of both mutations in kae1-E292R,E295R cells almost completely inactivated Kae1p (and presumably EKC/KEOPS) function, as growth was comparable with the strongly affected null allele. Mutation of Asp236 (kae1-D236R) had a weaker effect, although growth was clearly affected at all temperatures and not supported at 37 °C. These phenotypes are not due to protein instability, as mutant proteins have similar stability as wild-type Kae1p (see below).
Figure 4.
Growth defect of mutations in yeast that were designed to disrupt the interaction between Bud32p and Kae1p. (A) Mutations that are predicted to affect the Kae1p/Bud32p interaction do not support wild-type growth. Growth at different temperatures of kae1Δ cells ectopically expressing wt or mutant kae1 (upper panel) or bud32Δ cells ectopically expressing wt or mutant bud32 (lower panel). Tenfold serial dilutions were spotted on YPDA and incubated for 3 days at the indicated temperatures. Note that bud32Δ/pCenbud32 cells grow less well than wild-type yeast (compare bud32 wt row to W303), as ectopic expression of wt Bud32p is toxic. Toxicity is partially lost in mutant bud32-R72E that expresses a partially defective Bud32p. (B) Compensation of the Kae1p E292R and E295R charge inversion mutants. Compensations were assayed by expressing the bud32-K57E,R60E allele in kae1Δ cells ectopically expressing wt or mutant kae1 (kae1-E292R, kae1-E295R) that also expressed endogenous Bud32p. Tenfold serial dilutions were spotted on YPDA and incubated for 3 days at the indicated temperatures. A strong compensation was observed for the E295R mutation in Kae1p.
Mutation of cognate Bud32p residues in the interaction patches overall confirmed the above data, although the analysis was somewhat complicated by the observation that ectopic expression of wild-type Bud32p results in a moderate growth phenotype that is exacerbated at 37 °C (Figure 4A, lower row panels, compare ectopic expression, bud32 WT row, with the wild-type expressed protein, W303 row). This is likely due to a toxic effect of the moderate overexpression of Bud32p. In these conditions, it is not possible to unambiguously conclude on the effect of mutating Arg72 (facing Glu292 from Kae1p), as the bud32-R72E mutant grows better than the ectopically expressed wt protein. We surmise that the compromised Bud32p–R72E mutant is less toxic than the wt protein when overexpressed, possibly due to a defective Kae1p/Bud32p interaction. However, mutation of the other two Bud32p residues in the same interaction patch (Lys57 and Arg60, bud32-K57E,R60E; Figure 4A, lower row panels) is detrimental to growth at all temperatures to similar levels as the bud32Δ mutant, which mirrored well the effect of mutating the opposing Glu292 and Glu295 in Kae1p. Similarly, mutation of Arg204 in Bud32p (bud32-R204E) had a similar effect on growth as the mutation of Asp236 in Kae1p with whom it interacts (kae1-D236R), indicating that alteration of the second interaction patch both on the Kae1p and Bud32p side also affects yeast growth, although to a lesser extent.
To substantiate these results, we attempted compensation of the Kae1p E292R and E295R charge inversion mutants by expressing the bud32-K57E,R60E allele in kae1-E292R and kae1-E295R cells. As shown in Figure 4B, the growth phenotype of kae1-E292R cells was only weakly suppressed by the presence of the Bud32-K57E,R60E. However, a strong suppression was observed for the E295R mutation in Kae1p, indicating that the presence of opposite charges in the facing Bud32 residues restored the interaction and virtually normal growth.
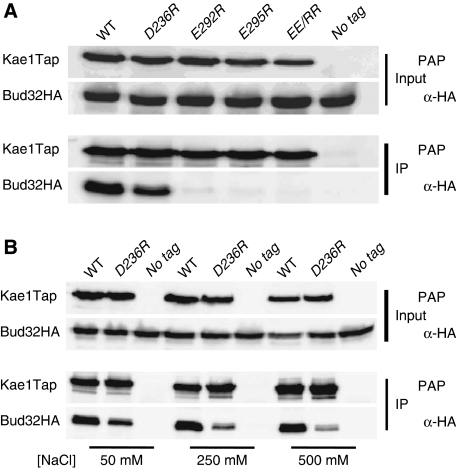
To confirm more directly that our mutations affected the Kae1p/Bud32p interaction, we analysed the ability of Kae1p mutants to bind Bud32p in vivo. We used yeast cells that harbour plasmid-borne, TAP-tagged versions of Kae1p (wt or mutants) and express chromosomally HA-tagged Bud32p (Bud32p–HA). Total protein extracts prepared from these strains were used for co-immunoprecipitation experiments. Note that by this assay we assessed the ability of mutant proteins to compete with endogenous Kae1p for Bud32p–HA association. Addition of the TAP tag does not significantly alter Kae1p function, as the tagged proteins behaved similarly to the non-tagged versions in complementation assays (data not shown). In vivo association of Bud32p–HA and Kae1p–TAP proteins strictly mirrored the growth proficiency of mutants. Mutation of Glu292, Glu295 or both impeded Bud32p–HA pull down by TAP-tagged Kae1p (Figure 5A). Mutant Kae1p–D236R was still able to bind Bud32p–HA, although to a lesser extent compared with wt Kae1p–TAP. This interaction was, however, salt sensitive, as it was almost completely lost when immunoprecipitation was performed at 500 mM salt concentration (Figure 5B).
Figure 5.
Mutations at the Kae1p/Bud32p interface affect the association of the two proteins in vivo. (A) Wild-type or mutant Kae1p–TAP (as indicated) were used to immunoprecipitate the complex from extracts expressing Bud32p–HA. Lanes are labelled according to the mutation introduced in Kae1p–TAP (e.g., D236R corresponds to the Kae1p–D236R mutant). Input (10% of the amount used in the immunoprecipitation reaction) or immunoprecipitate (IP) were probed by western blotting using peroxydase-anti-peroxydase complex (PAP) to reveal the Kae1p–TAP or anti-HA antibody as indicated. No tag: untagged Kae1p. (B) Salt sensitivity of the Bud32p/Kae1p–D236R interaction was probed by performing the immunoprecipitation reaction at different salt concentrations as indicated. No tag: untagged Kae1p.
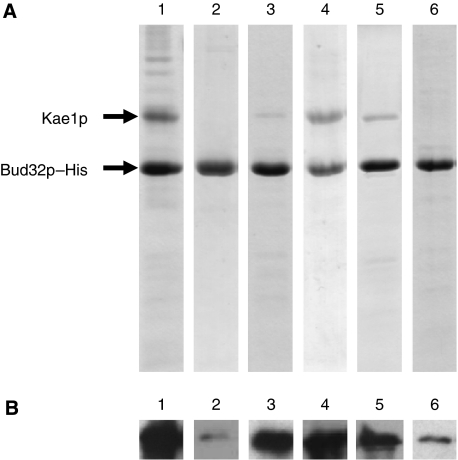
To confirm the results obtained in vivo with the mutants of yeast Kae1p and Bud32p, we decided to co-express and purify the two proteins using a polycistronic vector on which only Bud32p has been His-tagged (Bud32p–His). The SDS–PAGE analysis of the IMAC elution profile (Figure 6, lane 1) shows that wt Kae1p and Bud32p–His co-purify. Analytical gel filtration further proved that wt Kae1p and Bud32p–His form a 1:1 complex (data not shown). To analyse the interactions between wt Kae1p and Bud32p–His, we co-expressed the wt Kae1p with Bud32p–K57E,Y59E,R60E, Bud32p–R72E and Bud32p–R204E, and wt Bud32p with the Kae1p–D236R and Kae1p–E292R. Western blot analysis using antibodies raised against Kae1p demonstrated that all Kae1p mutants are expressed in soluble form (data not shown). Mutation Bud32p–R204E does not seem to affect significantly its interaction with wt Kae1p (Figure 6, lane 4). However, all other interface mutations significantly affect the amount of Kae1p that co-purifies with Bud32p–His (Figure 6, lanes 2, 3, 5 and 6). The Coommassie blue staining and western blot intensities suggest that Bud32p–K57E,Y59E,R60E and Kae1p–E292R are the two mutants for which the interaction with the wild-type partner is drastically affected (see lanes 2 and 6, respectively, of Figure 6). This observation is in good agreement with the in vivo and functional results obtained in yeast where we can show that the more destabilizing effect of the interaction is observed for the Bud32p–K57E,R60E and Kae1p–E292R mutants. These results obtained using recombinant yeast Kae1p and Bud32p mutants expressed in Escherichia coli confirm that the altered interaction observed between mutants from Kae1p and Bud32p in yeast is due to the disruption of direct physical interactions between both proteins and not due to the destabilization of interactions with other partners of the EKC/KEOPS complex. Together, these results are consistent with the notion that yeast Bud32p and Kae1p interact directly and that this interaction is required for the growth-limiting function of the EKC/KEOPS complex. Our data indicate a more prominent function of the interaction patch opposing Glu292/Glu295 in Kae1p to Lys57/Arg60 (and possibly Arg72) in Bud32p. The second patch (Asp236 in Kae1p and Arg204 in Bud32p) is not essential and contributes to a lesser extent to the interaction.
Figure 6.
Mutations in the Kae1p/Bud32p interaction region affect their direct interaction. (A) Coomassie staining SDS–PAGE of eluted fractions of WT or mutants of Kae1p and Bud32p–His purified on NiIDA. Only the 100 mM imidazole elution fractions are shown. (B) Western blot probed against Kae1p (same fractions and same volumes loaded as in A). Lane 1, wild-type Bud32p–His and Kae1p; lane 2, Bud32p–His–K57E,Y59E,R60E and wild-type Kae1p; lane 3, Bud32p–His–R72E and wild-type Kae1; lane 4, Bud32p–His–R204E and wild-type Kae1p; lane 5, Kae1p–D236R and wild-type Bud32p–His; lane 6, Kae1p–E292R and wild-type Bud32p–His.
Altogether, these elements strongly support that the structure of the MjKae1/Bud32 fusion protein can be considered as an accurate model of the Kae1p/Bud32p subcomplex of EKC/KEOPS.
The interaction between Bud32p and Kae1p is required for both the transcription and the telomere maintenance function of the EKC/KEOPS complex in yeast
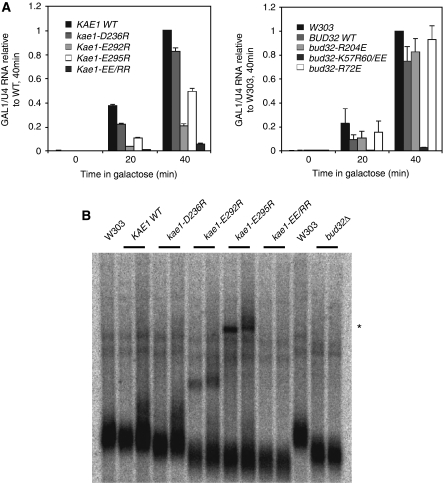
The EKC/KEOPS has been shown to have an important function in transcription and to be required for telomere maintenance. We therefore investigated whether the Kae1p/Bud32p interaction is also required for one or both functions. To this end, we first monitored the activation kinetics of the GAL1 gene after exposure of cells to galactose-containing medium by real-time RT–PCR. The galactose regulon has been shown to depend on the integrity of the EKC/KEOPS for full expression (Kisseleva-Romanova et al, 2006). As shown in Figure 7A, disruption of the Kae1p/Bud32p interaction correlated well with defective GAL1 induction. The mutants displaying the strongest defect in transcription are the ones that more efficiently disrupt the Kae1p/Bud32p interaction (i.e., mutation of Glu292 and Glu295 in Kae1p and that of Lys57 and Arg60 in Bud32p). Mutants that have a more modest effect on the interaction and growth rate affect transcription only to a limited extent (e.g., kae1-D236R) or behave as the wild type (e.g., bud32-R204E). Note that ectopic expression of wild-type Bud32p also slightly affects GAL1 expression, which mirrors the moderate growth defect of this strain compared with bona fide wild-type cells (Figure 7A).
Figure 7.
The association of Kae1p and Bud32p is required both for transcription and telomere homeostasis. (A) Real-time RT–PCR analysis of GAL1 mRNA production in kae1 (left panel) and bud32 (right panel) mutant strains. Cells were exponentially grown in raffinose-containing medium and shifted to galactose for the indicated times. For a better visual appraisal of the differences, GAL1 mRNA signals (normalized to U4 snRNA) are expressed relative to the levels attained after 40 min induction in wild-type strains. In the right panel, the strain expressing endogenous Bud32p was used as a reference strain to evaluate the effect of moderate Bud32p overexpression on GAL1 transcription. (B) Southern blot analysis of telomere length in Kae1p mutants. Genomic DNAs from two independent clones of the indicated strains were digested with Xho1 that cuts within a repeated region present on most telomeres. The telomeric fragments were separated by gel electrophoresis and detected with specific telomeric probes. The bud32Δ strain was loaded as control for shortened telomeres. The band marked with * is of unknown origin.
We also studied the effects of Kae1p mutations in telomere maintenance. It was shown that inactivation of EKC/KEOPS components induces telomere shortening (Downey et al, 2006). As shown in Figure 7B, telomeres are shorter in mutants that strongly affect the Kae1p/Bud32p interaction (i.e., kae1-E292R, kae1-E295R and kae1-E292R,E295R). On the other hand, telomeres of kae1-D236R cells are only marginally smaller, which parallels a moderate effect on the Kae1p/Bud32p interaction.
From these data, we conclude that the interaction between Kae1p and Bud32p is required for both the transcription and the telomere homeostasis function of the EKC/KEOPS.
Discussion
The recently discovered EKC/KEOPS complex has been shown to partake in transcription and telomere homeostasis in yeast. The EKC/KEOPS yeast complex consists of five proteins: Bud32p, Kae1p, Gon7p, Cgi121p and Pcc1p. All these proteins (except Gon7p, which is fungi specific) have homologues in all eukaryotic genomes, suggesting that EKC/KEOPS carries important and conserved functions in the eukaryotic domain. This hypothesis is also supported by functional complementation between the yeast and human proteins (Facchin et al, 2003). However, understanding the molecular mechanisms that underlie these functions remains a major challenge.
Kae1p and Bud32p are the only proteins of the complex whose sequences and structures carry putative functional signatures that can be connected to enzymatic activities. Kae1p and Bud32p are also the only two EKC/KEOPS proteins with clear-cut homologues in other cellular domains. Kae1 is a universal protein, present in all completely sequenced genomes, whereas the kinase Bud32 has no bacterial homologues but is present in all archaea (Galperin and Koonin, 2004; Hecker et al, 2007). Interestingly, the juxtaposition or fusion of KAE1 and BUD32 into a single gene in most archaeal genomes suggests a close functional relationship conserved between archaea and eukaryotes and extending beyond the physical connection.
The interaction between Kae1p and Bud32p was first demonstrated in yeast by two-hybrid studies (Ho et al, 2002; Lopreiato et al, 2004). Their presence in the multiprotein EKC/KEOPS complex was detected both by large-scale studies and directed biochemical analysis (Gavin et al, 2002; Downey et al, 2006; Kisseleva-Romanova et al, 2006). Direct interaction between the two proteins and its requirement for the function of EKC/KEOPS had, however, not been demonstrated. In this study, we describe the structure of the M. jannaschii MJ1130 protein, whose N- and C-terminal halves are homologous to yeast Kae1p and Bud32p, respectively. This provides the first structure of a Bud32p orthologue. The Kae1 and Bud32 moieties of MJ1130 interact through a large and conserved surface area. Using the MJ1130 structure as a model, we designed mutations that disrupt the Kae1p/Bud32p interface in the orthologous yeast complex. In vivo and in vitro analysis of mutants demonstrated that the integrity of the Kae1p/Bud32p interaction (and presumably of the EKC/KEOPS complex) is crucial for its function of telomere maintenance and transcription in yeast.
Bud32p is characterized by its small size and low sequence similarity compared with other members of the protein kinase family. It was demonstrated that recombinant yeast Bud32p is able to autophosphorylate and to phosphorylate casein (Stocchetto et al, 1997). The human homologue PRKK also has kinase activity (Abe et al, 2001), strongly suggesting that the Bud32p protein family encodes for catalytically active kinases. The importance for activity of some of the catalytic and nucleotide binding residues was sampled through site-directed mutagenesis (Facchin et al, 2002), and their effects are confirmed by their location in the ATP-binding pocket of the MjBud32 structure. We show here that the in vitro kinase activity of Bud32p is strongly inhibited by Kae1p. On the other hand, the structure of the MjKae1/Bud32 complex revealed that the P-loop of MjBud32 is in an inactive conformation and that no AMPPNP is bound. As the MjBud32 ATP-binding site is not sterically blocked by MjKae1, the inhibitory effect of MjKae1 is probably due to conformational changes induced upon complex formation with MjBud32. The binding of physiological activators or inhibitors to protein kinases very often modifies their 3D structure. For instance, the p16INK4a tumour suppressor binds to both terminal lobes of cyclin-dependent kinase 6, close to the ATP-binding cleft (Russo et al, 1998). This binding induces a relative reorientation of the N- and C-terminal lobes of Cdk6, indirectly preventing cyclin binding. This movement also distorts the active site cleft, interfering with ATP binding.
Comparison of the two copies of the complexes in the asymmetric unit showed that the MjKae1 and Bud32 have slightly different relative orientations. We therefore suspect that the detailed structure of the interface between Bud32 and Kae1 may be dynamic and influenced by the binding of protein factors (the other EKC/KEOPS partners in yeast for instance). The phosphorylation state of Bud32 and/or Kae1, or still, the occupation of the nucleotide-binding pocket of Bud32 and/or Kae1 may all be factors that influence their interaction and conformation. The active site conformation and kinase activity of Bud32 may be steered by protein–protein interactions. Our previously determined structure of PaKae1 in complex with AMPPNP is virtually the same as MjKae1 in complex with MjBud32 (Hecker et al, 2007), and therefore Bud32 does not induce substantial conformational rearrangements in Kae1.
Although the EKC/KEOPS complex has been implicated in both transcription and telomere homeostasis, it is still unclear whether the complex functions in two distinct pathways or whether a single activity affects both processes in a direct or indirect manner. The mechanism of action of the complex and its enzymatic activities are still a matter of speculation, but the findings reported here underscore the essential requirement for the interaction of Bud32p and Kae1p for the function of this complex. Phosphorylation of specific substrates by Bud32p might alter the structure of chromatin and therefore may impact both transcription and telomere maintenance. The possibility exists that the bifunctional archaeal protein is the prototypical functional unit of the complex that is reconstituted in eukaryotes by the physical interaction of the separated polypeptides. In this perspective, the Kae1p/Bud32p complex as a whole might carry the essential activity of the EKC/KEOPS. In all of these scenarios, the intimate connection of the two proteins contributes some ‘added value' for full function, which is consistent with the outcome of our experiments.
The conservation of Kae1p/Bud32p raises several questions when the function of the EKC/KEOPS is considered in an evolutionary perspective. In fact, the absence of telomeres and the different structure of chromatin in archaea with respect to eukaryotes seemingly suggests differences in the function of the complex in the two kingdoms. This might reflect a divergent evolution from a prototypical function, possibly accompanied by the appearance of eukaryotic-specific ancillary subunits such as Gon7p, Cgi-121 and Pcc1p. Alternatively, both the archaeal and eukaryotic EKC/KEOPS have a similar primary function that only impacts telomere metabolism (and possibly chromatin remodelling/transcription) in eukaryotes. Further progress in the characterization of the biochemical activities of the complex will be required to shed light on the function and mechanism of action of these important factors.
In conclusion, we have clearly shown here that the direct interaction between Kae1p and Bud32p is required for both the transcription and the telomere homeostasis function of the EKC/KEOPS. We were guided by the present structure of the archaeal fusion protein to study the effect of interface mutations in the context of the yeast complex. The fact that these mutations have dramatic phenotypic effects, similar to those observed with deletion mutations of the respective components, strongly suggests that MJ1130 is a good model for the EKC/KEOPS subcomplex and should be a very useful tool for a more detailed study of the function of EKC/KEOPS complex in eukaryotes.
Materials and methods
Crystallization and resolution of the structure
Cloning, recombinant expression and purification of all archaeal and yeast constructs are reported as Supplementary data. SeMet-labelled protein was prepared using standard protocols and stored in 20 mM Tris–HCl, pH 8, and 200 mM NaCl. Crystals were grown at 19 °C from a 2:0.5 μl mixture of a 15 mg/ml (0.24 mM) protein solution incubated previously with 2.4 mM AMPPNP with 0.1 M sodium citrate, 5–10% isopropanol and 0.1 M sodium citrate, pH 5.6. For data collection, the crystals were transferred into a cryoprotectant crystallization solution with progressively higher glycerol concentrations up to 30% v/v. The diffraction data were recorded at the Se-edge on beam line ID14-EH4 (ESRF, Grenoble, France).
The structure was determined by the SAD method using the anomalous signal from the selenium element. Data were processed using the MOSFLM package (Leslie, 1992). The space group was P21212 with two molecules per asymmetric unit. All the expected Se sites (15 per protein) were found with the program SHELXD in the 50-4 Å resolution range (Schneider and Sheldrick, 2002). Refinement of the Se atom positions, phasing and density modification were performed with the program SHARP (Bricogne et al, 2003). Two copies of the N-terminal domain of the protein (residues 1–323) were positioned by molecular replacement using the coordinates of the P. abyssi Pab1159 crystal structure (Hecker et al, 2007) with the program MOLREP (Vagin and Isupov, 2001). The C-terminal domain (residues 340–530) was manually built into the experimental electron density map generated by SHARP at 3.2 Å resolution. This model was then refined against the 3.05 Å data set using REFMAC (Murshudov et al, 1997) and rebuild with the ‘TURBO' molecular modelling program (http://www.afmb.univ-mrs.fr/rubrique113.html).
The final model contains residues 1–31 and 43–530 from monomer A and residues 1–31, 43–325 and 340–532 from monomer B as well as AMPPNP bound to the N-terminal domain from both copies. With the exception of Ala 10 from monomer A, all residues display main-chain dihedral angles that fall within allowed regions of the Ramachandran plot as defined by the program PROCHECK (Laskowski et al, 1993). Statistics for all the data collections and refinement of the different structures are summarized in Table I. The atomic coordinates and structure factors have been deposited into the Brookhaven Protein Data Bank under the accession number 2vwb.
Table 1.
Crystallographic data
| SeM | Native | |
|---|---|---|
| Resolution (Å) | 150–3.2 (3.37–3.2) | 20–3.05 (3.21–3.05) |
| Space group | P21212 | P21212 |
| Cell parameters (Å) | a=147.6; b=149.5; c=65.2 | a=147.4; b=148.9; c=65.1 |
| Total number of reflections | 97 167 | 132 557 |
| Total number of unique reflections | 24 853 | 26 424 |
| Rsym (%)a | 11.7 (62.8) | 9.9 (55.6) |
| Completeness (%) | 99.9 (100) | 94.7 (96.2) |
| I/σ(I) | 4.4 (1.1) | 12.4 (2.8) |
| Redundancy | 3.9 (4) | 5 (5.1) |
| Refinement | ||
| Resolution (Å) | 20–3.05 | |
| R/Rfree (%)b | 22/30.1 | |
| R.m.s.d. bonds (Å) | 0.011 | |
| R.m.s.d. angles (deg) | 1.478 | |
| Ramachandran plot | ||
| Most favoured (%) | 85 | |
| Allowed (%) | 15 | |
| PDB code | 2VWB | |
| Values in parentheses are for highest resolution shell. | ||
| aRsym=∑h∑i∣Ihi−〈Ih〉∣/∑h∑iIhi, where Ihi is the ith observation of the reflection h, whereas 〈Ih〉 is the mean intensity of reflection h. | ||
| bRfactor=∑∥Fo∣−∣Fc∥/∣Fo∣. Rfree was calculated with a small fraction (5%) of randomly selected reflections. | ||
Phosphotransferase assay
The phosphotransferase activity of yeast Bud32p was essayed with either recombinant wild-type or D161A mutant His-tagged protein on Kae1p, Grx4p or dephosphorylated casein as substrates (Lopreiato et al, 2004). Incubations were performed at 30 °C for 15 min in 20 μl of buffer containing 20 mM Tris–HCl, 100 mM NaCl, 1 mM DTT, 10 mM MnCl2 and 25 μM [γ-32P]ATP (1 μCi per reaction, specific activity of 6000 Ci/mmol). After incubation, reactions were stopped by the addition of loading buffer and the samples were analysed by electrophoresis (SDS–PAGE). The gels were then soaked in 16% TCA for 10 min at 90 °C and neutralized in 100 mM phosphate buffer pH 7.4. This procedure eliminates most unincorporated [32P], non-protein material, phosphorylation on His and Asp residues and enriches samples for O-phosphorylated proteins (Levine et al, 2006). The treated gels were finally stained in neutralization buffer supplemented with 5% TCA and 0.1% Coomassie blue and radiolabelled proteins visualized by autoradiography.
Yeast strains construction and manipulation genetics
Yeast strains used in this study are listed in Supplementary Table S2 and are derived from W303-1A (Thomas and Rothstein, 1989). Yeast cultures and genetic manipulations have been performed accordingly to standard methods (Adams et al, 1997; Longtine et al, 1998). Site-directed mutagenesis of Kae1p and Bud32p sequences has been performed by standard PCR-based strategy and extensive sequencing was performed to verify that unwanted changes were not introduced. Wild-type or mutant genes were cloned in pCM185 (Gari et al, 1997) by homologous recombination directly in yeast cells. For immunoprecipitation experiments, Tap-tagged version of these proteins (C-terminal) were also constructed and cloned in pCM185 vectors. Strains ectopically expressing the modified proteins were generated after transformation of the appropriate shuffle strains and 5-FOA selection (Boeke et al, 1984). Growth at different temperatures was assayed by spotting serial dilutions on YPDA plates.
Co-immunoprecipitation and western blotting
See Supplementary data.
RNA analysis
Yeast cells were grown to OD600 0.4 in Raffinose-containing medium and then shifted to galactose-containing medium to activate the GAL regulon. Samples were taken after 20 and 40 min of induction and RNAs have been prepared as described previously (Kisseleva-Romanova et al, 2006). GAL1 RNAs were analysed by real-time RT–PCR (Light-Cycler, Roche) with primers 5′-CTGCAAATGTTTTAGCTGCCACGTA-3′ and 5′-CATCTTTGTTAACCGTTCGATGCC-3′. Primers 5′-ATCCTTATGCACGGGAAA-3′ and 5′-CACCGAATTGACCATGAG-3′ were used to amplify U4snRNA. Amplification efficiencies were calculated from serial dilutions for every set of amplification reactions. RNA levels were normalized to the levels of U4 snRNA.
Southern blot analysis of telomere length
Yeast strains were grown in YPDA to exponential phase (OD600 0.8–1.0) and cells were collected. Genomic DNAs have been purified from yeast cells with standard methods and approximately 50 μg were digested with XhoI and separated on 1.2% agarose gel. After transfer onto nitrocellulose membrane, filters have been hybridized with a specific probe (telomeric Y′-TG1–3 repeats) as described (Schramke et al, 2004).
Supplementary Material
Supplementary Material
Acknowledgments
We are indebted to N Ulryck, Dr P Dorlet and Dr N Leulliot for their technical assistance. This work was supported by the Centre National de la Recherche Scientifique, by the Human Frontier Science Program (HFSP), the Association pour la Recherche sur le Cancer (ARC), la Ligue contre le Cancer, the Agence Nationale de la Recherche (ANR) and by the European 3D-repertoire program (LSHG-CT-2005-512028). RL was supported by a fellowship from the Association pour la Recherche sur le Cancer (ARC).
References
- Abe Y, Matsumoto S, Wei S, Nezu K, Miyoshi A, Kito K, Ueda N, Shigemoto K, Hitsumoto Y, Nikawa J, Enomoto Y (2001) Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J Biol Chem 276: 44003–44011 [DOI] [PubMed] [Google Scholar]
- Adams AG, Dawson DE, Kaiser CA, Stearns T (1997) Methods in Yeast Genetics. Cold Spring Harbor, NY: CSHL Press [Google Scholar]
- Aravind L, Koonin EV (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol 287: 1023–1040 [DOI] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197: 345–346 [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Buss KA, Cooper DR, Ingram-Smith C, Ferry JG, Sanders DA, Hasson MS (2001) Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J Bacteriol 183: 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, Emili A, Greenblatt JF, Harrington L, Lydall D, Durocher D (2006) A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168 [DOI] [PubMed] [Google Scholar]
- Facchin S, Lopreiato R, Ruzzene M, Marin O, Sartori G, Gotz C, Montenarh M, Carignani G, Pinna LA (2003) Functional homology between yeast piD261/Bud32 and human PRPK: both phosphorylate p53 and PRPK partially complements piD261/Bud32 deficiency. FEBS Lett 549: 63–66 [DOI] [PubMed] [Google Scholar]
- Facchin S, Lopreiato R, Stocchetto S, Arrigoni G, Cesaro L, Marin O, Carignani G, Pinna LA (2002) Structure-function analysis of yeast piD261/Bud32, an atypical protein kinase essential for normal cell life. Biochem J 364: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV (2004) ‘Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res 32: 5452–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, Dorlet P, Brochier C, Quevillon-Cheruel S, Le Cam E, van Tilbeurgh H, Forterre P (2007) An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res 35: 6042–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hurley JH (1996) The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct 25: 137–162 [DOI] [PubMed] [Google Scholar]
- Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, Hofmann K, Namane A, Mann C, Libri D (2006) Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J 25: 3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laronde-Leblanc N, Guszczynski T, Copeland T, Wlodawer A (2005) Structure and activity of the atypical serine kinase Rio1. FEBS J 272: 3698–3713 [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A (2005) The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim Biophys Acta 1754: 14–24 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Leslie A (1992) Joint CCP4 and EACMB Newsletter Protein Crystallography. Warrington, UK: Daresbury Laboratory [Google Scholar]
- Levine A, Vannier F, Absalon C, Kuhn L, Jackson P, Scrivener E, Labas V, Vinh J, Courtney P, Garin J, Seror SJ (2006) Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6: 2157–2173 [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein–protein recognition sites. J Mol Biol 285: 2177–2198 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lopreiato R, Facchin S, Sartori G, Arrigoni G, Casonato S, Ruzzene M, Pinna LA, Carignani G (2004) Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin. Biochem J 377: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D (1999) Detecting protein function and protein-protein interactions from genome sequences. Science 285: 751–753 [DOI] [PubMed] [Google Scholar]
- Mellors A, Lo RY (1995) O-sialoglycoprotease from Pasteurella haemolytica. Methods Enzymol 248: 728–740 [DOI] [PubMed] [Google Scholar]
- Murshudov G, Vagin A, Dodson E (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP (1998) Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395: 237–243 [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Geli V (2004) RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet 36: 46–54 [DOI] [PubMed] [Google Scholar]
- Stocchetto S, Marin O, Carignani G, Pinna LA (1997) Biochemical evidence that Saccharomyces cerevisiae YGR262c gene, required for normal growth, encodes a novel Ser/Thr-specific protein kinase. FEBS Lett 414: 171–175 [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Vagin AA, Isupov MN (2001) Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr 57: 1451–1456 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material