Abstract
The budding yeast Cdc13, Stn1 and Ten1 (CST) proteins are proposed to function as an RPA-like complex at telomeres that protects (‘caps') chromosome ends and regulates their elongation by telomerase. We show that Stn1 has a critical function in both processes through the deployment of two separable domains. The N terminus of Stn1 interacts with Ten1 and carries out its essential capping function. The C terminus of Stn1 binds both Cdc13 and Pol12, and we present genetic data indicating that the Stn1–Cdc13 interaction is required to limit continuous telomerase action. Stn1 telomere association, similar to that of Cdc13, peaks during S phase. Significantly, the magnitude of Stn1 telomere binding is independent of telomere TG tract length, suggesting that the negative effect of Stn1 on telomerase action might be regulated by a modification of CST activity or structure in cis at individual telomeres. Genetic analysis suggests that the Tel1 kinase exerts an effect in parallel with the Stn1 C terminus to counteract its inhibition of telomerase. These data provide new insights into the coordination of telomere capping and telomerase regulation.
Keywords: CST complex, Stn1, telomerase, telomere capping, telomere length regulation
Introduction
Telomeres in the budding yeast Saccharomyces cerevisiae, and nearly all other eukaryotes examined to date, comprise short repeated sequences (TG1–3 in yeast and T2AG3 in metazoans) that serve as a platform for the binding of proteins required for their replication and protection from degradation or end joining (reviewed in Verdun and Karlseder, 2007). The TG-rich DNA strand of telomeres, which forms the 3′ chromosome terminus, exists as a short single-stranded (ss) extension whose length increases during S phase through a regulated process of 5′ end resection of the CA-rich strand (reviewed in Chakhparonian and Wellinger, 2003). This 3′ TG-rich overhang serves as a binding site for the Cdc13 protein, first identified for its essential role in telomere protection, or ‘capping' (Garvik et al, 1995). In the absence of Cdc13 function, telomeres undergo extensive degradation, leading to a checkpoint-dependent cell cycle arrest in G2/M (Weinert and Hartwell, 1988; Lydall and Weinert, 1995). This essential capping function of Cdc13 appears to be carried out together with two other proteins, Stn1 and Ten1, both of which interact with Cdc13, as temperature-sensitive (Ts) lethal mutations in either gene also lead to telomere uncapping and a G2/M cell cycle arrest (Grandin et al, 1997, 2000, 2001). It is thus presumed that Cdc13, Stn1 and Ten1 act together in a complex (CST) that caps telomere ends.
Genetic analysis of CDC13 has revealed a complex role for Cdc13 protein in telomere maintenance. Cdc13 has a direct function in the recruitment of telomerase, through an interaction with the accessory factor Est1 (Evans and Lundblad, 1999; Pennock et al, 2001; Bianchi et al, 2004), that can be genetically separated from its capping function (Nugent et al, 1996). As such, cells carrying a cdc13-2 mutation undergo progressive telomere shortening and senescence, but their telomeres are not initially uncapped. The capping function of Cdc13 instead appears to be carried out by Stn1 (presumably together with Ten1), because fusion of the Cdc13 DNA-binding domain to Stn1 is sufficient for capping (Pennock et al, 2001). In addition to its essential role as a positive effector of telomerase function, Cdc13 is also implicated in negative regulation of telomere elongation, as removal of a C-terminal domain of the protein leads to extensive telomere elongation (Chandra et al, 2001).
The precise role of Stn1 in telomere maintenance, though less clear than that of Cdc13, also appears to be complex. As mentioned above, genetic and hybrid protein studies clearly indicate a role for Stn1 in telomere capping, probably carried out through direct interactions with Cdc13. However, a recent study shows that Stn1, when overexpressed, can provide a capping function in the absence of Cdc13 (Petreaca et al, 2006). STN1 mutants also display a telomere elongation phenotype (Grandin et al, 1997), and Stn1 protein has been proposed to negatively regulate telomerase action by competing with Est1 for binding to Cdc13 (Chandra et al, 2001). Interestingly, Stn1 also interacts with the Pol12 subunit of the Pol α–primase complex (Grossi et al, 2004), itself implicated in telomere length regulation and capping (Carson and Hartwell, 1985; Adams Martin et al, 2000; Qi and Zakian, 2000). Finally, studies in the related yeast Kluyveromyces lactis implicate Stn1 in the repression of recombination-based telomere tract elongation and rapid deletion (Iyer et al, 2005).
Here, we report the results of a series of experiments designed to dissect the role of Stn1 in both telomere length regulation and capping. We begin by defining regions and specific residues of Stn1 required for interactions with Ten1, Cdc13 and Pol12, which are then correlated with specific telomere functions. To address in molecular detail the role of Stn1 in telomere length regulation, we employ chromatin immunoprecipitation (ChIP) of Stn1 itself, or of Cdc13 and Est1, in cells carrying STN1 mutations. Finally, we use genetic epistasis analysis to explore the relationship between Stn1 and other regulators of the telomerase pathway, in particular the MRX (Mre11, Rad50 and Xrs2) complex and Tel1 (the yeast ATM homologue). Our results are discussed in terms of recent findings concerning the TG tract length-dependent regulation of telomerase action.
Results
Two functionally distinct interaction domains in Stn1
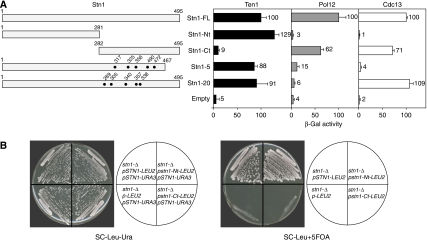
Two-hybrid analysis has shown previously that Stn1 interacts with both Cdc13 and Ten1 (Grandin et al, 1997, 2001; Chandra et al, 2001), two other proteins required for telomere capping, as well as with Pol12, the B subunit of the DNA polymerase α–DNA primase complex (Grossi et al, 2004). To determine whether Stn1 interacts with these three proteins through different or common regions, we generated two Stn1 hybrids with the Gal4 activation domain (GAD). GAD–stn1-Nt expresses the first 281 amino acids of Stn1 fused to GAD, whereas GAD–Stn1-Ct expresses a C-terminal Stn1 fragment (amino acids 282–495). These Stn1 hybrids were tested against two full-length Gal4 DNA-binding domain (GBD) fusions (GBD–Cdc13 and GBD–Ten1), and a fusion of LexA to an N-terminal domain of Pol12 (LexA–Pol12-Nt), in the appropriate two-hybrid reporter strains.
We found that GAD–stn1-Nt failed to interact with either GBD–Cdc13 or LexA–pol12-Nt, but displayed a strong interaction with GBD–Ten1, comparable to that of full-length GAD–Stn1 (Figure 1A). Conversely, GAD–stn1-Ct interacted with both GBD–Cdc13 and LexA–pol12-Nt to levels near that of full-length GAD–Stn1, but barely at all with GBD–Ten1 (Figure 1A). These data suggest that the C-terminal part of Stn1 (amino acids 282–495) is largely responsible for the Stn1 interaction with both Cdc13 and Pol12, whereas an N-terminal domain (1–281) binds to Ten1. To determine whether the stn1-Ct interaction with Pol12 and Cdc13 can be distinguished genetically, we mutagenized STN1 and used the two-hybrid assay to screen for mutations defective in the LexA–pol12-Nt interaction. This allowed us to identify one allele of STN1 (stn1-5) that displays a severely reduced interaction with both LexA–pol12-Nt and GBD–Cdc13. Interestingly, a second allele (stn1-20) fails to interact with LexA–pol12-Nt, but interacts normally with GBD–Cdc13 (Figure 1A). The functional consequences of these mutations will be described below.
Figure 1.
Identification of two interaction domains in Stn1. (A) Stn1 fusions to the Gal4 activation domain (GAD) are expressed from the pACT2 vector; Ten1 and Cdc13 fusions to the Gal4 DNA-binding domain are expressed from the pAS2 and pGBD vectors, respectively; a Pol12 fusion to LexA is expressed from the pLexA vector. All fusions are under the control of the ADH1 promoter. Data are the average of two independent β-galactosidase measurements normalized to stn1-FL, arbitrarily set to 100. (B) GAD/stn1-Nt rescues the lethality of an stn1-null strain. In the diagrams at the right of each plate photograph, the plasmids present in each strain are indicated (all strains carry a full deletion of the genomic STN1 gene). The names indicate both the STN1 allele and the genetic marker present in each plasmid (STN1-URA3=YCpLac33-STN1, STN1-LEU2=pACT2-STN1, stn1-Nt-LEU2=pGAD/stn1-Nt, stn1-Ct-LEU2=pGAD/stn1-Ct, empty=pGAD-LEU2; see Table II for details).
Essential capping function of the Stn1 N terminus
The specificity of interaction shown by the two different Stn1 domains in two-hybrid assays (Figure 1A) prompted us to determine whether the C and N termini of Stn1 execute different functions. To test this hypothesis, we expressed either GAD–stn1-Nt or GAD–stn1-Ct in a strain in which the endogenous copy of STN1 was deleted (stn1-Δ). This strain was kept alive by a centromeric plasmid containing STN1 and the URA3 gene (YCpLac33-STN1). After selecting for loss of YCpLac33-STN1 by growing cells on plates containing 5-FOA, we observed (Figure 1B) that GAD–stn1-Nt was able to rescue the lethality of a genomic stn1-Δ mutation, indicating that the N terminus of Stn1 (amino acids 1–281) is sufficient to carry out its essential function. Notably, GAD–stn1-Ct was unable to complement the lethality of the stn1-Δ mutation (Figure 1B). These data suggest that the stn1 interaction with Pol12 and Cdc13 is not necessary for cell viability, whereas its interaction with Ten1 is essential.
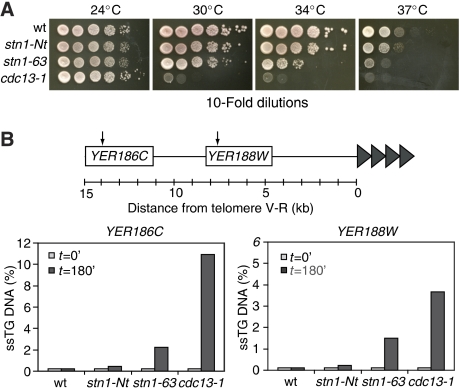
The observation that the Stn1 N terminus is indispensable for cell viability suggests that this portion of Stn1 carries out the essential function of telomere capping. To test this idea more directly, we generated a new STN1 allele, stn1-63. Charbonneau and colleagues have shown that the stn1-154 mutant displays a telomere elongation phenotype and a strong growth defect at 37°C, consistent with a Ts capping defect (Grandin et al, 2001). The stn1-154 allele possesses three different point mutations, one of which (D99E) maps to a highly conserved portion of the N terminus, whereas the other two map to a variable portion very near the C terminus (Supplementary Figure S1A). To test whether the D99E substitution is responsible for the Ts phenotype of stn1-154, we generated an allele of STN1 (stn1-63) containing only this point mutation in an otherwise wild-type STN1 gene. We found that this mutant allele (either on a centromeric plasmid or overexpressed as a GAD fusion from a multicopy plasmid) conferred a severe growth defect at elevated temperatures in an stn1-Δ strain, comparable to that of the stn1-154 allele (Figure 2A and data not shown). Furthermore, the stn1-63 and stn1-154 mutations display indistinguishable telomere elongation phenotypes at both 24°C and following a shift to 37°C (Supplementary Figure S1B).
Figure 2.
Temperature sensitivity and capping function of stn1 mutant alleles. (A) Strains were grown in YPAD and 10-fold serial dilutions were spotted on YPAD media at 23, 30, 34 and 37°C to test for viability. (B) Quantitative measurement of single-stranded DNA (QAOS) in the indicated (arrows) subtelomeric regions of wild-type, stn1-Nt, stn1-63 and cdc13-1 strains (YAP46, YAP42, YAP73 and DLY1108, respectively). Samples were collected at the permissive temperature (24°C; t=0) and after 180 min of growth at the non-permissive temperature (37°C; t=180).
To define more clearly the molecular defect caused by the stn1-63 mutation, and the roles of the two different Stn1 domains in telomere maintenance, we quantified the extent of CA strand erosion at a single telomere in stn1-Nt and stn1-63 strains, using an assay (QAOS; quantitative amplification of ss DNA) previously used to characterize the capping-defective Ts-lethal mutant cdc13-1 (Booth et al, 2001). This method provides a measure of ss DNA in a subtelomeric region (in this case, approximately 8 and 14 kb internal to the Chr. V-R telomere) and is thus a direct indicator of uncapping. The results in Figure 2B show that the stn1-Nt mutation had little or no influence on subtelomeric DNA structure, whereas the stn1-63 Ts allele, at the non-permissive temperature, caused a significant increase in ss DNA at the two internal sites at Chr. V-R. The amount of ss DNA generated in the stn1-63 mutant strain is less than that found in a cdc13-1 mutant, consistent with the fact that the latter strain is considerably more thermo-sensitive (Figure 2A).
To determine whether the Stn1 N terminus is sufficient for normal telomeric DNA and chromatin structure, we performed additional analysis of the stn1-Nt strain. Using a sensitive hybridization assay to detect telomeric ss TG-repeat DNA (i.e. ss DNA at the telomere itself), we indeed found elevated levels in the stn1-Nt strain compared with wild type (Supplementary Figure S2A). This ss DNA is largely terminal as hybridization is effectively eliminated by treatment with exonuclease I. Despite this alteration in terminal DNA structure, we found that the stn1-Nt strain (as well as stn1-63) displayed normal levels of subtelomeric gene silencing, or telomere position effect (TPE, see Supplementary Figure S2B). As TPE relies upon the duplex TG-repeat binding factor Rap1 for its establishment (Kyrion et al, 1993; Moretti et al, 1994), this finding supports the notion that the telomere structure defect in the stn1-Nt strain is restricted to the very end of the telomere.
Stn1 C terminus is required for telomere length control
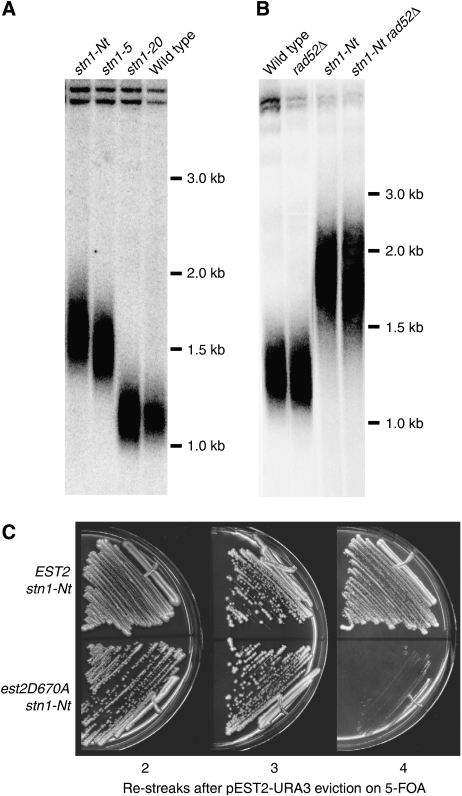
Genetic and two-hybrid data led Chandra et al (2001) to propose that the Stn1–Cdc13 interaction negatively regulates telomerase recruitment in cis. This model predicts that a reduction in association between Stn1 and Cdc13, such as that observed for stn1-Nt in a two-hybrid assay (Figure 1), should result in relaxed telomere length control. Indeed, we found that the expression of GAD–stn1-Nt in an stn1-Δ strain led to over-elongated telomeres (Figure 3A). Furthermore, stn1-Δ cells expressing GAD–stn1-5 (which fails to interact with Cdc13; see Figure 1A) display a telomere elongation phenotype similar to that of stn1-Nt, whereas the stn1-20 allele, which interacts normally with Cdc13, but not with Pol12 (see Figure 1A), has little or no effect on telomere length (Figure 3A). Taken together, these data suggest that an interaction between the Stn1 C terminus and Cdc13 exerts a negative effect on telomere elongation.
Figure 3.
Analysis of telomere length in different stn1 mutant strains. (A) Southern blot of XhoI-digested genomic DNA hybridized with a Y′ probe. The stn1-Nt, stn1-5 and stn1-20 strains (YAP73, YAP70 and YAP71, respectively) were streaked five times prior to DNA extraction. (B) Telomere elongation in the presence of the stn1-Nt allele is recombination independent (rad52Δ=YAP162; stn1-Nt rad52Δ=YAP172) and (C) telomerase dependent (stn1-Nt est2D670A=YAP115).
The telomere over-elongation observed in the stn1-Nt strain could be the result of partial deregulation of telomerase, or of recombination-dependent effects. To test the latter possibility, we deleted RAD52, required for most homologous recombination events in yeast, in the stn1-Nt strain. This had no effect on telomere elongation caused by stn1-Nt (Figure 3B). In contrast, inactivation of telomerase in an stn1-Nt strain, by the introduction of the est2D670A mutation, caused telomere shortening and senescence (Figure 3C). These results indicate that telomere over-elongation in the stn1-Nt strain is due to an effect on telomerase.
Stn1 binds telomeres in a cell cycle-regulated but TG tract length-independent manner
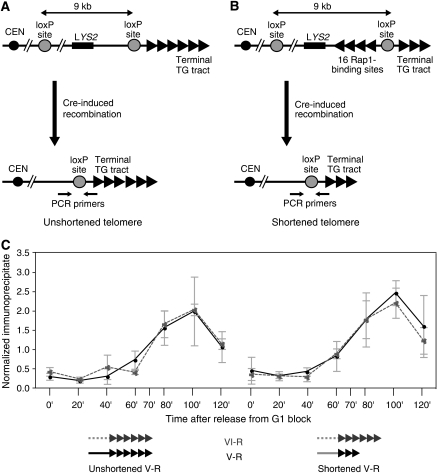
Although the results described above suggest that Stn1 has a direct function in regulating both telomerase action and telomere capping, there has so far been no evidence reported to indicate if or when Stn1 is physically associated with telomeric chromatin. We thus used a ChIP assay to measure binding of a 13 × Myc epitope-tagged version of Stn1 (Stn1–Myc) as a function of cell cycle progression. In addition, we performed these experiments in a pair of strains that differ in TG tract length at a single marked telomere (Bianchi and Shore, 2007a, 2007b), thus allowing us to test the idea that binding of the negative regulator Stn1 might be increased at longer TG tracts, where telomerase action is disfavoured (Teixeira et al, 2004). In one strain, Cre–LoxP recombination leads to a shortened TG tract at an engineered Chr. V-R telomere (Figure 4B), whereas in a control strain the recombination event leaves the same Chr. V-R telomere tract unchanged (Figure 4A). In both strains, Stn1–Myc binding at a different telomere (Chr. VI-R) served as an internal control. Cre-mediated recombination was induced by the addition of galactose to cell cultures that were immediately afterwards arrested in the G1 phase of the cell cycle by the addition of mating pheromone (α factor). Following release of cells into a synchronous cell cycle by removal of α factor, protein binding (Stn1–Myc) was measured by ChIP at 20-min intervals.
Figure 4.
A system for the generation of a single shortened telomere on Chr. V-R to measure the association of Myc-tagged Stn1. (A, B) Cre-induced recombination of the terminal part of Chr. V-R generates an unshortened telomere (A) or a shortened telomere (B) depending on the absence or presence of 16 Rap1-binding sites immediately internal to the distal LoxP site. (C) Quantification by chromatin immunoprecipitation (ChIP) of the association of Stn1–Myc to the unshortened (left) and shortened (right) Chr. V-R telomere (continuous line), normalized to the signal obtained at the reference Chr. VI-R telomere (dashed line) in each strain. Data shown are the average of three independent experiments.
This experiment demonstrated that Stn1 associates specifically with both telomeres examined, with a clear peak of binding occurring at 100 min following α factor release (Figure 4C), which under these conditions corresponds to late S phase (data not shown). Similar binding profiles have been observed by us and others for epitope-tagged versions of Cdc13, Est1 and Est2 (Taggart et al, 2002; Schramke et al, 2004; Takata et al, 2005; Bianchi and Shore, 2007a, 2007b). Significantly, we observed no temporal or quantitative difference between Stn1 association at the shortened Chr. V-R telomere compared with either the internal Chr. VI-R telomere or the unshortened V-R telomere in the control strain. A similar result is observed for Cdc13, whereas both Est1 and Est2 binding is markedly increased (∼2-fold) at a shortened telomere (Bianchi and Shore, 2007b).
Stn1 C terminus influences the efficiency of telomerase action at an elongating telomere
To analyse further the dynamics of telomerase-dependent TG addition in the stn1-Nt mutant, we turned to a de novo telomere formation assay (Figure 5A). We and others have shown previously that the presence of an 80-bp telomeric repeat sequence adjacent to a DNA double-strand break (DSB; generated by induced expression of the HO endonuclease) is sufficient to provoke rapid and efficient telomerase-dependent TG-repeat addition at the break (Diede and Gottschling, 1999, 2001; Bianchi et al, 2004). This telomere-‘healing' reaction depends on both Cdc13 and Est1, and in particular on an interaction between these two proteins characterized genetically by Lundblad and colleagues (Evans and Lundblad, 1999; Pennock et al, 2001) and Bianchi et al (2004). To avoid complications that might arise from variable expression of plasmid-borne copies of mutant stn1-Nt, we have performed these experiments in strains where the stn1-Nt mutant allele is integrated into the chromosome (indicated as stn1-Nt-i). It should be noted that in these experiments both the wild-type and the truncated STN1 gene are expressed from the strong ADH1 promoter.
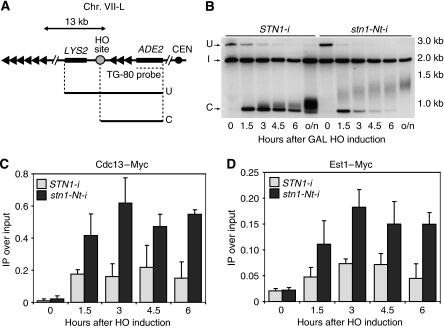
Figure 5.
Increased efficiency of telomerase elongation at an HO-induced DNA double-strand break (DSB) in strains carrying a chromosomal stn1-Nt allele. (A) Schematic representation of the modified subtelomeric region of Chr. VII-L. (B) Southern blot monitoring the healing at Chr. VII-L after DSB induction ‘I' indicates a band from the NMD5 locus that serves as an internal loading standard, ‘U' denotes the uncut fragment hybridizing to an ADE2 probe and ‘C' indicates the band resulting from ‘U' after HO cleavage. Telomere formation at the DSB was detectable as a smeared band just above the ‘cut' (C) fragment (see Materials and methods for details). (C, D) Analysis by ChIP of the binding of Cdc13–Myc (STN1-i=YAP210; stn1-Nt-i=YAP211) and Est1–Myc (STN1-i=YAP214, stn1-Nt-i=YAP215) following induction of a DSB.
We thus decided to compare, in wild-type and stn1-Nt-i strains, the efficiency and extent of telomere elongation, as well as the binding of Cdc13 and Est1 proteins to the forming telomere. As shown in Figure 5B, the wild type and mutant display remarkably different behaviour in this telomere-‘healing' assay. Thus, the smear of lower mobility species (elongating ends) appears earlier in the mutant, where the overall rate of end elongation is clearly higher. Furthermore, the band resulting from the HO cut (‘C' in Figure 5B) clearly disappears at a higher rate in the mutant cells (∼2-fold higher in several independent experiments, data not shown), consistent with an increase in the frequency of telomerase action. An alternative explanation of this increased rate would be that the TG ends in the stn1-Nt-i mutant cells are more prone to degradation, perhaps through 5′ end resection. To test this idea, we first quantified the efficiency of stable telomere formation in mutant and wild-type cells, using a simple biological assay (measurement of Ade+, Lys− colonies by replica plating). By this measure, telomere formation in the mutant cells is 80% that of wild type (data not shown). We next used a ChIP assay to measure RPA binding at the break site, which serves as a highly sensitive measure of ss DNA production following HO cutting (Hirano and Sugimoto, 2007). This assay revealed a slight (∼2-fold) increase in the mutant relative to the wild type at the TG-80 end of the break, though to levels still 10-fold below that of an end not containing TG repeats (Supplementary Figure S5). Taken together, these data suggest that the TG-flanked HO cut site in the stn1-Nt-i mutant cells may be more permissive to both exonucleolytic attack and telomerase action, though the precise rate of either is at present difficult to determine.
To address the mechanism by which the Stn1 C terminus negatively regulates telomere elongation, we used the ChIP assay to quantify the level of both Cdc13 and Est1 binding at the forming telomere (Bianchi et al, 2004; Negrini et al, 2007). As shown in Figure 5C and D, binding of both Cdc13 and Est1 at the elongating TG-80 end was approximately two-fold higher in an stn1-Nt-i strain than in a wild-type strain. This increase in Cdc13 and Est1 binding might result from the action of telomerase, which generates additional TG-repeat sequence, and thus more potential binding sites for Cdc13 (and, indirectly, Est1). We therefore conducted similar ChIP experiments in strains expressing a catalytically inactive version of Est2 (est2D670A), where TG-repeat addition at the break does not occur (Supplementary Figure S6). However, we found that Cdc13 binding remained high in the stn1-Nt-i strain, relative to the STN1 wild-type control (as did that of Est1), even in the absence of telomerase action (Supplementary Figure S3), suggesting that it might be largely due to the apparent increase in ss DNA at the break site in these cells.
Continuous telomere elongation in stn1-Nt cells
Mutations in other negative regulators of telomere elongation, such as Rif proteins, result in a new steady-state telomere length (Hardy et al, 1992; Wotton and Shore, 1997). This new equilibrium is reached relatively rapidly (probably <25 generations) due to active telomerase-dependent elongation (Diede and Gottschling, 1999; Levy and Blackburn, 2004). Surprisingly, we found instead that TG repeat addition an stn1-Nt strain continues for at least ∼340 generations (17 re-streaks, ∼20 generation per streak), thus resulting in telomeres with TG tracts that measured ∼4 kb in length (Figure 6A). Remarkably, the average rate of this continuous elongation was about 130 bp per re-streak and appeared to be largely independent of TG tract length. Thus, even at the later generations, the elongation rate is maintained at the same average rate as in the earlier ones (∼7 bp per cell division). In addition, we note that late generation cells (200 and beyond) display apparent amplification of the short Y′ element as well as an increase in a sub-population of shorter telomeres that may be the result of telomere rapid deletion (Bhattacharyya and Lustig, 2006).
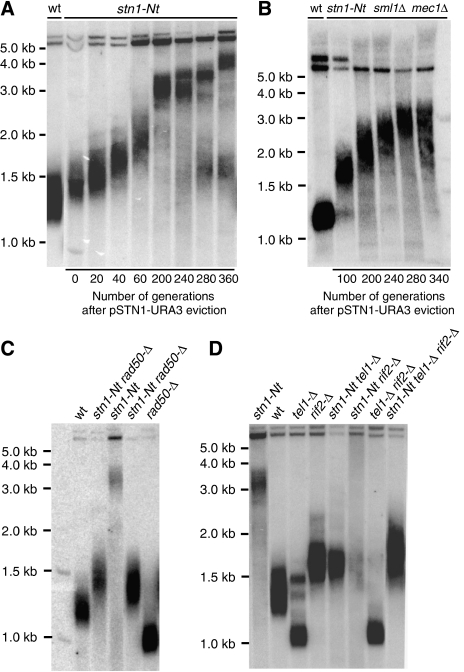
Figure 6.
Telomere length and genetic interaction analysis of stn1-Nt. (A, B) Southern blot of XhoI-digested genomic DNA hybridized with a Y′ probe. Following YCpLac33-STN1 eviction, cells were grown on YPAD medium at 30°C for the indicated number of generations before preparation of genomic DNA. (C, D) Southern blot analysis, using a Y′ probe, of XhoI-digested genomic DNA from the indicated single-, double- and triple-mutant strains. After tetrad dissection of a heterozygous diploid strain, the indicated mutant segregants were re-streaked at least 15 times on YPD medium at 30°C prior to DNA preparation.
Our two-hybrid analysis indicates that the C- and N-terminal truncations of Stn1 can independently interact with Ten1 and Cdc13–Pol12, respectively (Figure 1). This observation led us to investigate whether the apparently unlimited telomere elongation caused by the absence of Stn1 C terminus could be reversed by the co-expression of the C-terminal portion of Stn1. To test this hypothesis, a multicopy plasmid carrying the Stn1 C-terminal domain driven by a GAL1 promoter was constructed. The galactose-induced overexpression of this portion of Stn1 is lethal in the stn1-Nt mutant strain (as in an STN1 wild-type strain). However, under repressing condition (growth on glucose-containing medium), where the Stn1 C-terminal fragment is weakly expressed (data not shown), cell viability was unaffected and, as shown in Supplementary Figure S4, the telomere elongation defect conferred by the stn1-Nt mutation was significantly reduced. This result indicates that the Stn1 C terminus, which can bind to Cdc13 but not to Ten1, is alone able to negatively regulate telomere elongation.
MRX and Tel1 are required for continuous telomere elongation in stn1-Nt cells
To understand better the role of the Stn1 C-terminal domain in telomerase regulation, we generated strains carrying the stn1-Nt allele in combination with mutations in other factors involved in telomere length regulation. Several studies have shown that telomere length equilibrium results from a balance of positive and negative regulatory networks. Mutations in RIF1 and RIF2, part of the so-called ‘protein-counting' mechanism (Marcand et al, 1997, 1999), lead to telomere over-elongation, whereas mutations in components of the MRX complex (Mre11, Rad50 or Xrs2) or Tel1 (the yeast ATM homologue) have been shown to result in short, but stable telomeres (reviewed in McEachern et al, 2000). Genetic epistasis analysis suggests that MRX and Tel1 act in a common pathway downstream from both Rif1 and Rif2 (Ritchie and Petes, 2000; Chan et al, 2001; Grossi et al, 2004). As shown Figure 6C and D, gene disruption of either RAD50 or TEL1 caused telomere shortening, whereas disruption of RIF2 induced telomere lengthening, as previously reported. Notably, however, both stn1-Nt tel1-Δ and stn1ΔNt rad50-Δ double mutants, and an stn1-Nt tel1-Δ rif2-Δ triple mutant, had telomeres that were longer than the wild-type strain, indicating that the stn1-Nt can completely reverse the telomere shortening effect of both tel1-Δ and rad50-Δ. Nevertheless, both tel1-Δ and rad50-Δ abolish the continuous elongation phenotype conferred by stn1-Nt. By contrast, stn1-Nt sml1-Δ mec1-Δ cells (lacking the yeast ATR homologue Mec1) do display telomere over-elongation (Figure 6B). Finally, the double mutant stn1-Nt rif2-Δ shows very long and heterogeneous telomeres.
Discussion
The results described here demonstrate that the telomere capping and telomere length control functions of Stn1 are to a large extent executed by two independent domains that can function separately from each other. These functional domains are associated with discrete protein–protein interactions: the Stn1 N terminus interacts with Ten1 to cap the telomere, whereas the C terminus of Stn1 interacts with both Cdc13 and Pol12, with the Cdc13 interaction having a critical function in the regulation of telomerase action. One possible caveat to these conclusions is that the stn1-Nt mutant allele must be expressed from a strong promoter (ADH1) for the cells to be viable, as has also been observed by Petreaca et al (2007). Although it is difficult to rule out the possibility that this high level of expression might in and of itself influence the stn1-Nt phenotype, we note that de novo telomere formation and equilibrium telomere length are similar or identical when comparing wild-type STN1 expressed from either its native promoter or the ADH1 promoter (Figure 5 and data not shown).
Capping function of the Stn1 N terminus
Our data show that an N-terminal domain (amino acids 1–281) of Stn1 fulfills its essential function of telomere end protection, consistent with recent reports on both the S. cerevisiae protein (Gao et al, 2007; Petreaca et al, 2007) and its homologue in the related yeast K. lactis (Iyer et al, 2005). This capping function of Stn1 may derive solely from its ability to interact with Ten1, as the capping defect in stn1-154, which we show is due to an Asp to Glu substitution at amino acid 99, is associated with an abolished Ten1 interaction and is suppressed by fusion of the mutant Stn1 protein with Ten1 (Grandin et al, 2001). Interestingly, the failure of stn1-Nt to interact with Cdc13 in a two-hybrid assay (Figure 1) suggests that telomere capping may not require a direct interaction between Stn1 and Cdc13, as proposed previously (Pennock et al, 2001). Delivery of stn1-Nt to telomeres may also be promoted by Ten1, which itself interacts with Cdc13 (Grandin et al, 2001; Gao et al, 2007), and with the telomeric TG-rich single-strand overhang, through its own OB fold (Gao et al, 2007). An alternative interpretation of our data is that stn1-Nt and Ten1 are able to bind the telomere tip independently of Cdc13, when overexpressed, thus bypassing the normal Cdc13 requirement for telomere capping (Petreaca et al, 2006). Consistent with this possibility, an OB fold-containing region of stn1-Nt (amino acids 69–199) and full-length Ten1 (which also contains an OB fold) have recently been shown to bind specifically in vitro to TG-containing ss DNA (Gao et al, 2007).
Our finding that Pol12 fails to interact with the Stn1 N terminus in two-hybrid assays is in contradiction to in vitro studies reported previously (Petreaca et al, 2006). We do not know the reason for this discrepancy, but would point out that it is unlikely to be due to a trivial artefact, because the GAD–stn1-Nt hybrid used in our experiments is functional: it complements an STN1 deletion for growth and interacts well with GBD–Ten1. In any event, we have confirmed by a GST–Pol12 pull-down assay that both the Stn1 N and C termini, expressed independently in yeast, can bind to Pol12 (Supplementary Figure S7). We would therefore suggest that both halves of Stn1 contribute in vivo to Pol12 binding. As our conclusion that the Stn1–Cdc13 interaction (and not the Stn1–Pol12 interaction) has a predominant function in telomere length regulation is based on a defect in stn1-20 binding to Pol12, we have tested this in vitro. We observe a reduction of at least 25% in binding of the mutant compared with wild type (Supplementary Figure S7). This result is consistent with the two-hybrid findings, though weaker in magnitude, perhaps because the in vitro studies were performed with full-length Stn1 protein. Taken together, the data support our conclusion that the Stn1–Cdc13 interaction has a predominant function in telomere length regulation, and highlight the need for further studies of the Pol12 interaction with Stn1, and its functional consequences, particularly in telomere capping (Grossi et al, 2004; Petreaca et al, 2006).
Finally, it is worth noting that Iyer et al (2005) have described a different point mutation in the conserved N terminal region of the K. lactis Stn1 protein that leads to high levels of recombinogenic telomere elongation, reminiscent of that seen in human tumour cells with an active alternative lengthening of telomeres pathway. It will be interesting to see whether this or other mutations in the S. cerevisiae Stn1 protein will have similar effects.
Telomerase regulation and coordination of C-strand synthesis by Stn1
Telomere length homoeostasis is achieved through a balance between telomere elongation and shortening. Recent studies by Lingner and colleagues (Teixeira et al, 2004) show that telomeres in budding yeast are not elongated by telomerase at every cell cycle, and that the probability of elongation is inversely related to TG tract length. These and other data indicate that a mechanism(s) operates in cis that can sense TG tract length and use this information to control telomerase access and/or activity. Rap1 and its interacting Rif factors appear to be sensors of telomere length and negative regulators of telomerase action, whereas Cdc13, which is required to recruit telomerase to chromosome ends through an interaction with Est1, would appear to be an important regulatory target. Indeed, several recent studies indicate that the recruitment of the telomerase holoenzyme (including Est1) is a key step regulated by telomere tract length (Bianchi and Shore, 2007b; Hector et al, 2007; Sabourin et al, 2007).
Data presented here suggest that the Stn1 C terminus, which interacts with Cdc13 in two-hybrid assays, has a key and perhaps unique function as a negative regulator of telomerase action. Thus, cells lacking the Stn1 C terminus display an unusual telomere elongation phenotype, where elongation occurs at a constant rate (essentially independent of TG tract length) for at least 350 generations, yielding extremely elongated telomeres that are unusually homogenous in length. This phenotype is quite unlike that of cells lacking both Rif1 and Rif2 or the Rap1 C terminus, where elongation is more rapid and telomere length in a population of cells is extremely heterogeneous. Furthermore, in cells deleted for either RIF1 or RIF2, telomeres reach a new average length, unlike what is observed in stn1-Nt mutants. Significantly, the effect of stn1-Nt on telomere elongation is additive with that of rif2-Δ, which has been shown to increase the probability of telomerase action but not its processivity (Teixeira et al, 2004), arguing either that stn1-Nt affects telomerase accessibility through a different pathway than Rif2, or that it effects the activity (processivity) of telomerase already bound at the chromosome end. We propose that the more rapid elongation that occurs following HO cutting in the stn1-Nt strain reflects repeated rounds of telomerase binding and synthesis as these cells remain blocked in G2/M during the course of the experiment (Negrini et al, 2007), suggesting that the nascent telomere is either more accessible to telomerase, or in a state that more efficiently promotes telomerase synthesis once bound (or both). Although we cannot rule out an increase in telomerase processivity brought about by the deletion of the Stn1 C terminus, we note that telomeres in normally cycling stn1-Nt cells elongate at a modest pace (approximately 7 bp per cell division) and maintain a remarkably tight length distribution in the population, arguing against the idea that telomerase addition becomes unregulated under these conditions.
Our two-hybrid and ChIP data, together with the earlier findings of Chandra et al (2001), are consistent with a model in which the Stn1 C terminus and Est1 compete directly for Cdc13 binding. One caveat to this conclusion, though, is our finding that Cdc13 association increases concomitantly with that of Est1 in stn1-Nt strains. We are thus unable to determine whether increased Est1 binding is simply a consequence of increased Cdc13 telomere association or instead due to increased accessibility of telomere-bound Cdc13 to Est1 (or both). Why does Cdc13 telomere association appear to increase as a consequence of deletion of the Stn1 C terminus? Our data suggest that this might be due to the slight increase in ss TG-rich DNA detected in the stn1-Nt strain, which would increase the number of potential Cdc13-binding sites there. This increased single-strandedness might stem from weakened (or altered) recruitment of the polymerase α–primase complex to telomere ends, which is also promoted by the Stn1 C terminus, in this case through its interaction with Pol12 (Figure 1A; Grossi et al, 2004), leading to defective C-strand synthesis at or near the telomere terminus. Alternatively, the absence of the Stn1 C terminus in the CST complex may expose an additional region of Cdc13 to the ss overhang, increasing our ability to crosslink the protein to telomeric sequences. These two explanations are not mutually exclusive and additional work will be required to resolve this issue.
Stn1 and the regulatory system that monitors TG tract length
As pointed out above, several recent studies have demonstrated increased association of both telomerase holoenzyme (Est1 and Est2 subunits) and Tel1 kinase, but not Cdc13, at short telomeres (Bianchi and Shore, 2007b; Hector et al, 2007; Sabourin et al, 2007). One explanation for these observations is that the telomere length signal (still poorly defined, but presumably generated by the Rap1–Rif complex) modulates Tel1 binding, which in turn affects the phosphorylation state of the CST complex and, consequently, its ability to recruit telomerase. Consistent with this scenario, Tel1 kinase can phosphorylate Cdc13 in vitro at residues within the Cdc13 telomerase recruitment domain that are required for telomere length maintenance in vivo (Tseng et al, 2006).
We show here that Stn1 telomere association, similar to that of Cdc13, is quantitatively indistinguishable at short versus long telomeres. This finding suggests that the negative effect of the Stn1 C terminus on telomerase action is either constant (i.e. independent of telomere length) or modulated by telomere length through a modification of Stn1 at the telomere, rather than a change in the amount of protein associated there. Our finding that stn1-Nt strains display considerable telomere elongation even in the absence of Tel1 function (or that of the MRX complex; Figure 6), suggests that the Stn1 C terminus exerts an effect in parallel with Tel1 to control telomere length, probably by directly modulating telomerase association. The data also raise the possibility that the Stn1 C terminus is itself one of several downstream targets of the Tel1 kinase. In this scenario, phosphorylation of Stn1 would be promoted at short telomeres and would weaken its interaction with Cdc13, thus promoting Est1 (and telomerase enzyme) association. However, we found that mutation of three potential Tel1 target sites in Stn1 has no detectable effect on telomere length (unpublished data), arguing against this model. We thus favour a model in which Stn1 provides a constitutive inhibitory signal, independent of telomere tract length, whose effectiveness is modulated by changes in its partner protein Cdc13, and possibly other factors.
In summary, the data presented here suggest that the Stn1 C terminus, through an interaction with Cdc13, has a direct function in inhibiting telomerase association with the chromosome end, a process that recent work suggests is highly dynamic (Chang et al, 2007). Our analysis suggests that this ‘brake' on telomere elongation exerted by the Stn1 C terminus is counteracted by the positive arm of a ‘protein-counting' telomere length-regulatory mechanism involving the Tel1 kinase. Although the latter mechanism is modulated by telomere length (Bianchi and Shore, 2007b; Sabourin et al, 2007), we show that the Stn1 inhibition is likely not to be. We note that recent work in mammalian cells indicates that increasing telomerase levels also promotes a constant, tract length-independent telomere elongation phenotype (Cristofari and Lingner, 2006). The proposed similarity of the CST complex to the well-studied RPA complex (Gao et al, 2007) raises the interesting possibility that structural changes in CST, analogous to those proposed to occur in RPA (Fanning et al, 2006), might have an important function in regulating its interactions with both telomerase and the polymerase α–primase complex. A challenge for future studies will be to design experiments that allow one to characterize directly CST, telomerase holoenzyme and polymerase α–primase interactions in vitro, and, ultimately, during the dynamic process of telomere replication in living cells. The recent discovery of Stn1-like proteins in many organisms (Gao et al, 2007; Martin et al, 2007), including mammals, suggests that the findings reported here may have relevance beyond the budding yeasts.
Materials and methods
Yeast strains and plasmids
The yeast strains used in this study are listed in Table I, and are isogenic to W303 (Thomas and Rothstein, 1989), apart from specific point mutations or gene deletion/disruptions, or where otherwise indicated. Double-mutant strains were obtained by crossing of single-mutant strains followed by sporulation and tetrad dissection, unless indicated otherwise. Growth and manipulation of yeast were carried out according to standard procedures (Adams et al, 1997). CDC13, EST1 and STN1 were tagged using PCR products generated from pFA6a-13Myc-His3MX6 (Longtine et al, 1998). All strains used for the telomere healing assay are derived from W303-based YAB53 (Bianchi et al, 2004), and carry a deletion of the HO site at the MAT locus; a galactose-inducible copy of the HO endonuclease gene at the LEU2 locus; a cassette at the subtelomeric region of Chr. VII-L, between the ADH4 and MNT2 genes, containing the ADE2 gene, a sequence from the mouse Dbp gene (amplicon 7), a 80-bp TG-repeat sequence from a native telomere and the HO cut site (Figure 5A). The STN1 gene was deleted in a diploid YAB53 strain using a PCR-generated KanMX (G418 resistance) cassette (Longtine et al, 1998).
Table 1.
Yeast strains used in this study
| Name | Relevant genotype | Reference |
|---|---|---|
| W303-1A | MAT a ade2-1 trp1-1 can1-100 leu2-3, 112his3-11, 15 ura3-1rad5-535 | Thomas and Rothstein (1989) |
| W303-1B | MAT a ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3-1rad5-535 | Thomas and Rothstein (1989) |
| PJ69-4A | MAT a trp1-901 leu2-3,112 ura3-52 his3-200 gal4Dgal80D LYS′′∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ | James et al (1996) |
| CTY10-5D | MAT a ade2 trp1-901 leu2-3, 112 his3-200 gal4 gal80 URA3∷lexA op-lacZ | Bartel and Fields (1995) |
| YAB53 | W303 (RAD5, lys2), mata∷loxP, leu2∷pGALHO, mnt2∷LYS2 | Bianchi et al (2004) |
| DLY1108 | MAT a cdc13-1 | Booth et al (2001) |
| YAP34 | YAB53, CDC13-Myc13∷HIS3, VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP41 | MAT a stn1_LoxP+pGADstn1-154-LEU2 | This study |
| YAP42 | MAT a stn1_LoxP+pGADstn1-63-LEU2 | This study |
| YAP46 | MAT a stn1_LoxP+YCp-STN1-URA3 | Grossi et al (2004) |
| YAP70 | MAT a stn1_LoxP+pGADstn1-5-LEU2 | Grossi et al (2004) |
| YAP71 | MAT a stn1_LoxP+pGADstn1-20-LEU2 | Grossi et al (2004) |
| YAP73 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2 | Grossi et al (2004) |
| YAP100 | YAB53, EST1-Myc13∷HIS3, VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP115 | YAB53, est2D670A, CDC13-Myc13∷HIS3, VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP117 | YAB53, est2D670A, EST1-Myc13∷HIS3, VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP130 | YAB53, CDC13-Myc13∷HIS3, stn1∷KanR, pGADstn1-Nt-TRP1,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP131 | YAB53, EST1-Myc13∷HIS3, stn1∷KanR, pGADstn1-Nt-TRP1,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP150 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2, adh4∷URA3 | This study |
| YAP151 | MAT a stn1_LoxP+pGADstn1-63-LEU2, adh4∷URA3 | This study |
| YAP152 | MAT a sir2∷HIS3, adh4∷URA3 | This study |
| YAP153 | MAT a adh4∷URA3 | This study |
| YAP162 | MAT a rad52∷KanR | This study |
| YAP170 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2 | This study |
| YAP171 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2, rad50∷KanR | This study |
| YAP172 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2, rad52∷KanR | This study |
| YAP175 | MAT a tel1∷TRP1 | This study |
| YAP176 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2, tel1∷TRP1 | This study |
| YAP177 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2 rif2∷HIS3 | This study |
| YAP178 | MAT α te1∷TRP1, rif2∷HIS3 | This study |
| YAP179 | MAT a stn1_LoxP+pGADstn1-Nt-LEU2, te1∷TRP1, rif2∷HIS3 | This study |
| YAP180 | MAT α, rif2∷HIS3 | This study |
| YAP189 | YAB53, est2D670A, CDC13-Myc13∷HIS3, stn1∷KanR, pGADstn1-Nt-TRP1, VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP190 | YAB53, est2D670A, EST1-Myc13∷HIS3, stn1∷KanR, pGADstn1-Nt-TRP1, VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP204 | YAB53, CDC13-Myc13∷HIS3, stn1∷KanR, YCpLac33-STN1-URA,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP205 | YAB53, EST1-Myc13∷HIS3, stn1∷KanR, YCpLac33-STN1-URA,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP210 | YAB53, CDC13-Myc13∷HIS3, stn1∷KanR, GAD-STN1-HA-i∷TRP1,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP211 | YAB53, CDC13-Myc13∷HIS3, stn1∷KanR, GAD-stn1-Nt-HA-i∷TRP1,VII-L∷ADE2-Dbp(amp7)- TG-80-HO site | This study |
| YAP213 | YAB53, EST1-Myc13∷HIS3, stn1∷KanR, GAD-STN1-HA-i∷TRP1,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YAP214 | YAB53, EST1-Myc13∷HIS3, stn1∷KanR, GAD-stn1-Nt-HA-i∷TRP1,VII-L∷ADE2-Dbp(amp7)-TG-80-HO site | This study |
| YG145 | W303 MAT α, yku70∷LEU2 | This study |
| YSG376 | YAB53, VII-L∷ADE2-Dbp(amp7) λ DNA-HO site | This study |
| YAP220 | W303 MAT α, GAD-STN1-HA-i | This study |
| YAP221 | W303 MAT α, GAD-stn1-20-HA-i | This study |
| YAP222 | W303 MAT α, GAD-stn1-Nt-HA-i | This study |
| YAP223 | W303 MAT α, GAD-stn1-Ct-HA-i | This study |
The plasmids used in this study are listed in Table II. pGAD-stn1-Nt was generated by subcloning an NcoI–EcoRI fragment from pACT2-STN1 (Grandin et al, 1997) in NcoI–EcoRI-digested pGAD (Moretti et al, 1994). pGAD-stn1-Ct was generated by subcloning the EcoRI fragment from pACT2-STN1 in EcoRI-cleaved pGAD. The pstn1-63 plasmid was generated by subcloning the NcoI–BsmI fragment from YCp111-stn1-154 (Grandin et al, 2001) in NcoI–BsmI-digested YCpLac33-STN1 (Grandin et al, 1997). Integrative plasmids (GAD-stn1-HA-i) carrying the different GAD-stn1-HA alleles were generated by cloning a SapI–XhoI fragment from the pGAD-stn1-HA plasmids into SapI–XhoI digested pRS304. These constructs were digested with EcoRV and integrated in the TRP1 locus of yeast strains deleted for endogenous copy of STN1 and kept alive with YCpLac33-STN1-URA plasmid. The cells were then streaked on 5-FOA to evict the YCpLac33-STN1-URA plasmid.
Table 2.
Plasmids used in this study
| Name | Description | Reference |
|---|---|---|
| YCpLac33-STN1 | CEN, URA3, STN1, native promoter | Grandin et al (1997) |
| pACT2-STN1-HA | 2 μm, LEU2, GAD-STN1, ADH1 promoter | Grandin et al (1997) |
| YCp111-stn1-154 | CEN, LEU2, stn1-154, native promoter | Grandin et al (2001) |
| pGAD-stn1-5-HA | 2 μm, LEU2, GAD-stn1-5, ADH1 promoter | This study |
| pGAD-stn1-20-HA | 2 μm, LEU2, GAD-stn1-20, ADH1 promoter | This study |
| pstn1-63 | CEN, LEU2, stn1-63, native promoter | This study |
| pGAD-stn1-63-HA | 2 μm, LEU2, GAD-stn1-63, ADH1 promoter | This study |
| pGAD-stn1-Nt-HA | 2 μm, LEU2, GAD-stn1-Nt, ADH1 promoter | This study |
| pGAD-stn1-Ct-HA | 2 μm, LEU2, GAD-stn1-Ct, ADH1 promoter | This study |
| pGAL-stn1-Ct | 2 μm, LEU2, GAL prom-stn1-Ct | This study |
| pAS2-TEN1 | 2 μm, TRP1, GBD-TEN1, ADH1 promoter | Grandin et al (2001) |
| pLexA-pol12-Nt | 2 μm, TRP1, pLexA-pol12-Nt, ADH1 promoter | Grossi et al (2004) |
| GBD-CDC13 | 2 μm, TRP1, pGBD-CDC13, ADH1 promoter | Bianchi et al (2004) |
| pGBD | 2 μm, TRP1, pGBD, ADH1 promoter | James et al (1996) |
| pGAD | 2 μm, LEU2, GAD, ADH1 promoter | Grandin et al (1997) |
| PLexA | 2 μm, TRP1, LexA, ADH1 promoter | Bartel and Fields (1995) |
| pGAD-STN1-HA-i | Integrative, TRP1, ADH1 promoter, GAD-STN1 | This study |
| pGAD-stn1-Nt-HA-i | Integrative, TRP1, ADH1 promoter, GAD-Stn1-Nt | This study |
| pGAD-stn1-Ct-HA-i | Integrative, TRP1, ADH1 promoter, GAD-Stn1-Ct | This study |
| pGAD-stn1-20-HA-i | Integrative, TRP1, ADH1 promoter, GAD-Stn1-20 | This study |
PCR mutagenesis (Cadwell and Joyce, 1994) was used to generate STN1 C terminus mutations, and gap repair was used to generate pACT2-STN1 mutant plasmids. EcoRI-linearized pACT2-STN1 was co-transformed in the CTY10-5D yeast strain (already containing pLexA-Pol12-Nt) together with the mutagenized STN1 C-terminal PCR product. After transformation, cells were grown in liquid YAPD for 4 h and then plated on SC-Trp-Leu solid media containing X-gal.
Two-hybrid assays
The yeast two-hybrid assays were performed using CTY10-5D (Bartel and Fields, 1995) or PJ69-4A (James et al, 1996) strains harbouring pLexA/pGAD or pGBD/pGAD fusion plasmids, respectively. Strains containing the test plasmids were grown for 24 h in SC-Trp-Leu liquid medium, and β-galactosidase was measured by liquid assay (Moretti et al, 1994).
Telomere Southern blots
For Southern blot analysis of telomere length, yeast genomic DNA was isolated from overnight cultures and 2 μg was digested with XhoI. DNA fragments were separated by electrophoresis in 1.0% agarose gels (TBE 0.5 × ), transferred to HyBond N+ membranes, and hybridized with a random-primed, radiolabelled Y′ probe (Craven and Petes, 1999) by standard procedures. The membranes were then autoradiographed with a PhosphorImager (Bio-Rad Molecular Imager FX).
Southern analysis of DSB induction and telomere healing assay
For galactose induction of a DSB, overnight saturated cultures were diluted in YPE lactate to a final concentration of 5 × 106 cells/ml and grown for additional 3 h at 30°C. The HO endonuclease was then induced by the addition of galactose to a final concentration of 2%. DNA was isolated from 40 ml of cell culture and digested with EcoRV. DNA fragments were separated by electrophoresis in 0.8% agarose gels, transferred to HyBond N+membranes, and hybridized to ADE2 and NMD5 random-primed probes.
QAOS assay
ss DNA formation at subtelomeric regions of Chr. V-R was measured by the quantitative real-time PCR method QAOS (Booth et al, 2001). Briefly, cell cultures were grown in YPD at 24°C until saturation, then diluted to a final concentration of 5 × 106 cells/ml. These asynchronous cell cultures were grown for an additional 4 h at 24°C, at which point cells were centrifuged and resuspended in pre-warmed (37°C) YPD. Aliquots were then processed as described (Booth et al, 2001).
ChIP
ChIP experiments at the HO-generated DSB (Figure 5; Supplementary Figures S3 and S5) were performed with asynchronous cell cultures as described previously (Bianchi et al, 2004). ChIP experiments using the Cre–LoxP system (Figure 4) were performed as described previously (Bianchi and Shore, 2007a, 2007b). Despite very similar overall binding patterns in the two strains in each experiment, due to variations in the overall efficiency of immunoprecipitation between different experiments, results (calculated as a percentage of input present in the immunoprecipitates) were first normalized against a value obtained by averaging the signal for the Chr. VI-R telomere within each experiment.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Information
Acknowledgments
This paper is dedicated to the memory of Stephan B Schawalder, a great friend and brilliant colleague. We thank M Charbonneau and D Lydall for gifts of plasmids and/or strains, K Mishra for advice on the native Southern technique, N Roggli for expert graphics work and members of the Shore lab for helpful advice throughout the course of this study and comments on the paper. This study was supported by grants from the Swiss National Science Foundation and the Swiss Cancer League (OncoSuisse), by the NCCR program ‘Frontiers in Genetics' (sponsored by the Swiss National Science Foundation), and by the Canton of Geneva. AP is an NCCR ‘Frontiers in Genetics' doctoral student. LL was supported by the ‘Fondation Recherche Médicale' (Paris, France).
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T (1997) Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Adams Martin A, Dionne I, Wellinger RJ, Holm C (2000) The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol Cell Biol 20: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Fields S (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol 254: 241–263 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya MK, Lustig AJ (2006) Telomere dynamics in genome stability. Trends Biochem Sci 31: 114–122 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D (2004) Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell 16: 139–146 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2007a) Early replication of short telomeres in budding yeast. Cell 128: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2007b) Increased association of telomerase with short telomeres in yeast. Genes Dev 21: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Griffith E, Brady G, Lydall D (2001) Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res 29: 4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF (1994) Mutagenic PCR. PCR Methods Appl 3: S136–S140 [DOI] [PubMed] [Google Scholar]
- Carson MJ, Hartwell L (1985) CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42: 249–257 [DOI] [PubMed] [Google Scholar]
- Chakhparonian M, Wellinger RJ (2003) Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet 19: 439–446 [DOI] [PubMed] [Google Scholar]
- Chan SW, Chang J, Prescott J, Blackburn EH (2001) Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol 11: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J (2007) Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD (1999) Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152: 1531–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Lingner J (2006) Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE (2001) Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol 11: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol 20: 8397–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2001) Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CFJ, Sussel L, Shore D (1992) A RAP1-interacting protein involved in silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Sugimoto K (2007) Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell 18: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Chadha AD, McEachern MJ (2005) A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol 25: 8064–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ (1993) RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7: 1146–1159 [DOI] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae (in process citation). Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T (1995) Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270: 1488–1491 [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E (1999) Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P (2007) Protection of telomeres by a conserved Stn1–Ten1 complex. Proc Natl Acad Sci USA 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH (2000) Telomeres and their control. Annu Rev Genet 34: 331–358 [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D (2007) DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI (2006) Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol 8: 748–755 [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu H-C, Nugent CI (2007) The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics 177: 1459–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD (2000) The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA (2007) Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Geli V (2004) RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet 36: 46–54 [DOI] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Takata H, Tanaka Y, Matsuura A (2005) Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol Cell 17: 573–583 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC (2006) The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2007) Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Information