Abstract
DNA oxidation plays a substantive role in the pathophysiology of human diseases such as cancer. While the chemistry of nucleobase lesions has dominated studies of DNA damage, there is growing evidence that oxidation of 2-deoxyribose in DNA plays a critical role in the genetic toxicology of oxidative stress. As part of an effort to define the spectrum of 2-deoxyribose oxidation products arising in vitro and in vivo, we now describe methods for quantifying products arising from 4′-oxidation of 2-deoxyribose in DNA. The chemistry of 4′-oxidation partitions between either of two pathways to form either a 2-deoxypentos-4-ulose abasic site (oxAB), or a strand break comprised of a 3′-phosphoglycolate (3PG) residue and a 5′-phosphate, with release of either malondialdehyde and free base or a base propenal. Highly sensitive gas chromatography-mass spectrometry (GC/MS) methods were developed to quantify both lesions. The abasic site was converted to a 3′-phosphoro-3-pyridazinylmethylate derivative by treatment of the damaged DNA with hydrazine, which was released from DNA as 3-hydroxymethylpyridazine (HMP) by enzymatic hydrolysis. Similarly, 3PG was released as 2-phosphoglycolic acid (PG) by enzymatic hydrolysis. Following HPLC pre-purification, both PG and HMP were silylated and quantified by GC-MS with limits of detection of 100 and 200 fmol, and sensitivities of 2 and 4 lesions per 106 nucleotides (nt) in 250 μg of DNA, respectively. Following validation of the methods with oligodeoxynucleotides containing the two lesions, the methods were applied to DNA damage produced by bleomycin and γ-radiation. As expected for an agent known to produce only 4′-oxidation of DNA, the quantities of 3PG and oxAB accounted for all 2-deoxyribose oxidation events, as indicated by slopes of 0.8 and 0.3, respectively, in plots of lesion frequency against total 2-deoxyribose oxidation events, the latter determined by a plasmid nicking assay. 3PG residues and oxAB were produced at the rate of 32 and 12 lesions per 106 nt per μM, respectively. For γ-radiation, on the other hand, 4′-oxidation was found to comprise only 13% of 2-deoxyribose oxidation chemistry, with 3% oxAB (4 per 106 nt per Gy) and 10% 3PG (13 per 106 nt per Gy).
Keywords: Oxidatively damaged DNA, 2, deoxyribose oxidation, bleomycin. ionizing radiation, analytical method
INTRODUCTION
DNA oxidation by a variety of damaging agents, such as oxidizing, alkylating and nitrosating chemicals, plays a significant role in the pathophysiology of cancers, aging, and inherited diseases (1–3). While the chemistry of nucleobase oxidation has dominated studies of DNA damage, there is growing evidence that oxidation of 2-deoxyribose in DNA plays a critical role in the genetic toxicology of oxidative stress, including involvement in complex DNA lesions (4), cross-linking with DNA repair proteins (5,6) and the formation of endogenous DNA adducts (7–11). As part of an effort to define the spectrum of 2-deoxyribose oxidation products arising in vitro and in vivo, we now describe methods for quantifying products arising from 4′-oxidation of 2-deoxyribose in DNA.
The chemistry of 2-deoxyribose oxidation in DNA has its roots in the literature of ionizing radiation, as summarized in numerous reviews and texts (e.g., ref. 12). As the subject of the current studies, the chemistry of 4′-oxidation pathway partitions along either of two pathways to form a variety of stable and electrophilic products (Scheme 1). One pathway involves formation of an oxidized abasic site containing a 2-deoxypentos-4-ulose residue (oxAB1), as originally observed by Dizdaroglu et al. (13,14). The other pathway results in a break in the DNA strand with an invariant 3′-phosphoglycolate residue (3PG) and, depending on the oxidant (2,11,15,16), either a base propenal, or malondialdehyde and free base. The former arises from oxidation produced by a chemical mediator of inflammation, peroxynitrite, and bleomycin (2,15). Recent studies have revealed that γ-radiation and Fe+2-EDTA generate malondialdehyde and free nucleobase by a mechanism that does not involve simple degradation of a base propenal (11,16).
Scheme 1.
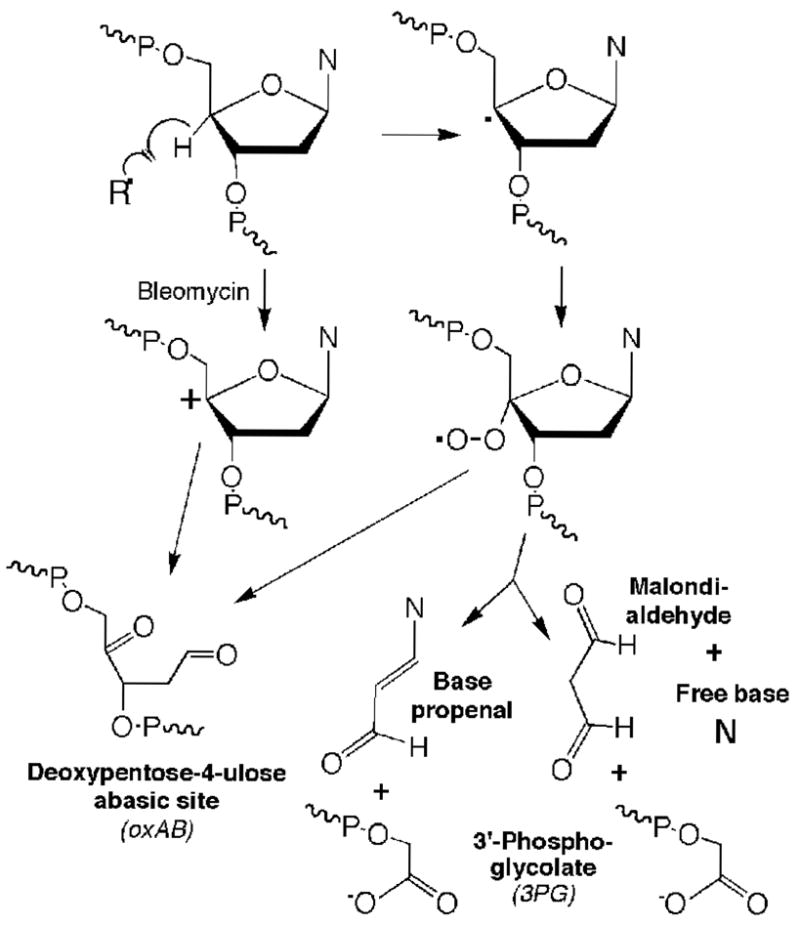
Chemistry of 4′-oxidation of 2-deoxyribose in DNA
There is emerging appreciation that the chemistry of 2-deoxyribose oxidation plays an important role in the cellular response to oxidative stresses. For example, the 2′-deoxyribonolactone abasic site arising from 1′-oxidation of 2-deoxyribose in DNA forms toxic protein-DNA cross-links with DNA repair proteins (5,6). There is also evidence for the formation of DNA adducts with several products of 2-deoxyribose oxidation in DNA. For example, base propenals react with dG to form the mutagenic pyrimidopurinone adduct, M1dG (9,11,17), while glyoxal adducts of dG can arise from reactions with 3′-phosphoglycolaldehyde residues generated by 3′-oxidation of 2-deoxyribose in DNA (7) and bicyclic adducts of dC, dG and dA are generated in reactions with the 2-phosphoryl-1,4-dioxobutane, products of 5′-oxidation of 2-deoxyribose (8,10,18–20). Finally, the repair of the various base lesions and terminal sugar fragments derived from 2-deoxyribose oxidation is hindered when the lesions are present in bistranded or complex DNA lesions, such as those produced by ionizing radiation (4,21) and by enediyne and bleomycin DNA cleaving antibiotics (22,23).
To better define the biological impact of 2-deoxyribose oxidation chemistry, we and others have endeavored to develop methods to quantify the various 2-deoxyribose oxidation products with the goal of identifying the predominant chemistries in the cellular environment and of establishing steady-state levels in cells and tissues (for example, see refs. 11,24–36). Here, we describe sensitive analytical methods to quantify the 3PG and oxAB products representative of 4′-oxidation of 2-deoxyribose in DNA, with application to DNA damage produced by γ-radiation and bleomycin.
EXPERIMENTAL METHODS
Materials
All chemicals and reagents were of highest purity available and were used without further purification unless noted otherwise. Calf thymus DNA, bleomycin, nuclease P1 (P. citrinum), phosphorus oxychloride and hydrazine were purchased from Sigma Chemical Co. (St. Louis, MO). Alkaline phosphatase (calf intestine) and plasmid pUC18 were purchased from New England Biolabs (Beverly, MA) and phosphodiesterase I (C. adamanteus venom) from USB Corp. (Cleveland, OH). Silica gel-60 (pore size, 0.040–0.063 mm) for flash chromatography was purchased from Merck and Co. (Gibbstown, NJ). [15N2]-Hydrazine sulfate was purchased from Cambridge Isotope Laboratories (Andover, MA). Solutions of calf thymus DNA and plasmid pUC18 DNA were dialyzed four-times against potassium phosphate buffer at 4 °C (50 mM, pH 7.4, Chelex-treated and autoclaved; ref. 37).
Instrumental Analyses
All HPLC analyses were performed on an Agilent model 1100 equipped with an Agilent 1040A diode array detector. UV spectra were obtained using a Beckman CU640 UV-visible spectrophotometer. GC-MS analyses were performed with a Hewlett-Packard 6890 series GC system equipped with a Hewlett-Packard 5973 mass selective detector using electron impact ionization at 70 eV. In all studies, the carrier gas was He and the column used was DB-35MS, mid-polar capillary column (30 m × 0.25 mm × 0.25 μm film thickness, cross-linked 35% phenyl-methylpolysiloxane).
Synthesis of hydroxymethylpyridazine (HMP; 3-pyridazinemethanol; CAS 37444-46-5)
Hydroxymethylpyridazine was synthesized by the method of Clauson-Kaas and Limborg (38), which involves acid-catalyzed hydrolysis of dimethoxydihydrofurfuryl and subsequent reaction with hydrazine. Following drying of ethyl acetate extracts with potassium carbonate and removal of ethyl acetate under vacuum, hydroxymethylpyridazine was purified by flash chromatography (10% hexane in ethyl acetate) to yield (55%) pale yellow crystals (m. p. 59–61 °C). NMR analysis: (DMSO-d6, 300 MHz) δ = 8.19 (1H, d, Ar-H), 6.79–6.77 (2H, m, Ar-H), 4.71 (1H, s, OH), 1.58 (2H, s, CH2) ppm. Positive ion mode MS: m/z 111.0 (M+1, 100%), 93.0 (M-OH, 6.7%); calculated for C5H6N2O: m/z 110.1. [15N2]-Hydroxymethylpyridazine was prepared as described above using [15N2]-hydrazine, with similar characteristics (data not shown).
Synthesis of 2-phosphoglycolic acid (PG; CAS 13147-57-4)
To a solution of phosphorous oxychloride (0.28 ml, 3.05 mmol) in 2 ml of trimethyl phosphate cooled to 0 °C was added glycolic acid (76 mg, 1 mmol) or [2H2]-glycolic acid (78 mg, 1 mmol). The mixture was stirred at 0 °C for 3 hr. The resulting clear solution was poured into 10 ml of ice water and the pH raised to 1.5 with ice-cold 2 N sodium hydroxide. 2-Phosphoglycolate was purified by reversed phase HPLC (Vydac C18 column, 250 × 3 mm, 5 μm; detection at 218 nm) using a mobile phase of 1:1 water:acetonitrile with 0.1% trifluoroacetic acid at a flow rate of 0.3 ml/min, which resulted in a retention time of 11 min. Following removal of HPLC solvent under vacuum, the resulting solid white powder had the following characteristics: 1H NMR (D2O): δ 4.50 (d, 2H); GC/EI-MS following derivatization with N,O-bis-(trimetylsilyl)trifluoroacetamide (BSTFA): m/z 372 (M); m/z 357 (M-CH3, base peak). Due to the unavoidable presence of contaminating salts, the concentration of PG was determined by [1H]-NMR spectroscopy with a known amount of dimethylsulfoxide as an internal standard (10,34) following repeated drying under vacuum and resuspension in [2H2]O (data not shown). The concentration of [2H2]-PG was determined by mixing with a known amount of unlabeled PG standard followed by GC-MS analysis.
Synthesis and characterization of an oligodeoxynucleotide containing a site-specific oxAB
As a standard for calibrating the analytical method, an oligodeoxynucleotide 5′-CTGAGCTCCAAAGYACCGGG-3′ (Y= oxAB; 1) was synthesized as previously described (39). The oligodeoxynucleotide was purified by HPLC (39) and the quantity of intact abasic sites determined by a [14C]-oxime derivatization method (36). Briefly, a 5 uM solution of 1 in 50 mM potassium phosphate (pH 7.4) was treated with 2 mM [14C]-methoxylamine (specific activity diluted to 5.5 mCi/mol with unlabeled methoxylamine) for 30 min at 37 °C. Unreacted methoxylamine was removed by two successive passages over Sephadex G-25 spin columns (Pharmacia). Following determination of the concentration of derivatized 1 spectroscopically by A260 and the quantity of [14C]-label by liquid scintillation counting, 1 equivalent of derivatized 1 was found to contain 1.9 equivalents of oxime-reactive carbonyl groups. This suggested that ~5% of 1 underwent hydrolysis during storage and handling, which was accounted for in subsequent analyses involving 1.
Synthesis of an oligodeoxynucleotide containing a 3PG
An oligodeoxynucleotide 5′-TGACTAGGGCCCGCAG-3′-PG (2) containing a 3PG was prepared from the corresponding 3′-phosphoglycerol-ended oligonucleotide (synthesized by Glen Research) by the method of Urata and Akagi (40). The final purity was determined to be >95% by denaturing gel electrophoresis and reversed phased HPLC analysis (data not shown), and the concentration was determined spectroscopically by A260.
Quantification of the hydroxymethylpyridazine derivative of the oxAB in oxidized DNA
Bleomycin treatments were performed in silylated glass vials containing Chelex-treated potassium phosphate buffer (50 mM, pH 7.4; 0.5 ml), calf thymus DNA (500 μg/mL) and 0–60 μM bleomycin, with 0.9 equivalents of freshly prepared Fe(NH4)2(SO4) solution added to initiate the reaction. Following 30 min at ambient temperature, [15N2]-HMP (400 pmol) was added as an internal standard. For treatment with ionizing radiation, silylated glass vials containing Chelex-treated potassium phosphate buffer (50 mM, pH 7.4; 0.5 ml) and calf thymus DNA (500 μg/mL) were exposed to 0–100 Gy of 60Co γ-radiation delivered at 1.35 Gy/min in a Gammacell-220 (MDS Nordion, Ottawa, ON, Canada) at ambient temperature in the presence of [15N2]-HMP, with samples incubated at ambient temperature for total exposure and incubation period of 30 min. Derivatization reactions were performed by adding hydrazine (20 μl, 1 M aqueous solution, 38 mM final concentration) and allowing the reaction to proceed at 25 °C for 30 min; the optimization of the hydrazine derivatization conditions was performed as previously described (data not shown; ref. 34). The pH was then adjusted to 5 by addition of sodium acetate buffer (33 μl, 3 M, pH 5.0) and the DNA hydrolyzed by the addition of zinc chloride (20 μl, 10 mM) and nuclease P1 (10 units), and incubation at 37 °C for 1.5 hr. Sodium acetate buffer (30 μl of 3 M, pH 7.4) and sodium hydroxide (20 μl of 2 M, adjusting pH to 8) were then added and phosphate groups were removed with alkaline phosphatase (20 units) and phosphodiesterase I (0.1 units) during incubation at 37 °C for 6 hr. The solution was filtered (Microcon YM-30 column) to remove enzymes and the pH was adjusted to 7 with 10 μl of 6 N HCl, followed by extraction with ethyl acetate (3 × 0.5 ml). The combined organic layers were dried under vacuum and the residue redissolved in 30 μl of 1:1 mixture of ethyl acetate and BSTFA containing 1% of trimethylchlorosilane, followed by incubation at ambient temperature for 30 min. The samples were transferred to 2 ml screw cap vials containing a 100 μl glass insert and from which 1 μl was subjected to GC/MS analysis with the following parameters: injector temperature, 250 °C; and column temperature, 70 °C for 4 min, followed by a linear gradient to 310 °C at 25 °C/min. The mass spectrometer was operated in the selective ion monitoring mode with m/z 167 for silylated HMP and m/z 169 for silylated [15N2]-HMP. Quantities of the oxAB were determined from the ratio of the areas of the HMP signal to that for [15N2]-HMP on the basis of linear calibration curves performed daily from samples containing from 0–200 pmol of HMP (m/z 167) plus 400 pmol of [15N2]-HMP (m/z 169).
Quantification of 3PG residues in oxidized DNA
DNA damage reactions with bleomycin and γ-radiation were performed exactly as described earlier for analysis of the oxAB. The samples were desalted by washing the DNA three times with 30 mM of ammonium acetate on YM-30 columns and resuspending the DNA in 0.45 mL of water. Following addition of [2H2]-glycolic acid internal standard (500 pmol), sodium acetate buffer (33 μl, 3 M, pH 5.0) and zinc chloride (20 μl, 10 mM), the DNA was hydrolyzed with nuclease P1 (10 units) at 37 °C for 1.5 hr. Protein was removed by filtration (Microcon YM-30 columns) followed by adjustment of the pH to 1.5 with 20 μl of 6 N HCl. The released PG was pre-purified by HPLC using the system described for the synthesis of PG, with collection of material eluting between 10.5 and 12.5 min (PG eluted after the void volume and before the nucleotides). The 11 solvent and volatile salts were removed under vacuum and the residue was taken up in 20 μl of BSTFA containing 1% of trimethylchlorosilane followed by incubation at ambient temperature for 30 min. The solutions were then analyzed by GC/MS as described earlier, with analysis at m/z 357 (silylated PG) and 359 (silylated [2H2]-PG) using the following parameters: column temperature, 75 °C for 3 min followed by a linear gradient to 300 °C at 50 °C/min. Quantities of PG were determined from calibration curves determined daily from 0–500 pmol of PG and 500 pmoles of [2H2]-PG.
Assay for total 2-deoxyribose oxidation events
A plasmid nicking assay was used to quantify 2-deoxyribose oxidation events caused by bleomycin or γ-radiation as a means to normalize the quantities of oxAB and 3PG. A 250 μg/ml solution of plasmid pUC18 DNA in Chelex-treated 50 mM potassium phosphate buffer (pH 7.4; 30 μl) was treated with 0–1.2 μM Fe+2-bleomycin or 0–0.3 Gy of γ-radiation as described earlier for a total exposure/incubation time of 30 min at ambient temperature. Under the conditions employed in the reactions of Fe+2-bleomycin with both calf thymus and plasmid DNA, plasmid nicking analysis revealed that the damage reaction was complete within several minutes, with no further DNA damage apparent during sample work up (data not shown). Following treatment, one-tenth volume of 1 M putrescine dihydrochloride was added and the solution was incubated at 37 °C for 30 min to cleave all types of abasic sites (e.g., ref. 41). The plasmid topoisomers present in the DNA sample (0.4 μg) were resolved on a 1% agarose gel at 2.5 V/cm for 1.5 h in Tris-borate-EDTA buffer (42). The gel was stained in a 0.5 μg/mL ethidium bromide solution for 1 h and then destained for 30 min in water, followed by acquisition of a digital image of the fluorescent DNA bands in the gel subjected to UV illumination (315 nm). The quantities of single- and double-strand breaks were calculated from the net increase in the percentage of nicked (form II) plasmid or linear (form III) plasmid, respectively, with a correction factor of 1.4 applied to the signal from the supercoiled (form I) plasmid due to its lower affinity for ethidium bromide (43).
RESULTS
Development and optimization of a GC/MS method to quantify the 2-deoxypentos-4-ulose abasic site
The overall strategy for the isotope-dilution GC/MS quantification of the oxAB and 3PG damage products is shown in Scheme 2. The instability of the oxAB (t1/2 ~20 hr at 37 °C; refs. 39,44) necessitated chemical stabilization in quantitative fashion, so we chose to convert it to a stable 3′-pyridazine moiety by derivatization with hydrazine as originally described by Hecht and coworkers (45). The pyridazine derivative is then released from DNA as 3′-phosphoro-3-pyridazinylmethylate by nuclease digestion and converted to ethyl acetate-extractable HMP by enzymatic dephosphorylation (Scheme 2). Ethyl acetate extracts containing both released HMP and added [15N2]-HMP internal standard were then silylated and analyzed by electron impact ionization GC/MS with selective ion monitoring at m/z 167 for silylated HMP and m/z 169 for silylated [15N2]-HMP. With only a two mass unit difference between the internal standard and the DNA-derived HMP, isotope natural abundance produced a [M+2] signal for HMP that contributed 6.26% to the signal for the internal standard molecular ion [M]. All analyses were corrected for this contribution.
Scheme 2.
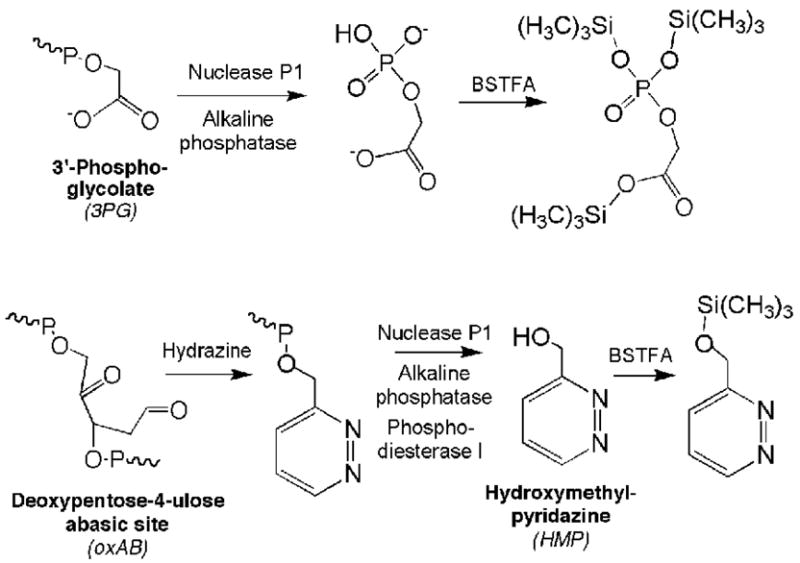
Approaches to analysis of 3′-phosphoglycolate (3PG) and the 2-deoxypentos-4-ulose abasic site (oxAB)
Each of the steps in the method was optimized for parameters such as concentration, reaction time, temperature and pH, as previously described (34). The ethyl acetate extraction of HMP (3X, pH 7) was 99.5% efficient and silylation was most efficient at 25 °C and 30 min reaction time with ratios of ethyl acetate to BSTFA between 50:50 to 70:30; there was a significant reduction in silylation when ethyl acetate was reduced below 30% (data not shown).
As shown in Supplementary Figure 1, linear calibration curves for the oxAB assay (r2 ≈ 0.99) were obtained under conditions identical to those used for sample analysis (50 mM Chelex-treated phosphate buffer, 500 μg/ml calf thymus DNA) by mixing varying amounts of HMP with 400 pmol of [15N2]-HMP followed by processing and GC/MS analysis. The calibration curves do not pass through the origin due to the fact that background peaks, introduced during work up, corresponding to a peak area ratio of 0.05, and having the same retention times as both of the HMP derivatives, were consistently observed for m/z 167. Since the quantity of HMP in samples is calculated from the equation of the best-fit straight line to the standard curve, this background level is subtracted. The limit of detection of the assay was determined to be 100 fmol, which translates to a sensitivity of 2 oxAB per 106 nucleotides in 250 μg of DNA.
The analytical method was further validated and the efficiency of HMP release determined with an oligodeoxynucleotide 5′-CTGAGCTCCAAAGYACCGGG-3′ with oxAB at position Y (39). Using a [14C]-oxime derivatization method (36), the freshly prepared oligodeoxynucleotide was found to be >95% pure in terms of intact abasic site. GC/MS analysis was performed on sets of samples in which varying amounts (1–400 pmol) of the oligodeoxynucleotide were mixed with 400 pmol of [15N2]-HMP and 250 μg of calf thymus DNA in a total volume of 0.5 ml. As shown in Figure 1, the amount of oxAB measured in the oligodeoxynucleotide was 72.6% (± 2.9) of the theoretical value, which represents the overall efficiency for the analytical method. The 27% difference may be due to incomplete derivatization of the abasic sites, incomplete enzymatic release of the HMP, or other factors. A correction factor of 0.73 was thus applied to all data.
Figure 1.
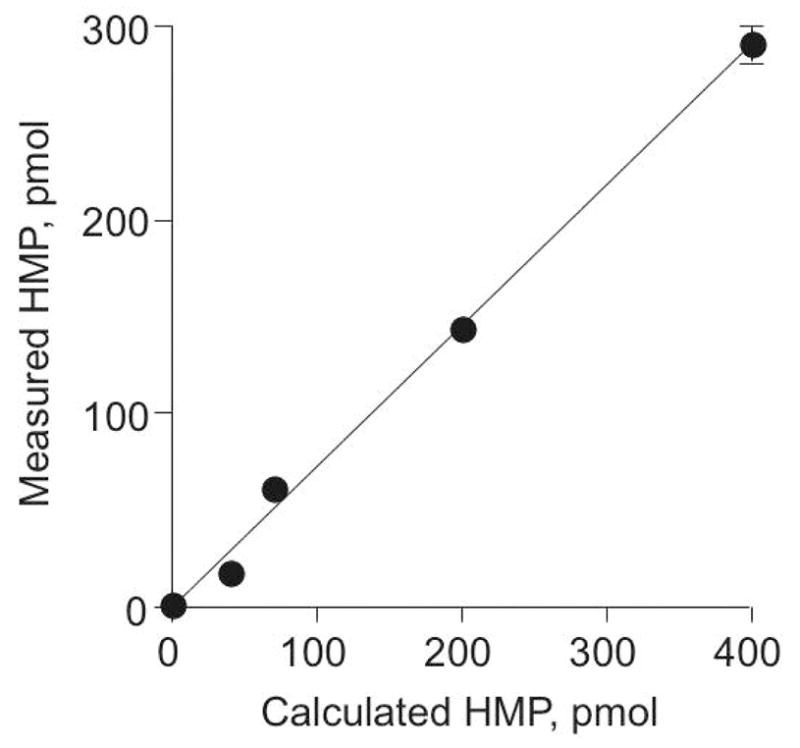
Validation of the GC/MS method for quantifying the oxAB using a synthetic oligodeoxynucleotide. Varying amounts (1–400 pmol) of an oligodeoxynucleotide containing a single oxAB were subjected to GC/MS analysis as described in Experiment Procedures, and the measured values compared to the expected quantity of oxAB. The data represent mean ± SD for samples analyzed in triplicate in three separate experiments and were fit by linear regression to an equation y = 0.726x + 0.163 with r2 = 0.996; the slope had SD = 0.029.
Development and optimization of a GC/MS method for the quantification of 3PG
As shown in Scheme 2, 3PG residues can be released from DNA by nuclease digestion and partially purified by HPLC prior to BSTFA silylation and GC/MS analysis, with [2H2]-PG added as internal standard prior to nuclease digestion. The enzymatic hydrolysis conditions were identical to those employed for the oxAB, while, on the basis of maximal MS signal, optimal silylation was found to occur when PG in the dried residue was treated with 100% BSTFA at ambient temperature (data not shown). Linear calibration curves for the GC/MS analysis of PG were obtained under conditions identical to those used for sample analysis (50 mM Chelex-treated phosphate buffer, 500 μg/ml calf thymus DNA) by mixing varying amounts of PG with 500 pmol of [2H2]-PG followed by processing and GC/MS analysis (Supplemental Figure 2). The limit of detection of the assay was 200 fmol or ~4 PG per 106 nt in 250 μg of DNA. The method was further validated using an oligodeoxynucleotide (5′-TGACTAGGGCCCGCAG-3′-PG) containing a 3PG, as described above for oxAB, with the amount of 3PG measured in the oligodeoxynucleotide >95% of the theoretical value (data not shown).
Formation of 3PG and oxAB in DNA treated with bleomycin and γ-radiation
The analytical methods were next applied to quantify the two chemical pathways of the 4′-oxidation of 2-deoxyribose in DNA. Samples of calf thymus DNA or plasmid pUC18 were treated with Fe+2-bleomycin or γ-radiation and oxAB and 3PG quantified by GC/MS. As shown in Figure 2, there is a linear dose-response for both DNA damage products and for both agents, with identical dose-response curves for calf thymus DNA and plasmid pUC18 (data not shown).
Figure 2.
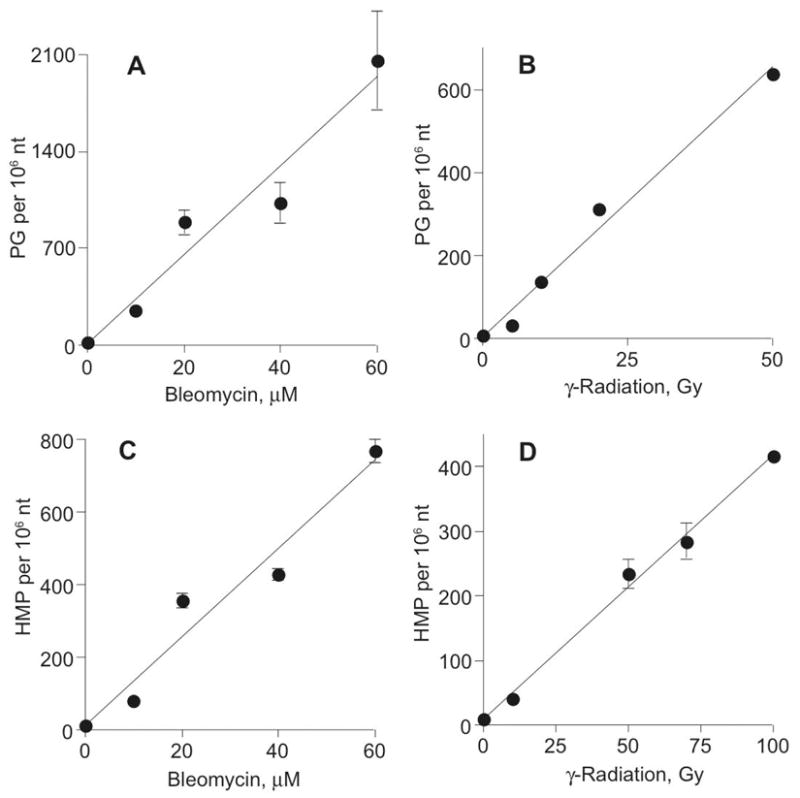
Quantification of oxAB and 3PG in DNA exposed to bleomycin (A, C) and γ-radiation (B, D). DNA was treated with varying doses of the oxidants and oxAB and 3PG were quantified as HMP (C, D) and PG (A, B), respectively, by GC/MS as described in Experimental Procedures. The data represent mean ± SD for three separate experiments; error bars are smaller than symbols in panel B. Linear regression of the data in each panel yielded lines with the following equations: A, y = 12.2x + 11.4 (r2 = 0.95); B, y = 4.10x + 8.5 (r2 = 0.99); C, y = 32.2x + 15.7 (r2 = 0.94); D, y = 13.0x + 10.7 (r2 = 0.99).
To determine the proportion of 2-deoxyribose oxidation events associated with 4′-oxidation chemistry by bleomycin and γ-radiation, we employed a plasmid nicking assay to estimate total 2-deoxyribose oxidation. This assay exploits the change in electrophoretic migration of plasmid DNA when a strand break relieves the negative supercoiling of a substrate plasmid molecule. Supercoiled plasmid pUC18 DNA was treated with bleomycin or γ-radiation under buffer and temperature conditions identical to those used for studies in calf thymus DNA, with abasic sites created by 2-deoxyribose oxidation (e.g., 2-deoxyribonolactone from 1′-oxidation, oxAB from 4′-oxidation; refs. 15,46) converted to strand breaks by treatment with putrescine dihydrochloride (22,37,41,47). Following gel electrophoretic separation of plasmid forms, the quantity of single-strand breaks and oxidized abasic sites was calculated from the proportion of nicked, circular DNA (form I) and the quantity of double-stranded lesions from the proportion of linearized plasmid (form III). As shown in Supplemental Figure 3, bleomycin was found to produce 43 (± 0.2) single-strand breaks and abasic sites per 106 nt per μM and 3 (± 0.3) double-strand breaks per 106 nt per μM. This ratio of 1 double-strand lesion for every 14 single-strand events with bleomycin is similar to previous observations (15,48). γ-Radiation produced 130 (±10) strand breaks and abasic sites per 106 nt per Gy and no double-strand breaks detectable in the plasmid nicking assay (data not shown), which is similar to the value of 141 determined previously in studies of radiation-induced 3′-phosphoglycolaldehyde formation (33).
The 2-deoxyribose oxidation data was then used to normalize the quantity of oxAB and 3PG produced by the oxidizing agents. As indicated by the slopes of lines fitted to data in Figure 3, bleomycin produced ~30 oxAB and ~80 3PG per 100 2-deoxyribose oxidation events. These values are similar to those determined by a plasmid nicking assay by Povirk and coworkers (49) and the sum of the two values is close to the 100% expected for bleomycin (48,50–52). For γ-radiation, the level of oxAB and 3PG fell to 3 and 10 per 100 2-deoxyribose oxidation events, respectively.
Figure 3.
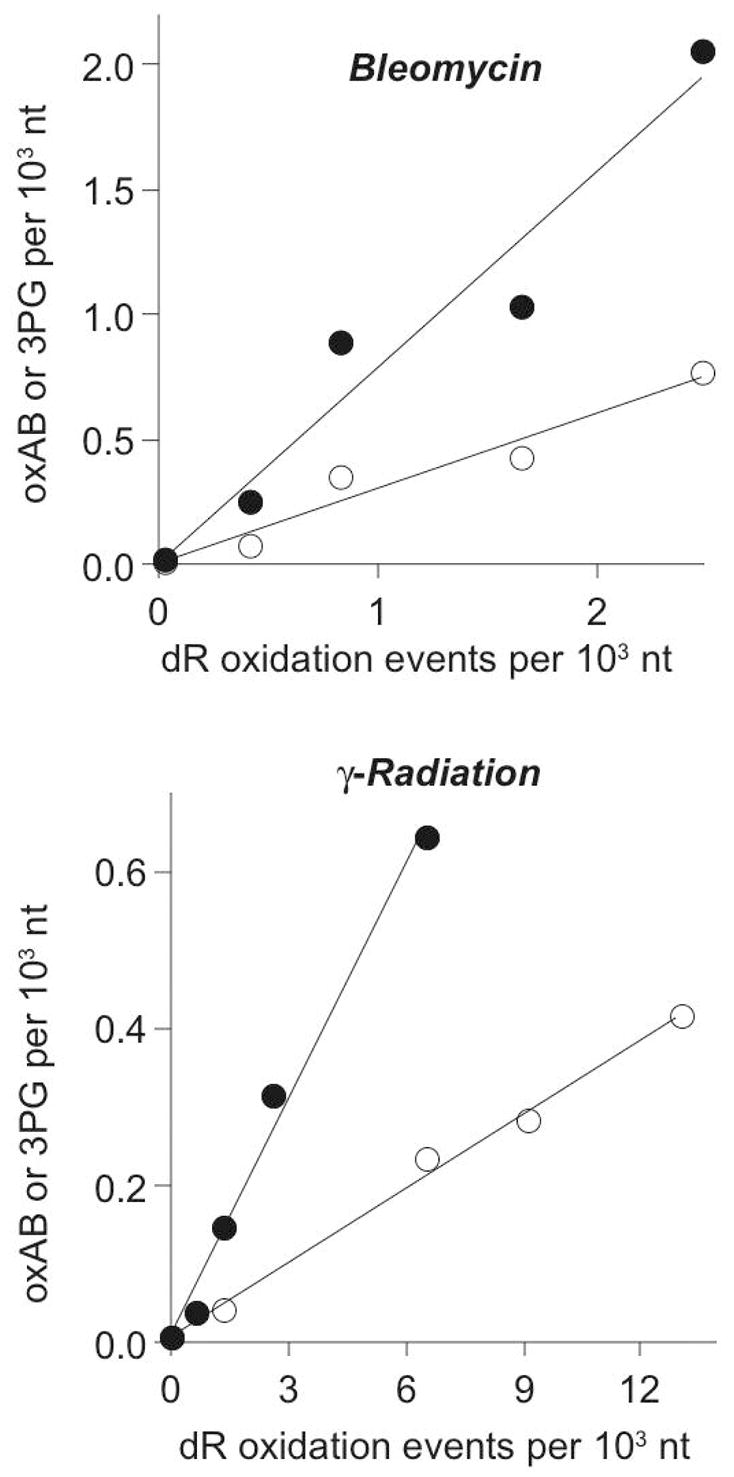
Frequency of 4′-oxidation products as a function of total 2-deoxyribose oxidation caused by bleomycin (upper) and γ-radiation (lower). The quantities of oxAB (○) and 3PG (●) were determined at varying doses of the oxidant and the data plotted relative to the total quantity of 2-deoxyribose oxidation measured by a plasmid nicking assay. Linear regression of the data in each panel yielded lines with the following equations: Upper Panel: oxAB, y = 0.3x + 0.006; 3PG, y = 0.79x + 0.007; Lower Panel: oxAB, y = 0.032x + 0.008; 3PG, y = 0.10x + 0.01.
DISCUSSION
There is emerging evidence pointing to 2-deoxyribose oxidation in DNA as an important determinant of the cellular response to oxidative stress, yet there are few tools available to characterize and quantify the various damage products under biological conditions or in cells. To this end, we have developed methods to quantify the 3PG and oxAB products arising from the two known pathways of 4′-oxidation of DNA to complement the growing number of methods to quantify 2-deoxyribose oxidation products (for example, see refs. 11,24–36). Previous efforts to develop sensitive analytical methods have focused solely on 3PG residues. For example, 3PG has been detected by [32P]-post-labeling of 3PG-containing nucleotides (28). While relatively sensitive, with fmol quantities of 3PG detected in 0.5 μg of DNA (28), the postlabeling method is semi-quantitative in nature due to the lack of internal standards and the need to analyze four separate signals representing 3PG residues attached to the four canonical nucleotides (28). Attempts to develop a GC/MS method for quantifying glycolic acid derived from 3PG residues in DNA (29,30) have been hampered by high background contamination of glycolic acid from a variety of sources (30).
The GC/MS methods developed here provide sensitive and specific assays for the two recognized pathways involved with 4′-oxidation of 2-deoxyribose in DNA, as shown in Scheme 1. The stabilization afforded by hydrazine derivatization of the oxAB, which was previously exploited in our studies of the 2-phosphoryl-1,4-dioxobutane residue derived from 5′-oxidation of 2-deoxyribose (34), provides for reproducible quantification of the oxAB with a limit of detection of 100 fmol and a sensitivity of ~2 lesions per 106 nt in 250 μg of DNA. Neither stability nor background contamination interfered with our approach to quantifying 3PG, for which the GC/MS method had a limit of detection of 200 fmol that translates into a sensitivity of 4 lesions per 106 nt in 250 μg of DNA. In both cases, the GC/MS assays for PG and HMP were validated with oligodeoxynucleotides containing site-specific 3PG and oxAB.
Several features of our approaches to developing GC/MS methods for the 3PG and oxAB have minimized the potential for adventitious oxidation damage that has been the subject of criticism in the use of GC/MS for biomarker quantification (53). For example, silylation of the analyte is often performed at high temperatures (70–90 °C) that can cause oxidatively damaged DNA. We have taken several steps to minimize adventitious DNA damage, first by performing the silylation reactions with BSTFA at ambient temperature. More importantly, however, is our use of HPLC pre-purification to isolate the PG and solvent extraction to purify the HMP derivative of the oxAB. The application of such precautions places GC/MS on par with LC/MS methods (54,55). Indeed, the accuracy and precision of our methods for quantifying 3PG and oxAB have been verified using oligodeoxynucleotides containing defined quantities of both lesions.
The combined application of the two analytical methods provides a window into the determinants of partitioning of 4′-oxidation chemistry in DNA. The results with bleomycin revealed that the oxAB and 3PG pathways occurred with a ratio of 3 to 8, which is consistent with the observations of Povirk and coworkers who used a plasmid nicking assay to show that ~40% of bleomycin-induced damage consisted of abasic sites (i.e., putrescine-sensitive lesions) and 60% direct strand breaks (49). By normalizing the 3PG and oxAB damage levels to the total quantity of 2-deoxyribose oxidation (Fig. 3), it was apparent that all of the 2-deoxyribose oxidation events were caused by 4′-oxidation. This is entirely consistent with previous studies that have identified 4′-oxidation as the sole chemistry performed by bleomycin (34,48,50–52).
The results with γ-radiation revealed a predominance of the 3PG pathway similar to bleomycin (Fig. 2). Indeed, our observation of 13 3PG lesions per 106 nt per Gy is in line with the 7 lesions per 106 nt Gy observed by Weinfeld and coworkers using a [32P] post-labeling method (28). However, we found that only 13% of all 2-deoxyribose oxidation events caused by γ-radiation involved 4′-chemistry (Fig. 3). This value is lower than expected from the solvent exposure model for hydroxyl radical reactivity with 2-deoxyribose in DNA proposed by Tullius and coworkers (56). In this model, which derives from observations made with Fe+2-EDTA, the 5′- and 4′-hydrogen atoms of 2-deoxyribose in B-DNA are significantly more solvent exposed and thus should be more reactive toward oxidation by hydroxyl radical (56). In light of the data compiled in Table 1, albeit an incomplete set of data, the reactivity of 2-deoxyribose toward γ-radiation-induced oxidants, which are primarily comprised of true hydroxyl radicals (12), is not solely determined by solvent accessibility, as is proposed to be the case for Fe+2-EDTA-generated oxidants (56). This is supported by the results of recent studies by Xue et al. (32) and by Roginskaya et al. (57), in which the 2-deoxyribonolactone arising from 1′-oxidation was found to account for the majority and >30%, respectively, of γ-radiation-induced 2-deoxyribose damage (32,57). However, Xue et al. also observed similarly high levels of the putative 2-deoxyribonolactone in DNA oxidized by Fe+2-EDTA using their biotinylated cysteine probe (32), a result that is at odds with the isotope effect studies (56). Explanations for the discrepancy include the lack of a primary isotope effect with 1′-oxidation arising from intramolecular oxidation caused by hydroxyl radical-induced nucleobase radicals (58,59) and possible lack of specificity of the probe due to reaction with other DNA oxidation products. Clearly, additional quantitative studies are needed to fill in the missing data and to provide a complete picture of the spectrum of products arising from 2-deoxyribose oxidation chemistry.
Table 1.
Quantities of γ-radiation-induced 2-deoxyribose oxidation products in DNA
| Species | Percent of Total 2-deoxyribose Oxidation | Reference | |
|---|---|---|---|
| 1′ | 2-Deoxyribonolactone abasic site | >30% | 32, 57 |
| 2′ | Erythrose abasic site? | ? | |
| 3′ | 3′-Phosphoglycolaldehyde | ~1% | 33, 35 |
| 3′-Oxo-nucleotide? | ? | ||
| 4′ | 3′-Phosphoglycolate | ~10% | Present studies |
| 2-Deoxypentos-4-ulose abasic site | ~3% | Present studies | |
| 5′ | 5′-Nucleoside-5′-aldehyde | ? | |
| 5′-(2-Phosphoryl-1,4-dioxobutane) | ~4% | 34 |
In conclusion, as part of a larger effort to systematically quantify 2-deoxyribose damage caused by different agents, we have developed and applied highly sensitive, accurate and precise GC/MS methods to measure lesions representing the two pathways involved in 4′-oxidation of 2-deoxyribose in DNA: 3PG and oxAB. While the application of the methods to damage produced by bleomycin and γ-radiation yielded similar proportions of the two damage pathways for the oxidants, the results considered in the context of total 2-deoxyribose oxidation revealed that 4′-oxidation accounted for only 13% of γ-radiation-induced 2-deoxyribose damage.
Supplementary Material
•Typical calibration curves for GC/MS analysis of phosphoglycolate and HMP.
•Graph of total 2-deoxyribose oxidation events produced by bleomycin in a plasmid nicking assay.
Acknowledgments
This work was supported by NIH grant CA26735. The authors thank Ms. Elaine Plummer for her assistance with GC/MS analyses performed in the Bioanalytical Facilities Core of the Center for Environmental Health Sciences at MIT, which is supported by NIEHS Center grant ES002109.
Footnotes
Abbreviations: BSTFA, N,O-bis-(trimetylsilyl)trifluoroacetamide; HMP, hydroxymethylpyridazine; oxAB, 2-deoxypentos-4-ulose abasic site; PG, 2-phosphoglycolate; 3PG, 3′-phosphoglycolate residues in DNA
References
- 1.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 2.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 4.Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto M, Greenberg MM, Kow YW, Hwang JT, Cunningham RP. The 2-deoxyribonolactone lesion produced in DNA by neocarzinostatin and other damaging agents forms cross-links with the base-excision repair enzyme endonuclease III. J Am Chem Soc. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- 6.DeMott MS, Beyret E, Wong D, Bales BC, Hwang JT, Greenberg MM, Demple B. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. J Biol Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 7.Awada M, Dedon PC. Formation of the 1, N2-glyoxal adduct of deoxyguanosine by phosphoglycolaldehyde, a product of 3′-deoxyribose oxidation in DNA. Chem Res Toxicol. 2001;14:1247–1253. doi: 10.1021/tx0155092. [DOI] [PubMed] [Google Scholar]
- 8.Gingipalli L, Dedon PC. Reaction of cis- and trans-2-butene-1,4-dial with 2′-deoxycytidine to form stable oxadiazabicyclooctaimine adducts. J Am Chem Soc. 2001;123:2664–2665. doi: 10.1021/ja0056421. [DOI] [PubMed] [Google Scholar]
- 9.Plastaras JP, Dedon PC, Marnett LJ. Effects of DNA structure on oxopropenylation by the endogenous mutagens malondialdehyde and base propenal. Biochemistry. 2002;41:5033–5042. doi: 10.1021/bi0113059. [DOI] [PubMed] [Google Scholar]
- 10.Bohnert T, Gingipalli L, Dedon PC. Reaction of 2′-deoxyribonucleosides with cis- and trans-1,4-dioxo-2-butene. Biochem Biophys Res Commun. 2004;323:838–844. doi: 10.1016/j.bbrc.2004.08.164. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X, Taghizadeh K, Dedon PC. Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J Biol Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 12.von Sonntag C. The chemical basis of radiation biology. Taylor & Francis; New York: 1987. [Google Scholar]
- 13.Dizdaroglu M, von Sonntag C, Schulte-Frohlinde D. Strand breaks and sugar release by gamma-irradiation of DNA in aqueous solution. J Am Chem Soc. 1975;97:2277–2278. doi: 10.1021/ja00841a051. [DOI] [PubMed] [Google Scholar]
- 14.Dizdaroglu M, Schulte-Frohlinde D, von Sonntag C. Strand breaks and sugar release by gamma-irradiation of DNA in aqueous solution. The effect of oxygen. Z Naturforsch. 1975;30C:826–828. [Google Scholar]
- 15.Dedon PC, Goldberg IH. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem Res Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 16.Rashid R, Langfinger D, Wagner R, Schuchmann HP, von Sonntag C. Bleomycin versus OH-radical-induced malonaldehydic-product formation in DNA. Int J Radiat Biol. 1999;75:101–109. doi: 10.1080/095530099140852. [DOI] [PubMed] [Google Scholar]
- 17.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc Natl Acad Sci U S A. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Vu CC, Byrns MC, Dedon PC, Peterson LA. Formation of 1,4-dioxo-2-butene-derived adducts of 2′-deoxyadenosine and 2′-deoxycytidine in oxidized DNA. Chem Res Toxicol. 2006;19:982–985. doi: 10.1021/tx0601197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrns MC, Vu CC, Neidigh JW, Abad JL, Jones RA, Peterson LA. Detection of DNA adducts derived from the reactive metabolite of furan, cis-2-butene-1,4-dial. Chem Res Toxicol. 2006;19:414–420. doi: 10.1021/tx050302k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrns MC, Predecki DP, Peterson LA. Characterization of nucleoside adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 2002;15:373–379. doi: 10.1021/tx0101402. [DOI] [PubMed] [Google Scholar]
- 21.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int J Radiat Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 22.Povirk LF, Goldberg IH. Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc Natl Acad Sci U S A. 1985;82:3182–3186. doi: 10.1073/pnas.82.10.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Povirk LF, Houlgrave CW. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: Specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry. 1988;27:3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhry MA, Dedon PC, Wilson DM, 3rd, Demple B, Weinfeld M. Removal by human apurinic/apyrimidinic endonuclease 1 (Ape 1) and Escherichia coli exonuclease III of 3′-phosphoglycolates from DNA treated with neocarzinostatin, calicheamicin, and gamma-radiation. Biochem Pharmacol. 1999;57:531–538. doi: 10.1016/s0006-2952(98)00327-x. [DOI] [PubMed] [Google Scholar]
- 25.Weinfeld M, Lee J, Ruiqi G, Karimi-Busheri F, Chen D, Allalunis-Turner J. Use of a postlabelling assay to examine the removal of radiation-induced DNA lesions by purified enzymes and human cell extracts. Mutat Res. 1997;378:127–137. doi: 10.1016/s0027-5107(97)00103-6. [DOI] [PubMed] [Google Scholar]
- 26.Jones GD, Boswell TV, Lee J, Milligan JR, Ward JF, Weinfeld M. A comparison of DNA damages produced under conditions of direct and indirect action of radiation. Int J Radiat Biol. 1994;66:441–445. doi: 10.1080/09553009414551431. [DOI] [PubMed] [Google Scholar]
- 27.Weinfeld M, Buchko GW. Postlabelling methods for the detection of apurinic sites and radiation- induced DNA damage. IARC Sci Publ. 1993;124:95–103. [PubMed] [Google Scholar]
- 28.Weinfeld M, Soderlind KJ. 32P-postlabeling detection of radiation-induced DNA damage: identification and estimation of thymine glycols and phosphoglycolate termini. Biochemistry. 1991;30:1091–1097. doi: 10.1021/bi00218a031. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Murugaiah V, Yeung B, Vouros P, Giese RW. 2-Phosphoglycolate and glycolate-electrophore detection, including detection of 87 zeptomoles of the latter by gas chromatography-electron-capture mass spectrometry. J Chomatogr. 1996;721:289–296. [Google Scholar]
- 30.Shao G, Giese RW. Trace detection of glycolic acid by electrophore labeling gas chromatography-electron capture mass spectrometry. Anal Chem. 2004;76:3049–3054. doi: 10.1021/ac0304267. [DOI] [PubMed] [Google Scholar]
- 31.Sato K, Greenberg ME. Selective detection of 2-deoxyribonolactone in DNA. J Am Chem Soc. 2005;127:2806–2807. doi: 10.1021/ja0426185. [DOI] [PubMed] [Google Scholar]
- 32.Xue L, Greenberg MM. Use of fluorescence sensors to determine that 2-deoxyribonolactone is the major alkali-labile deoxyribose lesion produced in oxidatively damaged DNA. Angew Chem Int Ed Engl. 2007;46:561–564. doi: 10.1002/anie.200603454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins C, Awada MM, Zhou X, Dedon PC. Analysis of 3′-phosphoglycolaldehyde residues in oxidized DNA by gas chromatography/negative chemical ionization/mass spectrometry. Chem Res Toxicol. 2003;16:1560–1566. doi: 10.1021/tx0341198. [DOI] [PubMed] [Google Scholar]
- 34.Chen B, Bohnert T, Zhou X, Dedon PC. 5′-(2-Phosphoryl-1,4-dioxobutane) as a product of 5′-oxidation of deoxyribose in DNA: elimination as trans-1,4-dioxo-2-butene and approaches to analysis. Chem Res Toxicol. 2004;17:1406–1413. doi: 10.1021/tx049818e. [DOI] [PubMed] [Google Scholar]
- 35.Collins C, Zhou X, Wang R, Barth MC, Jiang T, Coderre JA, Dedon PC. Differential oxidation of deoxyribose in DNA by gamma- and alpha-particle radiation. Radiat Res. 2005;163:654–662. doi: 10.1667/rr3344. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Liberman RG, Skipper PL, Margolin Y, Tannenbaum SR, Dedon PC. Quantification of DNA strand breaks and abasic sites by oxime derivatization and accelerator mass spectrometry: application to gamma-radiation and peroxynitrite. Anal Biochem. 2005;343:84–92. doi: 10.1016/j.ab.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 37.LaMarr WA, Sandman KM, Reeve JN, Dedon PC. Differential effects of DNA supercoiling on radical-mediated DNA strand breaks. Chem Res Toxicol. 1997;10:1118–1122. doi: 10.1021/tx970072c. [DOI] [PubMed] [Google Scholar]
- 38.Clauson-Kaas N, Limborg N. Nuclear oxidation of silvan and furfuryl alcohol. Acta Chem Scand. 1947;1:619–623. doi: 10.3891/acta.chem.scand.01-0884. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Stubbe J. Synthesis and characterization of oligonucleotides containing a 4′-keto abasic site. Biochemistry. 2004;43:5278–5286. doi: 10.1021/bi0495376. [DOI] [PubMed] [Google Scholar]
- 40.Urata H, Akagi M. A convenient synthesis of oligonucleotides with a 3′-phosphoglycolate and 3′-phosphoglycoaldehyde terminus. Tetrahedron Lett. 1993;34:4015–4018. [Google Scholar]
- 41.Dedon PC, Salzberg AA, Xu J. Exclusive production of bistranded DNA damage by calicheamicin. Biochemistry. 1993;32:3617–3622. doi: 10.1021/bi00065a013. [DOI] [PubMed] [Google Scholar]
- 42.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley and Sons; New York: 1989. [Google Scholar]
- 43.Milligan JR, Aguilera JA, Ward JF. Variation of single-strand break yield with scavenger concentration for plasmid DNA irradiated in aqueous solution. Radiat Res. 1993;133:151–157. [PubMed] [Google Scholar]
- 44.Kim J, Gil JM, Greenberg MM. Synthesis and characterization of oligonucleotides containing the c4′-oxidized abasic site produced by bleomycin and other DNA damaging agents. Angew Chem Int Ed Engl. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- 45.Sugiyama H, Xu C, Murugesan N, Hecht SM. Structure of the alkali-labile product formed during iron(II)-bleomycin-mediated DNA strand scission. J Am Chem Soc. 1985;107:4104–4105. [Google Scholar]
- 46.Pogozelski WK, Tullius TD. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen atom abstraction from the sugar moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 47.Kennedy LJ, Moore K, Jr, Caulfield JL, Tannenbaum SR, Dedon PC. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem Res Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- 48.Absalon MJ, Wu W, Kozarich JW, Stubbe J. Sequence-specific double-strand cleavage of DNA by Fe-bleomycin. 2. Mechanism and dynamics. Biochemistry. 1995;34:2076–2086. doi: 10.1021/bi00006a030. [DOI] [PubMed] [Google Scholar]
- 49.Bennett RO, Swerdlow PS, Povirk LF. Spontaneous cleavage of bleomycin-induced abasic sites in chromatin and their mutagenicity in mammalian shuttle vectors. Biochemistry. 1993;32:3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- 50.Kozarich JW, Worth L, Frank BL, Christner DF, Vanderwall DE, Stubbe J. Sequence-specific isotope effects on the cleavage of DNA by bleomycin. Science. 1989;245:1396–1399. doi: 10.1126/science.2476851. [DOI] [PubMed] [Google Scholar]
- 51.McGall GH, Stubbe J. Mechanistic studies of bleomycin-mediated DNA cleavage using isotope labeling. Nucleic Acids Mol Biol. 1988;2:85–104. [Google Scholar]
- 52.Stubbe J, Kozarich JW. Mechanisms of bleomycin-Induced DNA degradation. Chem Rev. 1987;87:1107–1136. [Google Scholar]
- 53.Cadet J, D’Ham C, Douki T, Pouget JP, Ravanat JL, Sauvaigo S. Facts and artifacts in the measurement of oxidative base damage to DNA. Free Radic Res. 1998;29:541–550. doi: 10.1080/10715769800300581. [DOI] [PubMed] [Google Scholar]
- 54.Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 55.Dizdaroglu M. Facts about the artifacts in the measurement of oxidative DNA base damage by gas chromatography-mass spectrometry. Free Radic Res. 1998;29:551–563. doi: 10.1080/10715769800300591. [DOI] [PubMed] [Google Scholar]
- 56.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci U S A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roginskaya M, Bernhard WA, Marion RT, Razskazovskiy Y. The release of 5-methylene-2-furanone from irradiated DNA catalyzed by cationic polyamines and divalent metal cations. Radiat Res. 2005;163:85–89. doi: 10.1667/rr3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong IS, Carter KN, Sato K, Greenberg MM. Characterization and mechanism of formation of tandem lesions in DNA by a nucleobase peroxyl radical. J Am Chem Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]
- 59.Tallman KA, Greenberg MM. Oxygen-dependent DNA damage amplification involving 5,6-dihydrothymidin-5-yl in a structurally minimal system. J Am Chem Soc. 2001;123:5181–5187. doi: 10.1021/ja010180s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
•Typical calibration curves for GC/MS analysis of phosphoglycolate and HMP.
•Graph of total 2-deoxyribose oxidation events produced by bleomycin in a plasmid nicking assay.


