Abstract
ADP ribosylation factor (ARF) is thought to play a critical role in recruiting coatomer (COPI) to Golgi membranes to drive transport vesicle budding. Yeast strains harboring mutant COPI proteins exhibit defects in retrograde Golgi to endoplasmic reticulum protein transport and striking cargo-selective defects in anterograde endoplasmic reticulum to Golgi protein transport. To determine whether arf mutants exhibit similar phenotypes, the anterograde transport kinetics of multiple cargo proteins were examined in arf mutant cells, and, surprisingly, both COPI-dependent and COPI-independent cargo proteins exhibited comparable defects. Retrograde dilysine-mediated transport also appeared to be inefficient in the arf mutants, and coatomer mutants with no detectable anterograde transport defect exhibited a synthetic growth defect when combined with arf1Δ, supporting a role for ARF in retrograde transport. Remarkably, we found that early and medial Golgi glycosyltransferases localized to abnormally large ring-shaped structures. The endocytic marker FM4–64 also stained similar, but generally larger ring-shaped structures en route from the plasma membrane to the vacuole in arf mutants. Brefeldin A similarly perturbed endosome morphology and also inhibited transport of FM4–64 from endosomal structures to the vacuole. Electron microscopy of arf mutant cells revealed the presence of what appear to be hollow spheres of interconnected membrane tubules which likely correspond to the fluorescent ring structures. Together, these observations indicate that organelle morphology is significantly more affected than transport in the arf mutants, suggesting a fundamental role for ARF in regulating membrane dynamics. Possible mechanisms for producing this dramatic morphological change in intracellular organelles and its relation to the function of ARF in coat assembly are discussed.
INTRODUCTION
ADP ribosylation factor (ARF) was originally identified as a protein cofactor required for efficient ADP ribosylation of heterotrimeric Gsα by cholera toxin (Kahn and Gilman, 1984). It was later shown to be a small GTP-binding protein with structural similarity to both monomeric and heterotrimeric G proteins (reviewed in Boman and Kahn, 1995). In eukaryotes, ARF is abundant and ubiquitous, representing approximately 0.03–1% of total cell protein, and is highly conserved across phylogenetic lines. For example, human and Saccharomyces cerevisiae ARF proteins share 74% amino acid sequence identity (Sewell and Kahn, 1988). In S. cerevisiae, ARF is encoded by two genes: ARF1 and ARF2, which encode proteins with 96% identity and are probably redundant in function (Stearns et al., 1990a). ARF3 encodes a protein 52% identical to Arf1p (Lee et al., 1994). Double arf1Δ arf2Δ mutants are inviable, indicating that these genes provide an essential function that cannot be carried out by ARF3. The Arf2 protein is only expressed at 10% the level of Arf1 protein, and strains deleted for the ARF2 gene show a wild-type phenotype. However, deletion of ARF1 causes strains to exhibit modest defects in protein secretion and modification. This is one initial line of evidence that suggested a role for ARF in the secretory pathway (Stearns et al., 1990a,b).
ARF proteins have been suggested to be important regulatory factors for several membrane trafficking events, including exocytic protein transport [endoplasmic reticulum (ER)-Golgi and intra-Golgi], endocytosis, and nuclear envelope assembly (Boman and Kahn, 1995). Among these processes, the best characterized function of ARF is to facilitate budding of nonclathrin-coated transport vesicles from purified Golgi membranes. Activation of ARF by a Golgi-localized guanine nucleotide exchange factor causes ARF-GTP to bind the Golgi membrane which leads to the recruitment of coatomer, a heptameric (α, β, β’, γ, δ, ε, ζ) complex of coat proteins (COPI), and budding of a vesicle (Rothman and Wieland, 1996). The ARF guanine nucleotide exchange factor is inhibited by brefeldin A (BFA) (Morinaga et al., 1996; Peyroche et al., 1996), a potent inhibitor of protein transport in mammalian cells (Takatsuki and Tamura, 1985) and yeast (Graham et al., 1993). ARF is also an activator of phospholipase D (PLD) (Brown et al., 1993; Cockcroft et al., 1994), an enzyme that cleaves phosphatidylcholine into choline and phosphatidic acid, and it has recently been suggested that PLD activation is primarily responsible for coatomer recruitment to Golgi membranes (Ktistakis et al., 1996). The precise mechanism that ARF plays in recruiting AP1/Clathrin coats to the trans-Golgi network for producing clathrin-coated vesicles (Stamnes and Rothman, 1993; Traub et al., 1993) is also unknown.
Although it seems clear that COPI is involved in vesicle-mediated protein transport, the direction these vesicles travel is controversial. Biochemical analysis using an in vitro protein transport assay suggested that COPI-coated vesicles mediate cis-Golgi to medial Golgi protein transport (Waters et al., 1991; Ostermann et al., 1993). However, a carboxy-terminal dilysine (KKXX) motif that mediates retrieval of type I ER membrane proteins from the Golgi back to the ER interacts specifically with COPI (Cosson and Letourneur, 1994), suggesting that COPI is involved in retrograde transport. Moreover, all coatomer mutants characterized thus far in yeast exhibit defects in KKXX protein retrieval (Letourneur et al., 1994; Cosson et al., 1996), but only a subset of these mutants are defective for anterograde (forward) transport (Hosobuchi et al., 1992; Duden et al., 1994). Surprisingly, mutants harboring temperature-sensitive alleles of SEC21 (γ-COP) were recently shown to exhibit dramatic protein-specific anterograde transport defects at the nonpermissive temperature. Cargo proteins such as carboxypeptidase Y (CPY) and pro-α-factor accumulated in the ER of sec21–3 cells, but invertase and HSP150 were secreted with normal transport kinetics from the same cells. These data suggest that the effect of COPI on anterograde transport is likely to be indirect, and that COPI mutants may fail to retrieve proteins back to the ER which are required to package specific cargo proteins into transport vesicles budding from the ER (Gaynor and Emr, 1997).
Established biochemical evidence linking the COPI complex with ARF (Orci et al., 1993; Ostermann et al., 1993), the fact that both arf1Δ and COPI mutants exhibit Golgi-specific glycosylation and ER to Golgi transport defects (Stearns et al., 1990b; Gaynor and Emr, 1997), and a previous report which indicated that sec21–1/arf1Δ double mutants were usually inviable (Stearns et al., 1990b) prompted us to investigate whether arf mutant strains (i.e., arf1Δ and arf1–3 ts arf2Δ strains) would also exhibit cargo-specific anterograde and/or retrograde transport defects. Unexpectedly, all anterograde cargo proteins assayed in the arf mutants exhibited similar kinetic defects in transport through multiple steps of the secretory pathway. Pulse-chase and genetic analyses indicated that the arf mutants were also partially defective for COPI-mediated Golgi to ER retrieval. Morphological studies revealed marked perturbations in Golgi, endosome, and vacuolar membranes in arf mutants. Most striking was the appearance of ring-like structures likely to correspond to abnormal Golgi and endosomal compartments. The dramatic morphological changes in multiple membrane structures in the arf mutants suggest that although ARF clearly affects COPI-mediated transport events, the primary role of ARF in the cell may instead be maintenance of normal membrane dynamics and composition.
MATERIALS AND METHODS
Strains, Plasmids, and Media
Yeast strains and plasmids used in this study are listed in Table 1. 6210arf1Δ and 6211arf1Δ were constructed by transforming SEY6210 and SEY6211 with the arf1::HIS3 allele carried on an EcoRI–SpeI fragment from pRB1293. Disruption of the chromosomal ARF1 gene was confirmed by polymerase chain reaction. A Western blot loaded with equal amounts of protein from SEY6210 and 6210 arf1Δ and probed with ARF antibodies confirmed an approximate 10-fold reduction of ARF protein in the 6210arf1Δ strain, as previously reported in a different strain background (Stearns, 1990a). The arf1–3 allele was a gift from Dr. Richard A. Kahn (Cavenagh and Kahn, unpublished data). All other strains were constructed by standard yeast genetic techniques. pKE2018 is a pSEY18-based plasmid (Herman and Emr, 1990) carrying KEX2 on a 4.71-kb EcoRI–BamHI fragment (gift from Paul Herman, Ohio State University, Columbus, OH). Yeast cells were grown on yeast extract, peptone, and dextrose (YPD) or synthetic minimal (SD) media supplemented as necessary (Sherman, 1991). Cells were grown in liquid SD media containing 0.2% yeast extract and required supplements for metabolic labeling experiments.
Table 1.
List of yeast strains and plasmids used in this study
| Strain/plasmid | Description | Reference |
|---|---|---|
| Strain | ||
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al. (1988) |
| 6210 arf1Δ | SEY6210 arf1::HIS3 | This study |
| SEY6211 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 | Robinson et al. (1988) |
| 6211 arf1Δ | SEY6211 arf1::HIS3 | This study |
| SEY6210.5 | SEY6210 × SEY6211 diploid | This study |
| 6210.5 arf1Δ | 6210 arf1Δ × 6211 arf1Δ diploid | This study |
| TGY122 | SEY6210 mnn1::LEU2 | This study |
| PSY315 | MATa leu2-3,112 his3-Δ200 lys2-801 | Kahn et al. (1995) |
| TT104 | MATa leu2-3,112 his3-Δ200 lys2-801 arf1::HIS3 | Kahn et al. (1995) |
| C156-1B | MATα arf1-3 arf2::LEU2 leu2-3,112 ura3-52 his3-Δ200 lys2-801 | R.A. Kahn |
| EGY101-16D | MATa ret1-1 leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 | Letourneur et al. (1994) |
| EGYA1011-3C | Matα ret1-1 leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 | This study |
| suc2-Δ9 arf1::HIS3 | ||
| EGY1211-6B | MATa sec21-1 leu2-3,112 ura3-52 his3-Δ200 suc2-Δ9 | This study |
| EGYA1211-1 | MATα sec21-1 leu2-3,112 ura3-52 his3-Δ200 lys2-801 suc2-Δ9 | This study |
| arf1::HIS3 | ||
| EGY103-19D | MATa sec21-2 leu2-3,112 ura3-52 his3-Δ200 lys2-801 suc2-Δ9 | Letourneur et al. (1994) |
| EGYA1212-12C | MATα sec21-2 leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 | This study |
| suc2-Δ9 arf1::HIS3 | ||
| EGY1271-7B | MATa sec27-1 leu2-3,112 ura3-52 his3-Δ200 lys-801 suc2-Δ9 | This study |
| EGYA1271-2D | MATα sec27-1 leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 | This study |
| suc2-Δ9 arf1::HIS3 | ||
| TGY413-6D | MATα ise1 leu2-3,112 ura3-52 his3-Δ200 suc2-Δ9 | Graham et al. (1993) |
| Plasmid | ||
| pRB1293 | arf1::HIS3 | Stearns et al. (1990) |
| pZV236 | MNN1 LEU2 2μ | Graham et al. (1994) |
| pKE2018 | KEX2 URA3 2μ | This study |
| pOH URA3 | OCH1::HA URA3 CEN | Harris and Waters (1996) |
| pEG1-KK | SUC2-WBP1-KK (URA3 CEN) | Gaynor et al. (1994) |
Cell Labeling, Subcellular Fractionation, Immunological Techniques, and Digital Imaging
Pulse-chase metabolic labeling of cells with 35S-amino acids, immunoprecipitations, and endoglycosidase H treatment of immunoprecipitates were done as described previously (Gaynor et al., 1994). Except where marked with an asterisk, the transport half-times reported in Table 2 were from pulse-chase experiments using strains SEY6210 (wild-type) and 6210arf1Δ (arf1) performed at 30°C with a 10-min labeling period and chase times of 0, 10, and 30 min for alkaline phosphatase (ALP) and carboxypeptidase S (CPS), and a chase of 0, 2.5, 5, and 10 min for invertase and HSP150. Those marked with a single asterisk were from pulse-chase experiments using strains PSY315 (wild-type) or C156–1B (arf1) preincubated for 30 min at 38°C, labeled for 5 min, and chased for 0, 2, 5, 10, and 20 min. To examine the secretion of invertase to the periplasmic space, cells were converted to spheroplasts after pulse-chase labeling as described previously (Gaynor and Emr, 1997). The pulse-chase procedure used to determine half-times for CPY and pro-α-factor are described in the legend to Figure 1. NIH Image software was used to quantitate band intensities from scanned, digitized images of autoradiograms. To determine half-times of transport, the log [100 (precursor/precursor + mature)] was plotted versus chase time for vacuolar proteins. The log [100 (internal/internal + external)] was used for invertase and HSP150, and the log [100 (glycosylated precursors/glycosylated precursors at 0 min)] was used for pro-α-factor.
Table 2.
Protein transport kinetics in wild-type and arf1 mutant cells
| Protein | Transport half-time (min)
|
Fold difference | |
|---|---|---|---|
| wild-type | arf1 | ||
| COPI-dependent | |||
| CPY | 5.5 | 14.5 | 2.6 |
| CPY* | 7.8 | 16.2 | 2.1 |
| ALP | 3.5 | 9.5 | 2.7 |
| CPS | 5.0 | 13.0 | 2.6 |
| Pro-α-factor** | 2.0 | 8.0 | 4.0 |
| COPI-independent | |||
| HSP150 | ∼1.0 | 2.5 | 2.5 |
| HSP150* | 1.4 | 4.8 | 3.4 |
| Invertase | ∼1.0 | 4.5 | 4.5 |
*Performed at 38°C in strain C156-1B (arf1-3).
**Performed at 20°C. The methods used for pulse-chase labeling and determining half-times are described in MATERIALS AND METHODS.
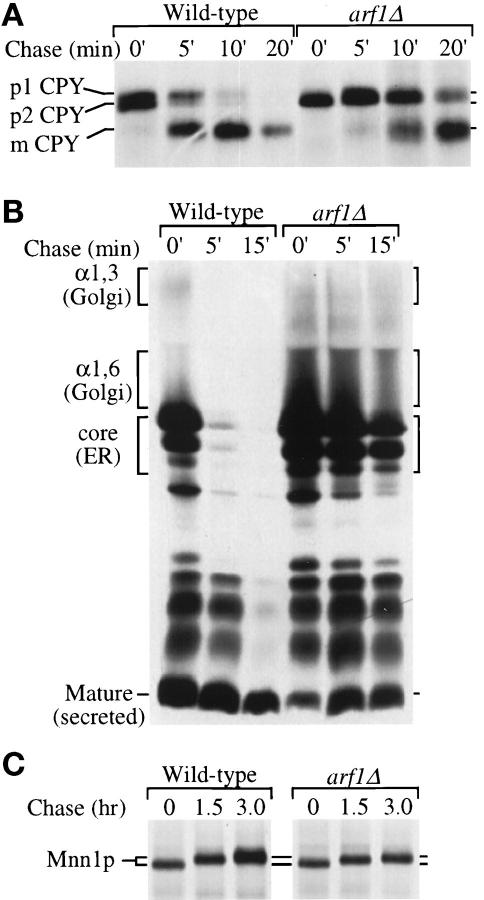
Figure 1.
Anterograde protein transport kinetics and modification in wild-type and arf1Δ cells. (A) Wild-type (PSY315) and arf1Δ (TT104) cells were labeled with 35S-amino acids for 5 min and chased for 20 min at 30°C. Equal aliquots of cells were removed at the times indicated and the chase was terminated by addition of trichloroacetic acid (TCA). CPY was recovered from the samples by immunoprecipitation and subjected to SDS-PAGE. The positions of the ER (p1), Golgi (p2), and vacuolar (m) CPY forms are indicated. Similar results were obtained using the isogenic strains SEY6210 (wild-type) and 6210 arf1Δ, which were used for the remainder of the pulse-chase experiments. (B) Cells were labeled for 10 min at 20°C and chased for 15 min. The chase was terminated at the times indicated, and α-factor was recovered by immunoprecipitation and subjected to SDS-PAGE. The position of ER (core), early Golgi (α1,6), medial Golgi (α1,3), and late Golgi-secreted (mature) α-factor forms are indicated. (C) Cells were labeled for 15 min and chased for 3 h at 30°C. The chase was terminated at the times indicated and Mnn1p was recovered by immunoprecipitation and subjected to SDS-PAGE.
Subcellular fractionation (Gaynor et al., 1994) (with modifications listed in Gaynor and Emr, 1997), Western blot analysis (Graham and Krasnov, 1995), immunofluorescence analyses (Graham et al., 1994), cell labeling with FM4–64 (Vida and Emr, 1995), BFA treatment of cells (Graham et al., 1993), and electron microscopy (Rieder et al., 1996) were performed as described previously. Any modification of these standard protocols are provided in the figure legends. Antisera to CPY (Klionsky et al., 1988), α-factor (Graham and Emr, 1991), α1–6 and α1–3 mannose linkages (Franzusoff and Schekman, 1989), Mnn1p (Graham et al., 1994), invertase (Gaynor et al., 1994), alkaline phosphatase (Seeger and Payne, 1992), HSP150 (Russo et al., 1992), and CPS (Cowles et al., 1997) have been described previously. Antiserum to Kex2p was prepared at Scantibodies Laboratory (Ramona, CA) against a bacterially expressed Kex2p-β-galactosidase fusion protein and affinity purified as described previously (Redding et al., 1991). Secondary antibodies were from Jackson Immunoresearch (West Grove, PA). Images of cells were acquired using a Zeiss Axioplan fluorescence microscope and a Hamamatsu C4880 cooled CCD camera controlled through MetaMorph software (Universal Imaging, West Chester, PA). Images were printed using a Tektronix Phaser IISDX printer.
RESULTS
arf Mutants Exhibit a General Kinetic Delay in Protein Transport
Previous studies of protein transport in arf1Δ mutants showed a slight intracellular accumulation and underglycosylation of invertase (Stearns et al., 1990b). Since the rate of invertase secretion is not affected in temperature conditional COPI mutants that exhibit a complete block in the transport of a subset of cargo proteins such as CPY and α-factor (Gaynor and Emr, 1997), we reasoned that arf1Δ mutants may show a more substantial transport defect for other proteins. To clarify the extent and specificity of protein transport defects in arf1Δ mutants, we monitored the transport kinetics of several marker proteins through the secretory pathway by pulse-chase and immunoprecipitation analysis.
The vacuolar hydrolase CPY is initially synthesized in the ER as a core-glycosylated p1 proenzyme, which is then converted to a p2 form in the Golgi complex by modification of core oligosaccharides and subsequently processed to the mature form (mCPY) upon arrival in the vacuole (Stevens et al., 1982). After a 5-min pulse-labeling period, an equivalent amount of labeled p1 and p2CPY was seen in the wild-type cells (Figure 1A, 0′). These CPY precursors were efficiently converted to the mature form during the chase period with a half-time of ∼5 min (Figure 1A and Table 2). In the arf1Δ mutant, only p1CPY was formed during the 5-min labeling period, suggesting that CPY had not yet been transported out of the ER. A CPY precursor form with slower mobility was formed during the chase, but fully glycosylated p2CPY was not observed (Figure 1A). The half-time for processing CPY to the mature form in the mutant was 14.5 min, approximately 2.5-fold slower than the half-time for CPY processing in wild-type cells (Table 2).
The transport kinetics of two integral membrane vacuolar proteins, ALP (Klionsky and Emr, 1989) and CPS (Spormann et al., 1992), and the secreted proteins invertase (Esmon et al., 1981) and HSP 150 (Russo et al., 1992) were also examined by pulse-chase analysis in wild-type and arf1Δ cells. The half-time was determined for proteolytic processing of ALP and CPS in the vacuole and for appearance of invertase and HSP150 in the media. Surprisingly, all of these proteins were transported through the secretory pathway of arf1Δ cells at a similar 2.5- to 4.5-fold slower rate than in wild-type cells (Table 2). The extremely rapid transport of HSP150 and invertase presented a problem in determining a precise half-time of secretion in wild-type cells, but these proteins were clearly transported more slowly in the arf1Δ cells. The primary conclusion from this analysis is that loss of Arf1p caused a general anterograde protein transport defect for both COPI-dependent (CPY, ALP, and CPS) and COPI-independent (invertase and HSP150) proteins.
Protein transport kinetics were also examined in a strain harboring a temperature conditional allele of ARF1 and deleted for ARF2 (arf1–3 ts arf2Δ), henceforth referred to as the arf1–3 ts strain. This strain grows well at temperatures up to 30°C but does not grow at 37°C (Kahn, unpublished observation). The kinetic defects in transport of CPY and HSP150 at the nonpermissive temperature (Table 2 CPY* and HSP150*) were remarkably similar to those observed in the arf1Δ strain. These data indicate that cells with insufficient ARF activity to support growth are still capable of transporting proteins through the secretory pathway, and that the transport defect observed in this strain is comparable for COPI-dependent and -independent cargo.
arf1Δ Mutants Exhibit Defects at Multiple Intercompartmental Transport Steps
A number of intercompartmental protein transport steps are required to move a protein from the ER through the Golgi complex to the extracellular space or vacuole. From the experiments described above, it was not possible to determine specifically the kinetically slow intercompartmental transport step(s) in the secretory pathway of arf1Δ cells. Therefore, the transport of yeast pro-α-factor was examined because the mobility of this protein in SDS gels undergoes substantial and well-characterized changes as it is transported through at least four functionally distinct compartments of the Golgi complex that are marked by 1) an initiating α1,6-mannosyltransferase; 2) an elongating α1,6-mannosyltransferase; 3) an α1,3-mannosyltransferase; and 4) Kex2p which initiates proteolytic processing of pro-α-factor to the mature form (Julius et al., 1983; Franzusoff and Schekman, 1989; Graham and Emr, 1991; Gaynor et al., 1994). Cells were labeled for 10 min at 20°C and chased for the times indicated in Figure 1B. Pro-α-factor was rapidly Golgi modified and converted to the mature 13-amino acid peptide hormone in wild-type cells. In contrast, a substantial fraction of both the ER core-glycosylated and Golgi-modified pro-α-factor forms were present throughout the chase period in the arf1Δ mutant. We measured the rate of pro-α-factor disappearance during the chase period to estimate its rate of transport from the ER through the trans-Golgi, where the glycosylated pro-piece is proteolytically removed. The half-time for pro-α-factor disappearance in the arf1Δ mutant was 8 min, which was fourfold slower than that measured in wild-type cells (Table 2, ∼2.0 min). Because both the ER- and Golgi-modified forms accumulated in the arf1Δ mutant, transport of pro-α-factor from the ER to the cis-Golgi and through the early Golgi compartments must both be defective.
The Golgi-specific glycosylation defect in arf1Δ mutants previously noted by others (Stearns et al., 1990b) was also apparent in our experiments (Figure 1A). Since the conversion of p1CPY to p2CPY is primarily due to the addition of α1,3-mannose residues (Graham and Krasnov, 1995), which is catalyzed by Mnn1p in the medial Golgi (Graham and Emr, 1991; Graham et al., 1994), we reasoned that the partial loss of this modification may result from underexpression or instability of Mnn1p in the arf1Δ mutant. To test this possibility, we performed a pulse-chase and immunoprecipitation analysis for Mnn1p, but extended the chase period to 3 h. In wild-type cells, Mnn1p undergoes a gradual posttranslational automodification, primarily on O-linked oligosaccharides, which is dependent on Golgi residence and causes a slow increase in relative molecular mass (Graham et al., 1994). Mnn1p was synthesized normally in the arf1Δ mutant and was stable for at least 3 h (Figure 1C). During the chase period, the relative molecular mass of Mnn1p increased gradually in both strains, suggesting that it had been transported out of the ER and localized in the Golgi complex. However, after a 3-h chase, the apparent molecular mass of Mnn1p in the wild-type strain increased 3.8 kDa, whereas it increased only 1.5 kDa in the arf1Δ mutant. These data suggest that the Mnn1p was expressed at normal levels, was localized to the Golgi complex, but was catalytically inefficient in the arf1Δ mutant. Consistent with the idea that Mnn1p is inactivated under ARF-deficient conditions, the arf1–3 ts mutant exhibited a near complete defect in α1,3-mannosylation of invertase after shift to nonpermissive temperature (our unpublished observations).
Retrograde Golgi-ER Protein Transport Is Inefficient in arf1Δ Cells
COPI/coatomer is required for Golgi to ER retrieval of type I membrane proteins containing a C-terminal cytoplasmic dilysine (KKXX) motif (Cosson and Letourneur, 1994; Letourneur et al., 1994). Given the intimate connection between ARF and coatomer (Ostermann et al., 1993), we reasoned that the arf1Δ mutant, like COPI mutants, may also be defective for dilysine-mediated Golgi to ER retrieval. We therefore analyzed retrieval of a chimeric protein containing the periplasmic enzyme invertase fused to the transmembrane domain and KKXX-bearing cytoplasmic tail of Wbp1p (Inv-Wbp1p) in the arf1Δ mutant. This ER-localized fusion protein is efficiently retrieved from the cis-most Golgi compartment wild-type cells, which contains initiating α1,6-mannosyltransferase (Och1p) activity but not elongating α1,6- or α1,3-mannosyltransferase activities. When the dilysine signal is destroyed by mutation or when the fusion protein is analyzed in COPI mutants, the 70-kDa chimera travels through more distal Golgi compartments, acquiring extensive outer chain carbohydrate modifications, and is then delivered to the vacuole, where it is proteolytically cleaved to produce a soluble 56-kDa invertase fragment (Gaynor et al., 1994; Letourneur et al., 1994).
To analyze Golgi-specific modification and vacuolar processing of Inv-Wbp1p, cells harboring the chimera were subjected to pulse-chase analysis. After 0 and 60 min of chase, the fusion protein was recovered by immunoprecipitation with invertase antiserum, then split into equal aliquots and subjected to a second immunoprecipitation with antiserum against either invertase, α1,6-mannose linkages, or α1,3-mannose linkages. Half of each resulting immunoprecipitate was treated with endoglycosidase H (endo H) to remove N-linked carbohydrates, which caused the protein to migrate as a discrete band and allowed the vacuolar processing event to be visualized. After 60 min of chase, more than 90% of the the Inv-Wbp1p recovered from both wild-type and arf1Δ mutant cells migrated as the intact 70-kDa form of the protein after endo H treatment (Figure 2A, +endo H), indicating that the fusion protein had not reached the vacuole but was retained in earlier compartments of the secretory pathway. The fusion protein was also quantitatively modified with α1,6- but not α1,3-linked mannose in both cell types (Figure 2A). Likewise, the Inv-Wbp1p was also not significantly mislocalized to the vacuole in the arf1–3 ts mutant preincubated for 1 h at nonpermissive temperature (our unpublished observations).
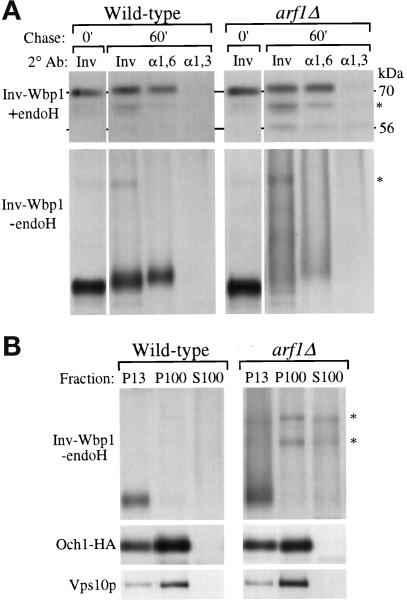
Figure 2.
Posttranslational modification and localization of a KKXX-bearing Inv-Wbp1 fusion protein in wild-type and arf1Δ cells. Wild-type (SEY6210) and arf1Δ (6210 arf1Δ) cells harboring pEG1-KK were labeled for 10 min and chased for 60 min at 30°C. (A) The chase was terminated at the times indicated by the addition of TCA, and Inv-Wbp1p was recovered from the samples by immunoprecipitation with anti-invertase serum. The primary immunoprecipitates were boiled in 1% SDS to dissociate the antibody–antigen complex, and the eluates were split into three equal aliquots that were subjected to a second immunoprecipitation with antiserum to either invertase (Inv), α1,6-linked mannose (α1,6) or α1,3-linked mannose (α1,3). Half of each immunoprecipitate was subsequently treated with endoglycosidase H (+endo H) to examine proteolytic processing of the polypeptide, and half was left untreated (−endo H) to examine the effect of glycosylation on the fusion protein’s mobility in SDS gels. At the 0-min chase point, none of the Inv-Wbp1p was precipitated with the linkage-specific serum; therefore, only the anti-invertase immunoprecipitate is shown. The position of the intact fusion protein (70 kDa) and vacuolar processed (56 kDa) forms are shown. Asterisks indicate nonspecific bands contaminating the immunoprecipitates. (B) Labeled spheroplasts harboring pEG1-KK or pOH were harvested after 60 min of chase, lysed, and subjected to differential centrifugation as described previously (Gaynor et al., 1994). Inv-Wbp1p and the Golgi proteins Och1-HA and Vps10p were recovered from the 13,000 × g pellets (P13), 100,000 × g pellets (P100), and 100,000 × g supernatants (S100) by immunoprecipitation.
When not treated with endo H, the glycosylated Inv-Wbp1p from wild-type cells migrated as a distinct band (Figure 2A, −endo H; Gaynor et al. 1994), indicating that it was efficiently retrieved from the cis-most Golgi compartment. In contrast, Inv-Wbp1p from arf1Δ cells migrated as a hyperglycosylated, high molecular mass smear (Figure 2A, −endo H). This migration pattern was most likely due to the elongating α1,6-mannosyltransferase, because this enzyme controls the extent of hyperglycosylation on endogenous invertase in wild-type cells (reviewed in Herscovics and Orlean, 1993), and because the fusion protein did not acquire significant α1,3-linked mannose in the arf1Δ mutant (Figure 2A, right panels). Modification of Inv-Wbp1p with α1,6-linked mannose required ER to Golgi transport, because in both a sec18–1 mutant [Sec18p/NSF is required for ER to cis-Golgi protein transport (Novick et al., 1980)] and a sec18–1/arf1Δ double mutant at nonpermissive temperature, the Inv-Wbp1p did not acquire α1,6-linked mannose (Gaynor et al., 1994 and our unpublished observations).
To determine the intracellular location of the hyperglycoslyated Inv-Wbp1p in the arf1Δ mutant, subcellular fractionation experiments were performed. Cells harboring the Inv-Wbp1p were converted to spheroplasts, pulse labeled, chased, and fractionated by differential centrifugation (see legend to Figure 2). In the arf1Δ mutant cells, as in wild-type cells, nearly all of the Inv-Wbp1p was found in the 13,000 × g pellet (Figure 2B, P13), which is where ER membrane proteins fractionate (Gaynor et al., 1994). In contrast, the trans-Golgi protein Vps10p and most of the cis-Golgi protein Och1-HA [Och1-HA is a hemagglutinin epitope-tagged form of Och1p (Harris and Waters, 1996)], were primarily found in the 100,000 × g pellet (Figure 2B, P100), which is where most Golgi membranes fractionate (Gaynor et al., 1994; Marcusson et al., 1994). Further support for ER localization of the hypermodified Inv-Wbp1p in arf1Δ cells was provided by resolving the P13 fraction on a two-step sucrose gradient, which revealed that the majority of the Inv-Wbp1p was found in a dense membrane, ER-enriched fraction. In contrast, both Vps10p and Och1-HA from the P13 primarily localized to the less dense membrane fraction, which is typical for Golgi proteins contaminating the P13 (our unpublished observations and Gaynor et. al., 1994). These analyses indicate that Golgi-ER retrieval of dilysine-containing proteins occurs even in the absence of Arf1p. However, because the Inv-Wbp1p acquired significant additional Golgi modification in arf1Δ cells compared with wild type, retrieval is likely to be less efficient, with hypermodification of Inv-Wbp1p reflecting a slightly increased residence time of the protein in the cis-Golgi and/or transport to more distal Golgi compartment(s) than normal prior to ER retrieval.
Genetic Interactions between ARF1 and COPI
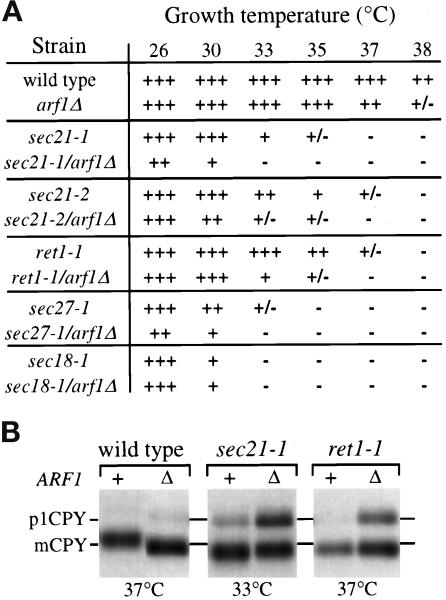
The pulse-chase and fractionation studies described above suggested that arf1Δ cells exhibit a modest defect in COPI-dependent retrograde transport of a KKXX-bearing fusion protein. This interpretation is supported by a previous study which reported that double mutant combinations of arf1Δ with sec21–1 (γ-COP) were usually inviable (or synthetically lethal), whereas arf1Δ did not exaggerate the growth defects of 11 other mutations that perturb the yeast secretory pathway (Stearns et al., 1990b). However, the sec21–1 mutant exhibits defects in both anterograde and retrograde protein transport (Hosobuchi et al., 1992; Letourneur et al., 1994). To determine whether arf1Δ is synthetically lethal with other COPI mutants that exhibit a slight (sec27–1) or undetectable (sec21–2, ret1–1) anterograde transport defect, we crossed the 6210arf1Δ strain with strains harboring mutations in SEC21 (γ-COP), RET1 (α-COP), and SEC27 (β′-COP). Surprisingly, all resulting COPI/arf1Δ haploid double mutants, even sec21–1/arf1Δ, were viable and occurred at the expected frequency after tetrad dissection. The dramatic increase in survival of our COPI/arf1Δ double mutants compared with the previous study (Stearns et al., 1990b) was most likely due to differences in laboratory strain backgrounds.
Because the COPI/arf1Δ mutants were in fact viable, we performed additional analyses to explore genetic interactions between COPI genes and ARF1. All of the double mutants exhibited synthetic growth defects that reduced the permissive growth temperatures 4–7°C relative to the corresponding COPI single mutants (Figure 3A). As a control, a sec18–1/arf1Δ double mutant was prepared which did not show a synthetic growth defect (Figure 3A), even though sec18–1 mutants exhibit a defect at each of the protein transport steps that are perturbed in arf1Δ cells (Graham and Emr, 1991; Figure 1). Anterograde transport of CPY was also assayed in these mutants. Cells were incubated at the indicated temperatures, pulse labeled for 10 min, and chased for 60 min. sec21–1/arf1Δ and ret1–1/arf1Δ are shown as representative examples. Interestingly, at temperatures where the single mutants exhibited little to no defect in transport of CPY, p1CPY was still present after a 60-min chase period in all double mutants (Figure 3B). Even the ret1–1/arf1Δ double mutant showed a substantial defect, which was surprising, because ret1–1 mutants have not been observed to exhibit defects in anterograde transport of CPY at any temperature or condition tested (Letourneur et al., 1994). The underglycosylation of CPY in the arf1Δ mutant was also observed for the COPI mutants but was not exaggerated in the double mutants (Figure 3B). Together, these analyses suggest that synthetic defects observed between arf1Δ and COPI mutants may represent a decreased capacity of these double mutants to recycle factors required for efficient ER export of proteins like CPY and further support a role for Arf1p in the Golgi-ER retrieval pathway.
Figure 3.
Synthetic defects in COPI/arf1Δ double mutants. (A) Growth of strains harboring the arf1Δ allele in combination with sec21–1 (γ-COP), sec21–2 (γ-COP), ret1–1 (α-COP), or sec27–1 (β′-COP) alleles on YPD plates was compared with the corresponding single mutants at the temperatures indicated. Complete genotypes of strains are provided in Table 1. +++, wild-type growth; ++, growth slower than wild type; +, very slow growth (minimal single colony formation); +/−, almost no growth (no single colony formation); and −, complete lethality. (B) COPI mutant strains harboring either the ARF1 wild-type (+) or arf1Δ (Δ) allele were labeled for 10 min and chased for 60 min at the temperature indicated. CPY was recovered by immunoprecipitation and subjected to SDS-PAGE. Positions of the ER (p1) and vacuolar (m) forms of CPY are indicated.
Immunofluorescent Localization of Golgi Enzymes to Large Ring Structures in arf1Δ Cells
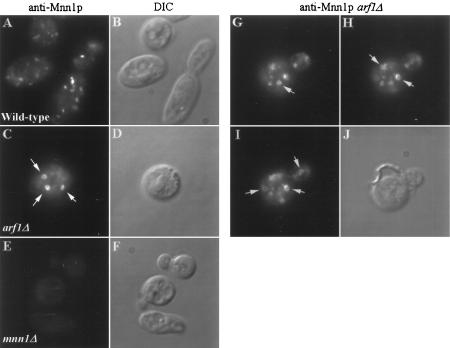
The Golgi-specific glycosylation defect in the arf1Δ mutant could not be attributed to underexpression or instability of at least one Golgi mannosyltransferase, and the increase in Mnn1p relative molecular mass observed by pulse-chase analysis in the arf1Δ mutant suggested that this protein was localized to the Golgi complex (Figure 1C). To examine Mnn1p localization and Golgi morphology in the arf1Δ mutant more directly, immunofluorescence analyses were performed. In wild-type cells, antibodies to Mnn1p stained several small, solid punctate structures (Figure 4A), which is typical of Golgi morphology in S. cerevisiae (Redding et al., 1991; Graham et al., 1994). No staining was seen in cells harboring a deletion of the MNN1 gene (Figure 4E), demonstrating the specificity of the antibody preparation. Remarkably, Mnn1p was localized to abnormally enlarged, ring-shaped structures in the arf1Δ mutant (Figure 4C). Ring structures were found in approximately 90% of the cells that stained well, and the number of rings varied from 1 to ∼7 per cell. Figure 4, G–H, shows a Z-axis series of a budding cell with five ring structures that appeared in different optical planes. The rings were typically one-tenth to one-fifth the diameter of the cell and appeared to be localized randomly throughout the cytoplasm. Most arf1Δ cells also exhibited normal-appearing Golgi structures in addition to the rings. The strains used for immunofluorescence studies are diploid and overexpress Mnn1p to allow easier detection; however, the haploid arf1Δ strains that express Mnn1p at wild-type levels also exhibited ring structures to a similar extent.
Figure 4.
Immunofluorescent localization of Mnn1p to large ring structures in arf1Δ cells. Immunofluorescent staining of wild-type (SEY6210.5 pZV236, A), arf1Δ (6210.5 arf1Δ pZV236, C and G–I), and mnn1Δ (TGY122, E) cells using affinity-purified antibodies to Mnn1p was performed as described previously (Graham et al., 1994). (B, D, F, and J) Images captured using DIC optics and correspond to the adjacent fluorescent image. (G–I) Images captured at three different optical planes while focusing down through the cell (a Z-axis series). Arrows indicate ring structures visible in each optical plane. Rings were visible in 90% of the arf1Δ cells stained for Mnn1p.
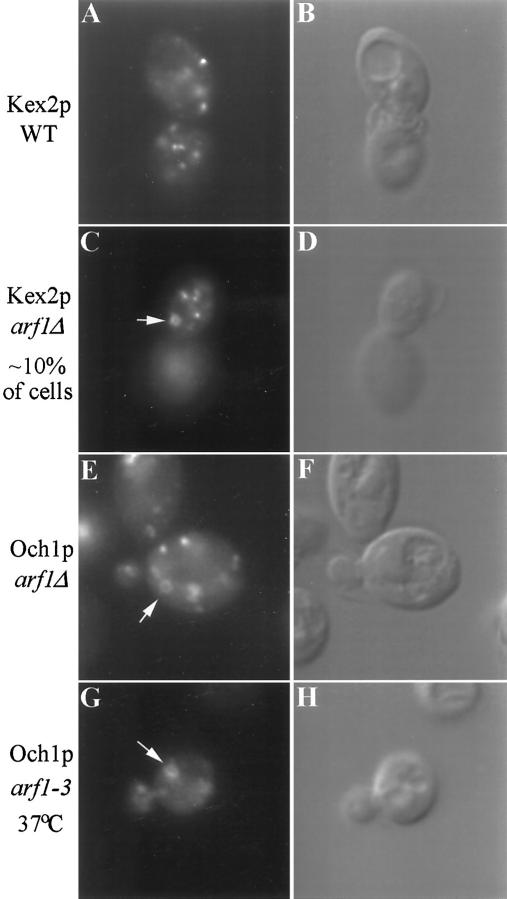
Mnn1p marks a medial Golgi compartment in wild-type yeast (Graham and Emr, 1991; Graham et al., 1994). We also examined the localization of proteins that mark the cis- and trans-Golgi by staining cells with antibodies to Och1-HA (an epitope-tagged initiating α1,6-mannosyltransferase) and Kex2p. The staining pattern observed for Kex2p and Och1-HA in wild-type cells was again typical of the yeast Golgi (shown for Kex2p only in Figure 5A). The Och1-HA staining pattern in the arf1Δ mutant was similar to that of Mnn1p: most of the cells exhibited ring-shaped structures (Figure 5E). However, only about 10% of the arf1Δ mutant cells exhibited ring structures when stained with antibodies to Kex2p (Figure 5C), suggesting that the morphology of the Kex2p compartment was much less affected than the earlier Golgi compartments and that the compartmental organization of the Golgi was maintained. In no case did the ring structures align with the vacuole, which appeared as a large depression when observed using differential interference contrast (DIC) optics, and costaining with antibodies to the vacuolar ATPase and Mnn1p revealed clearly distinct structures (our unpublished observations). The localization of Och1-HA was also examined in the arf1–3 ts strain incubated at the nonpermissive temperature (Figure 5G) with results comparable to those observed in the arf1Δ strain. Ring structures were also observed in arf1–3 ts cells incubated at permissive temperature, suggesting that this strain is partially deficient in ARF activity at the permissive temperature, a condition where growth and α1,3-mannosylation appear to be normal (our unpublished observations).
Figure 5.
Immunofluorescent localization of Kex2p and Och1p in arf mutant cells. Wild-type (SEY6210.5 pKE2018) and arf1Δ (6210.5 arf1Δ pKE2018) cells overexpressing Kex2p and arf1Δ cells expressing Och1-HA (6210.5 arf1Δ pOH URA3) were stained using affinity-purified antibody to Kex2p (A and C) or a monoclonal antibody to the HA epitope tag (E). (B, D, and F) Images captured using DIC optics correspond to the adjacent fluorescent image. Ring structures were apparent in most arf1Δ cells stained for Och1-HA, whereas only ∼10% of arf1Δ cells exhibited rings when stained for Kex2p. (G and H) An arf1–3 ts mutant (strain C156–1B, G–H) transformed with pOH was preincubated for 1 h at 37°C before fixation and immunofluorescence localization of Och1p.
FM4–64 Staining Reveals Large Ring-shaped Endosomal Structures in arf1Δ Cells
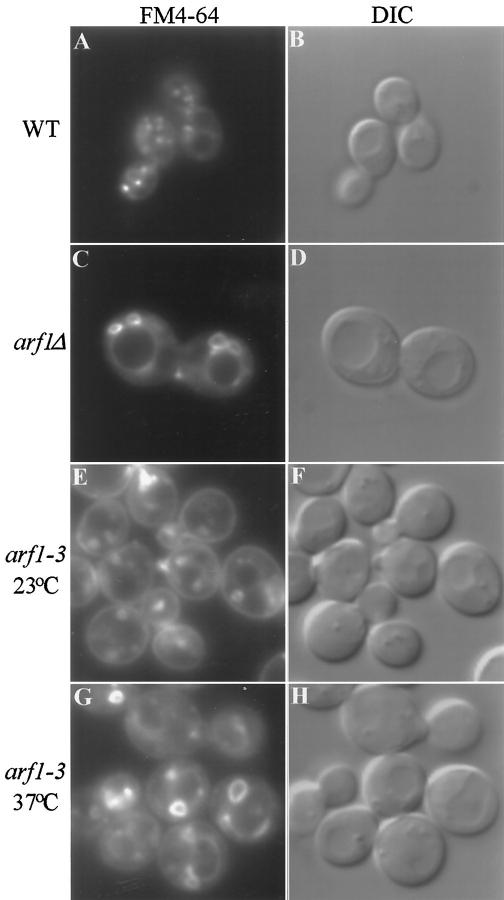
The morphology of organelles in the endocytic pathway was also examined by staining living cells with the lipophilic styryl dye FM4–64. This compound intercalates into the plasma membrane of cells and is delivered to the vacuole via small cytoplasmic intermediates that represent endocytic vesicles and endosomes (Vida and Emr, 1995). Wild-type and arf1Δ cells were stained with FM4–64 on ice for 45 min, allowing the dye to stain the plasma membrane, then the cells were warmed to 25°C in fresh media lacking the dye to initiate endocytosis. Cells were collected for fluorescence microscopy 10 min later, a chase point where endosomes and endocytic compartments would be prominently stained but where most of the dye would not yet have reached the vacuole. As previously described (Vida and Emr, 1995), a number of small punctate structures corresponding to endosomes and endocytic membranes were observed in the cytoplasm of wild-type cells (Figure 6A). In contrast, large ring structures were seen in the arf1Δ cells (Figure 6C) that were clearly distinct from the vacuole, which appears as a large depression in the DIC image (Figure 6, B and D). These ring structures were commonly found juxtaposed to the vacuole or plasma membrane and were larger (on average) than the rings observed by immunofluorescence of Golgi enzymes. A few small punctate structures were also seen in most arf1Δ cells. At later time points, the vacuoles were stained normally in both wild-type and arf1Δ cells, and there did not appear to be a substantial difference in the rate of FM4–64 endocytosis or in vacuole morphology between arf1Δ and wild-type cells in these experiments. These data suggest that the arf1Δ mutation markedly perturbs the structure of endosomes in vivo.
Figure 6.
FM4–64 staining of endocytic membranes in wild-type and arf mutant cells. (A–D) Wild-type (SEY6210) and arf1Δ (6210 arf1Δ) cells were incubated in YPD containing 30 μM FM4–64 on ice for 45 min and then warmed to 25°C to initiate endocytosis as described previously (Vida and Emr, 1995). Cells were collected and applied to concanavalin A-treated slides 10 min after the temperature shift, and the images were acquired ∼10 min after cells were applied to the slides. (E–H) The arf1–3 ts mutant (C156–1B) was grown at 23°C and then half of the culture was shifted to 37°C and incubated for 1 h. Cells were harvested and resuspended at ∼2 × 108 cells/ml in YPD media prewarmed to 23°C or 37°C and incubated for 10 min. FM4–64 was added to 30 μM and incubated for 7 min, then the cells were harvested and resuspended in fresh prewarmed media and the incubation was continued. Images of the cells were acquired ∼15–20 min after addition of fresh media.
The structure of endocytic compartments was also examined in the arf1–3 ts strain (Figure 6) grown at 23°C and preincubated for 1 h at either 23°C (Figure 6, E and F) or 37°C (Figure 6, G and H) before staining with FM4–64. Rings stained with FM4–64 were observed in cells incubated at both temperatures; however, the rings observed in cells stained at 37°C were typically larger and less uniform in size than the rings observed in cells stained at 23°C. As with the arf1Δ mutant, the ring structures observed in the arf1–3 ts mutant (Figure 6, E and G) are distinct from the vacuoles, which are apparent in the accompanying DIC images (Figure 6, F and H). The kinetics of FM4–64 delivery to the vacuole in the arf1–3 ts strain was not significantly different from wild-type cells at either temperature. Although the size of the endocytic ring structures observed by FM4–64 in the arf1–3 ts or arf1Δ cells varied, many cells exhibited rings that were approximately 1.5–2 μm in diameter (up to one-third the diameter of the cell), particularly in the arf1–3 ts cells incubated at 37°C. Ring structures this large were never observed by immunofluorescence localization of Golgi enzymes.
Endosome Structure and FM4–64 Endocytosis Are Perturbed in BFA-treated Cells
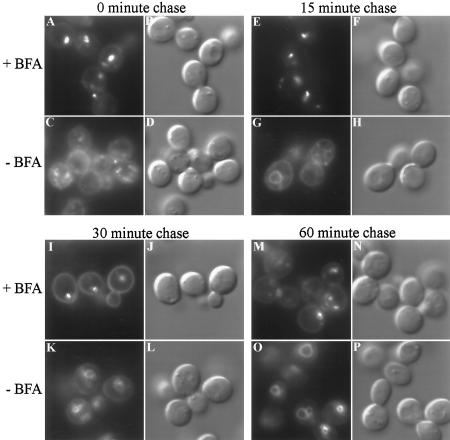
The dramatic difference in endosome morphology between wild-type and arf mutant cells strongly suggests a role for ARF in endocytosis. To test the effects of ARF inactivation on endosome morphology and endocytic membrane flux in a strain harboring wild-type ARF genes, we examined the effect of BFA on the uptake of FM4–64 using an erg6/ise1 strain of yeast that is sensitive to this drug. We had previously shown that BFA rapidly induces a block in protein transport early in the yeast secretory pathway (Graham et al., 1993; Gaynor and Emr, 1997), and others have shown that BFA blocks transport of proteins from early to late endosomes (Hicke et al., 1997). To test whether this drug can also perturb FM4–64 endocytosis immediately after addition, erg6 cells were incubated in the presence or absence of BFA for 5 min and then stained for 7 min with FM4–64 and resuspended in fresh media with or without BFA as appropriate for a chase period of 1 h. Cells were harvested at the time points indicated (Figure 7) and immediately examined by fluorescence microscopy. The FM4–64 was rapidly internalized in BFA-treated (Figure 7, A and B) and untreated cells (Figure 7, C and D). However, the BFA-treated cells exhibited a marked inhibition in delivery of FM4–64 to the vacuole (Figure 7, E, I, and M) as compared with untreated cells (Figure 7, G, K, and O), in which vacuolar staining was observed in 15 min. In addition, endocytic ring structures were observed in the BFA-treated cells (Figure 7, I and M) that appeared to be similar to what was observed in the arf mutants.
Figure 7.
BFA perturbs transport of FM4–64 from endosomes to the vacuole. Strain TGY413–6D was preincubated at ∼2 × 108 cells/ml in YPD containing 50 mM Na HEPES (pH 7.0) for 15 min at 23°C. The culture was split in half and BFA was added to 100 μM final concentration to one culture (+BFA) and an equivalent aliquot of ethanol was added to the control culture (−BFA). The cells were incubated for 5 min before staining with FM4–64 for 7 min. The cells were then harvested and resuspended in fresh media (+/− BFA) to chase the FM4–64. Cells were applied to conconavalin A-treated slides at the chase times indicated and the images were acquired immediately.
Electron Microscopy Reveals Rings of Interconnected Membrane Tubules in arf Mutant Cells
To examine the ultrastructural morphology of the arf1Δ mutant and to characterize further the ring structures we had observed by fluorescence microscopy, wild-type and arf mutant cells were prepared for electron microscopy (see MATERIALS AND METHODS). Large rings of seemingly discontinuous membrane were frequently observed in arf1Δ mutant cells (Figure 8, B–D, arrows and arrowheads). These rings were typically 0.3–0.8 μm in diameter, approximately one-tenth to one-fifth the diameter of the cell. Structures of this type were rarely observed in electron micrographs of wild-type cells (a representative cell is shown in Figure 8A). Most of the ring structures were formed from vesicular and tubular profiles in a circular arrangement (Figure 8D), although some sections, particularly those that appear to glance the surface of a ring, showed a reticular network of membrane comprising the rings (marked with asterisks in Figure 8, B and C). Even though the membrane appeared to be discontinuous in thin sections, the rings were nearly always complete and usually circular, consistent with a hollow sphere structure comprised of interconnected membrane tubules rather than a short cylindrical object. The interior of the spheres gave the same appearance as the surrounding cytoplasm and is likely continuous with the cytoplasm. The double rings marked with an arrowhead in Figure 8B may represent a sphere with one side folded inward perpendicular to the section plane. Comparable structures were found in the arf1–3 ts strain incubated for 1 h at the nonpermissive temperature before fixation, including some large ring structures that were approximately one-fifth to one-third the diameter of the cell. Attempts at immunogold labeling of ring structures that contain Mnn1p have not been successful; however, the similar size, structure, and prevalence in the cell strongly suggest that the rings observed in electron micrographs are the same structures observed by fluorescence microscopy.
Figure 8.
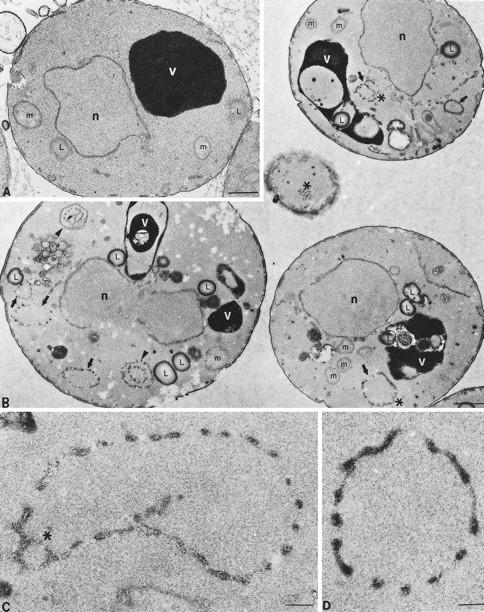
Electron microscopy of wild-type and arf1Δ cells. Cells were grown at 30°C and prepared for electron microscopy as described previously (Rieder et al., 1996). Representative thin sections through SEY6210 wild-type (A) and 6210arf1Δ (B) cells are shown. (A and B) Positions of the nucleus (n), electron-dense vacuole (v), mitochondria (m), and lipid droplets (L) are indicated, and the bar represents 0.5 μm. In B (arf1Δ cells), arrows point to single-ring structures, arrowheads point to double or multiple ring structures (i.e., rings within rings), and asterisks are positioned next to reticulated membrane, which is often visualized as part of the rings. (C and D) Higher magnification photographs of representative ring structures observed in arf1Δ cells; the asterisk indicates reticulated membrane, and the bar represents 0.05 μm.
The vacuoles often exhibited distorted shapes and appeared to have more internal membrane in the arf1Δ mutant when observed by electron microscopy. This may represent a more involuted vacuolar membrane, increased autophagy of other organelles, or an accumulation of lipid droplets within the vacuole. There also appeared to be more lipid droplets in the cytoplasm of arf1Δ cells, but we did not observe a marked accumulation of ER in this mutant. Clearly, the rings of discontinuous or reticular membrane and the unusual appearance of the vacuoles were the most dramatic morphological difference between arf1Δ and wild-type cells observed by electron microscopy, although the vacuoles observed in living cells by light microscopy appeared to be normal (Figure 6D).
DISCUSSION
To gain a better understanding for the role of ARF in vivo, we have taken a broad, multifaceted approach to analyze phenotypic defects in yeast arf mutants. Analysis of multiple secretory cargo proteins in these mutants revealed general defects in both anterograde and retrograde transport through the ER/Golgi system as well as Golgi-specific glycosylation abnormalities. Even though protein transport was only modestly affected, morphological studies demonstrated that the arf mutants exhibited substantial perturbations of the Golgi complex. In addition, we have found that the arf mutants exhibit a marked change in endosome structure and that BFA both perturbs endosome morphology and causes an immediate block in the delivery of FM4–64 from endosomes to the vacuole, together providing significant in vivo evidence that ARF plays a role in endocytosis in yeast. These data thus clearly indicate that ARF is required in vivo to maintain the normal structure and function of both Golgi and endocytic compartments. The comprehensive nature of our in vivo analysis combined with previous reports on the biochemical properties of ARF has allowed us to formulate a model to explain how ARF might regulate both the structure and function of multiple intracellular organelles.
Arf1p Is Required for Maintenance of Normal Golgi and Endosome Morphology
Fluorescence microscopy of the arf mutants revealed a particularly striking change in the structure of Golgi and endosomal compartments. Large ring structures were stained with antibodies to early and medial Golgi enzymes and with the endocytic marker FM4–64 (Figures 4–6). The simplest interpretation of these data is that Golgi and endocytic compartments adopt similar but distinct ring-shaped structures in these cells. It is possible that the same ring-shaped compartment is stained by antibodies to the Golgi enzymes and by FM4–64 in the arf mutant cells, which could occur by inappropriate fusion of Golgi and endosome compartments, mislocalization of Golgi enzymes to the endosome, or misrouting of FM4–64 through the Golgi. However, we favor the idea that the Golgi and endosome compartments retain separate identities in the arf mutants for the following reasons. First, the ring structures stained by FM4–64 were generally larger and more frequently found juxtaposed to the vacuole or plasma membrane than ring structures observed by immunofluorescent localization of Golgi enzymes. This was particularly evident for the arf1–3 ts mutant, in which many cells exhibited extremely large endocytic structures, some of which were more than twice the diameter as the largest rings observed when these cells were stained for Och1-HA by immunofluorescence. Second, the growth rate of arf1Δ cells was nearly indistinguishable from that of wild-type cells (Figure 3), and protein transport and modification were only modestly affected. These observations do not seem compatible with a complete breakdown of compartmental identity in the arf mutants through membrane fusion or protein mislocalization. Finally, distinct ARF proteins have been associated with the function of the cis-Golgi and endosomes in mammalian cells (Boman and Kahn, 1995), and we have shown that BFA perturbs transport through early Golgi compartments (Graham et al., 1993) and through the endocytic pathway (Figure 7) in yeast. For these reasons, we suggest that loss of ARF function affects both organelles in yeast and that these organelles remain physically and functionally distinct in arf mutant cells.
Thin-section electron microscopy of arf1 mutant cells provided insight into the structural basis of these membrane rings. We typically observed clusters of apparently vesicular and tubular profiles arranged with even spacing to form a ring. In some sections, the membrane that formed the rings presented a more reticular profile. Many arf mutant cells exhibited several ring structures within the same section, suggesting the presence of multiple, independent ring structures. We interpret these two-dimensional images to represent section planes through hollow spheres of interconnected membrane tubules.
Some insight into how these structures might form comes from previous structural analyses of both mammalian and wild-type yeast Golgi membranes. In mammalian cells, individual Golgi cisternae are arranged in contorted stacks of membrane ribbons with regions of compact zones, where cisternal membranes appear to be continuous and form a flattened saccule with few fenestrae, separated by tubular zones from which transport vesicles appear to bud (Rambourg and Clermont, 1990; Weidman et al., 1993). The Golgi complex of wild-type yeast has been characterized by immunoelectron microscopy and appears as isolated disk, or shallow bowl, structures with a diameter of ∼400 nm and a width of ∼50 nm (Preuss et al., 1992). Tubular zones continuous with the plate-like Golgi cisternae in wild-type cells were not described as such in this detailed report on the ultrastructure of the yeast Golgi. However, tubular zones would appear as small vesicular elements when lateral views of Golgi cisternae are observed in thin sections. Indeed, Preuss et al. (1992) noted that small vesicular profiles, which may represent tubular zones, tended to cluster around yeast Golgi cisternae observed in thin sections. Others have observed tubular networks of membrane in wild-type yeast that were suggested to represent Golgi membranes (Rambourg et al., 1993). The compact zones and tubular zones of a cisterna are unlikely to be static structures; rather, it is more plausible to imagine these structures in a dynamic equilibrium, with the relative amount of compact and tubular zones in constant flux within a ribbon of membrane. If tubular zones are precursors to transport vesicles, then these zones may normally be transient, particularly in yeast where vesicle-mediated protein transport through the cell is extremely rapid.
Potential Mechanisms for ARF-dependent Regulation of Organelle Morphology
The morphological data presented here suggest a novel role for ARF in regulating the equilibrium between compact and tubular zones of Golgi cisternae. We speculate that a yeast Golgi cisterna normally exists as a curved, disk-shaped compact zone with a modest tubular zone at the periphery that gives rise to transport vesicles, analogous to mammalian Golgi cisternae. In the arf mutants, the size of the compact zone would decrease by fenestration of this region, and most of the cisterna would become a tubular network. This would increase the area occupied by the Golgi disk and would likely make the membrane more flexible. Expansion of the curved Golgi membrane and greater flexibility could bring the edges in close enough proximity to allow tubular elements at the periphery to interconnect and form a hollow sphere, which is what the ring-like structures observed in arf mutant cells appear to represent. The initial fenestration event could result from extensive fusion of apposed membranes of the same cisterna initiated from the luminal leaflets of the bilayer. Flattened Golgi cisternae, in which the lipid bilayers are within 50 nm of each other, seem particularly amenable to this process. In the arf mutants, the luminal fusion events would not be balanced properly by the opposing reaction, homotypic fusion of membrane initiated from the cytoplasmic membrane surface of a tubular cisterna. Consistent with our observations in arf1Δ cells, tubular membrane networks have been observed by electron microscopy in cells treated with BFA (Rambourg et al., 1995).
How might ARF regulate these processes? Apart from its initially described role in ADP ribosylation, there are currently two biochemical activities attributed to ARF in normal cells: 1) recruitment of coats (COPI and AP1/clathrin) to sites of vesicle budding (Rothman and Wieland, 1996) and 2) activation of PLD (Brown et al., 1993; Cockcroft et al., 1994). ARF appears to be dispensable for coatomer recruitment in vitro if membranes are pretreated with PLD (Ktistakis et al., 1996), suggesting that the primary role of ARF may be to regulate lipid bilayer modification. A role for ARF in regulating the lipid composition of membrane bilayers is consistent with the pleiotropic phenotypes exhibited by the arf mutants in our study. For instance, altered membrane composition in the arf mutants may account for the inefficient function of both Golgi glycosyltransferases and proteins required for vesicular transport. Our morphological data can be explained if modulation of Golgi and endosome membrane composition by ARF influences the probability of homotypic membrane fusion events initiated from the luminal membrane surface versus those initiated from the cytoplasmic membrane surface. Yeast have two distinct PLD enzymes which are encoded by at least two genes (Waksman et al., 1996, 1997). Although it is not yet clear whether a PLD activity is required in the yeast secretory or endocytic pathways, the ARF-dependent activation of mammalian PLD provides an example of how ARF could influence the properties of the cytoplasmic leaflet of these organelles. It is also possible that ARF may regulate other lipid modifying enzymes responsible for normal composition of Golgi and endosome membranes.
In Vivo Evidence for Additional Functions for ARF beyond COPI Recruitment
The specific synthetic growth defects observed for all arf1Δ COPI double mutants constructed strongly implicates ARF activity in COPI function. Indeed, mutant COPI alleles that do not exhibit an anterograde transport defect still exhibit exaggerated growth and protein transport defects when combined with arf1Δ. Likewise, Golgi to ER retrieval of a fusion protein bearing a KKXX motif in arf1Δ cells was perturbed, since the fusion protein appeared to reach a more distal Golgi compartment than normal prior to being recycled. Although many observations have suggested additional roles for ARF aside from coat recruitment, there is little in vivo support for these observations that does not rely on overexpression of dominant-activated mutant forms of ARF or the inhibitory affect of GTPγS on in vitro systems. Our data are most consistent with the idea that ARF plays a more pleiotropic role in the cell rather than simply recruiting COPI to Golgi membranes. For instance, yeast COPI mutants exhibit dramatic protein-specific defects in anterograde transport, where proteins such as CPY, ALP, and α-factor require COPI function for ER export whereas invertase and HSP150 do not. If the only and/or primary function of ARF is to recruit coatomer to form COPI-coated vesicles, we would expect arf mutant cells to exhibit similar cargo-specific anterograde defects. However, the transport kinetics of all anterograde proteins assayed were two- to fourfold slower in the arf mutant cells as compared with wild-type cells. COPI mutants also have a clearly distinct effect on Golgi structure and function from what we observed in the arf mutants. For instance, in sec21–3 cells at the nonpermissive temperature, the normally punctate immunofluorescent staining pattern of Och1p-HA appeared to disperse, although the staining pattern of Mnn1p was unaffected (Gaynor and Emr, 1997). The Mnn1p staining pattern was also unaffected in a ret1–1 mutant (our unpublished observations), and the Golgi-specific glycosylation defects we and others observed in the arf1Δ mutant were significantly different from those observed in COPI mutants (Gaynor and Emr, 1997). The effects of the arf mutations on Golgi and endosome morphology are also substantially different from those previously described for clathrin or adaptin mutants in yeast (Payne and Schekman, 1989; Phan et al., 1994; Rad et al., 1995; Stepp et al., 1995).
As with most temperature-conditional alleles, we cannot determine with absolute certainty whether the arf1–3 ts mutant exhibits a complete loss of function phenotype at the nonpermissive temperature. However, it is clear that protein transport can continue relatively efficiently in this mutant under conditions of insufficient ARF activity to support growth and despite substantial morphological changes to organelles in the exocytic and endocytic pathways. Interestingly, strains harboring temperature conditional alleles of the ARF exchange factor (gea1–2 gea2Δ and gea1–4 gea2Δ) or deleted for an ARF-GAP protein (gcs1Δ) exhibit a partial block in protein transport which is comparable to what we have observed in the arf mutants (Peyroche et al., 1996; Poon et al., 1996). Collectively, these observations suggest that these partial transport defects may in fact represent a near-to-complete loss of ARF function phenotype, which is reflected in the much more profound effect ARF seems to have on organelle morphology than on transport.
Perhaps the most efficient method to examine the immediate loss of ARF function phenotype is through the inhibitory effect of BFA on the ARF exchange factor. We have previously shown that BFA inhibits export of COPI-dependent proteins from the ER (Graham et al., 1993; Gaynor and Emr, 1997). In contrast, COPI-independent proteins are efficiently transported out of the ER in the presence of BFA but are subsequently blocked in transport between early Golgi compartments (Gaynor and Emr, 1997). Here, we show that BFA also perturbs endocytosis and induces the formation of endosomal ring structures that stain with FM4–64 (Figure 7). Others have observed rings of discontinuous membrane by electron microscopy of BFA-treated cells (Rambourg et al., 1995) that appear to be identical to the structures we observed in the arf mutants (Figure 8). Because the arf1Δ, arf1–3 ts, and BFA-treated cells all exhibit similar morphological defects, it appears that this represents a loss of function phenotype rather than a unique effect of ARF underexpression. On the other hand, the effects of BFA on protein and lipid transport are much more substantial than those observed for either the existing arf or gea (ARF exchange factor) mutant alleles. BFA also clearly inhibits COPI-independent transport steps (Gaynor and Emr, 1997) and has recently been shown to block transport of internalized α-factor from early to late endosomes (Hicke et al., 1997). Together, these observations support the idea of additional target protein(s) for BFA.
The data presented in this manuscript are most consistent with the idea that arf mutant cells exhibit general defects in Golgi and endosome structure and function which are likely to arise from morphological and/or membrane composition changes. This therefore supports the idea that rather than simply regulating coat recruitment, ARF may instead function as a regulator of lipid bilayer composition, which in turn provides an appropriate membrane environment onto which coat proteins assemble. Consistent with this, other proteins known to affect phospholipid dynamics, such as Sec14p (Bankaitis et al., 1990) and Vps34p (Herman and Emr, 1990; Schu et al., 1993), have also been shown to affect protein trafficking. General protein transport defects in the arf mutants could result either directly from membrane defects and/or from the combined effects that these membrane pertubations might have on both COPI and clathrin/AP function. Interestingly, proteins which regulate ARF’s activation-deactivation cycle (ARF-GTP ←→ ARF-GDP) are themselves sensitive to the lipid composition of the membrane (Boman and Kahn, 1995). This may provide a feedback mechanism by which ARF responds to membrane composition changes in normal cells. Clearly, the regulation of ARF activation, the regulation of membrane phospholipid content, and the function of coat proteins in vesicular transport are highly interdependent, and it seems most logical to propose that perturbations in normal phospholipid composition of Golgi and endosomes accounts for the significant morphological membrane aberrancies and the protein trafficking defects observed in arf1Δ mutants.
ACKNOWLEDGMENTS
We are grateful to J. Michael McCaffery for his expert assistance in the electron microscopy work and Tom Pilgrim for photographic assistance; David Botstein, Tim Stearns, Sandra Harris, and Gerry Waters for strains and plasmids; and Randy Schekman, Marja Makarow, and Greg Payne for antibodies. We are particularly grateful to Rick Kahn for the arf1–3 ts allele and antibodies to ARF. We acknowledge Daryll DeWald for initially observing the underglycosylation of CPY in arf1Δ cells, Beverly Wendland for collaborative work on FM4–64 staining of BFA-treated cells, and thank members of the Graham and Emr laboratories for helpful discussion. This work was supported by grants from the National Institutes of Health (GM-32703 and CA-58689 to S.D.E.; GM-50409 to T.R.G.) and the National Science Foundation (MCB-9600835 to T.R.G.). The CCD camera and image analysis system was purchased with funds from a grant from the National Science Foundation (BIR-9419667).
REFERENCES
- Bankaitis V, Aitken J, Cleves A, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Boman AL, Kahn RA. Arf proteins: the membrane traffic police? Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Thomas GM, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty NF, Truong O, Hsuan JJ. Phospholipase D: a downstream effector of ARF in granulocytes. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- Cosson P, Demolliere C, Hennecke S, Duden R, Letourneur F. Delta- and zeta-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Hosobuchi M, Hamamoto S, Winey M, Byers B, Schekman R. Yeast beta- and beta′-coat proteins (COP). Two coatomer subunits essential for endoplasmic reticulum-to-Golgi protein traffic. J Biol Chem. 1994;269:24486–24495. [PubMed] [Google Scholar]
- Esmon B, Novick P, Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981;25:451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a sec18(NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Krasnov VA. Sorting of yeast alpha-1,3-mannosyltransferase is mediated by a lumenal domain interaction, and a transmembrane domain signal that can confer clathrin-dependent Golgi localization to a secreted protein. Mol Biol Cell. 1995;6:809–824. doi: 10.1091/mbc.6.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Scott P, Emr SD. Brefeldin A reversibly blocks early but not late protein transport steps in the yeast secretory pathway. EMBO J. 1993;12:869–877. doi: 10.1002/j.1460-2075.1993.tb05727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Seeger M, Payne GS, MacKay V, Emr SD. Clathrin-dependent localization of α1,3-mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J Cell Biol. 1994;127:667–678. doi: 10.1083/jcb.127.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics A, Orlean P. Glycoprotein synthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Hicke L, Zanolari B, Pypaert M, Roher J, Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol Biol Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosobuchi M, Kreis T, Schekman R. SEC21 is a gene required for ER to Golgi protein transport that encodes a subunit of a yeast coatomer. Nature. 1992;360:603–605. doi: 10.1038/360603a0. [DOI] [PubMed] [Google Scholar]
- Julius D, Blair L, Brake A, Sprague G, Thorner J. Yeast α-factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983;32:839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J Biol Chem. 1984;259:6228–6234. [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistakis NT, Brown HA, Waters MG, Sternweis PC, Roth MG. Evidence that phospholipase D mediates ADP ribosylation factor-dependent formation of Golgi coated vesicles. J Cell Biol. 1996;134:295–306. doi: 10.1083/jcb.134.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJ, Stevens LA, Kao YL, Moss J, Vaughan M. Characterization of a glucose-repressible ADP-ribosylation factor 3 (ARF3) from Saccharomyces cerevisiae. J Biol Chem. 1994;269:20931–20937. [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr S, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Morinaga N, Tsai SC, Moss J, Vaughan M. Isolation of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP ribosylation factor (ARF) 1 and ARF3 that contains a Sec7-like domain. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Orci L, Palmer DJ, Amherdt M, Rothman JE. Coated vesicle assembly in the Golgi requires only coatomer and ARF proteins from the cytosol. Nature. 1993;364:732–734. doi: 10.1038/364732a0. [DOI] [PubMed] [Google Scholar]
- Ostermann J, Orci L, Tani K, Amherdt M, Ravazzola M, Elazar Z, Rothman JE. Stepwise assembly of functionally active transport vesicles. Cell. 1993;75:1015–1025. doi: 10.1016/0092-8674(93)90545-2. [DOI] [PubMed] [Google Scholar]
- Payne GS, Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Phan HL, Finlay JA, Chu DS, Tan PK, Kirchhausen T, Payne GS. The Saccharomyces cerevisiae APS1 gene encodes a homolog of the small subunit of the mammalian clathrin AP-1 complex: evidence for functional interaction with clathrin at the Golgi complex. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer RA, Johnston GC. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad MR, Phan HL, Kirchrath L, Tan PK, Kirchhausen T, Hollenberg CP, Payne GS. Saccharomyces cerevisiae Apl2p, a homologue of the mammalian clathrin AP beta subunit, plays a role in clathrin-dependent Golgi functions. J Cell Sci. 1995;108:1605–1615. doi: 10.1242/jcs.108.4.1605. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y. Three-dimensional electron microscopy: structure of the Golgi apparatus. Eur J Cell Biol. 1990;51:189–200. [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Jackson CL, Kepes F. Effects of brefeldin A on the three-dimensional structure of the Golgi apparatus in a sensitive strain of Saccharomyces cerevisiae. Anat Rec. 1995;241:1–9. doi: 10.1002/ar.1092410102. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Kepes F. Modulation of the Golgi apparatus in Saccharomyces cerevisiae sec7 mutants as seen by three-dimensional electron microscopy. Anat Rec. 1993;237:441–452. doi: 10.1002/ar.1092370402. [DOI] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Russo P, Kalkkinen N, Sareneva H, Paakkola J, Makarow M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc Natl Acad Sci USA. 1992;89:3671–3675. doi: 10.1073/pnas.89.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell JL, Kahn RA. Sequences of the bovine and yeast ADP-ribosylation factor and comparison to other GTP-binding proteins. Proc Natl Acad Sci USA. 1988;85:4620–4624. doi: 10.1073/pnas.85.13.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:3–21. [PubMed] [Google Scholar]
- Spormann DO, Heim J, Wolf DH. Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J Biol Chem. 1992;267:8021–8029. [PubMed] [Google Scholar]
- Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990a;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Willingham MC, Botstein D, Kahn RA. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci USA. 1990b;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon SK. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Takatsuki A, Tamura G. Brefeldin A, a specific inhibitor of intracellular translocation of vesicular stomatitis virus G protein: intracellular accumulation of high mannose type G protein and inhibition of its cell surface expression. Agric Biol Chem. 1985;49:899–902. [Google Scholar]
- Traub LM, Ostram JA, Kornfeld S. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman M, Eli Y, Liscovitch M, Gerst JE. Identification and characterization of a gene encoding phospholipase D activity in yeast. J Biol Chem. 1996;271:2361–2364. doi: 10.1074/jbc.271.5.2361. [DOI] [PubMed] [Google Scholar]
- Waksman M, Tang X, Eli Y, Gerst JE, Liscovitch M. Identification of a novel Ca2+-dependent, phosphatidylethanolamine-hydrolyzing phospholipase D in yeast bearing a disruption in PLD1. J Biol Chem. 1997;272:36–39. doi: 10.1074/jbc.272.1.36. [DOI] [PubMed] [Google Scholar]
- Waters MG, Serafini T, Rothman JE. “Coatomer”: a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- Weidman P, Roth R, Heuser J. Golgi membrane dynamics imaged by freeze-etch electron microscopy: views of different membrane coatings involved in tubulation versus vesiculation. Cell. 1993;75:123–133. [PubMed] [Google Scholar]