Abstract
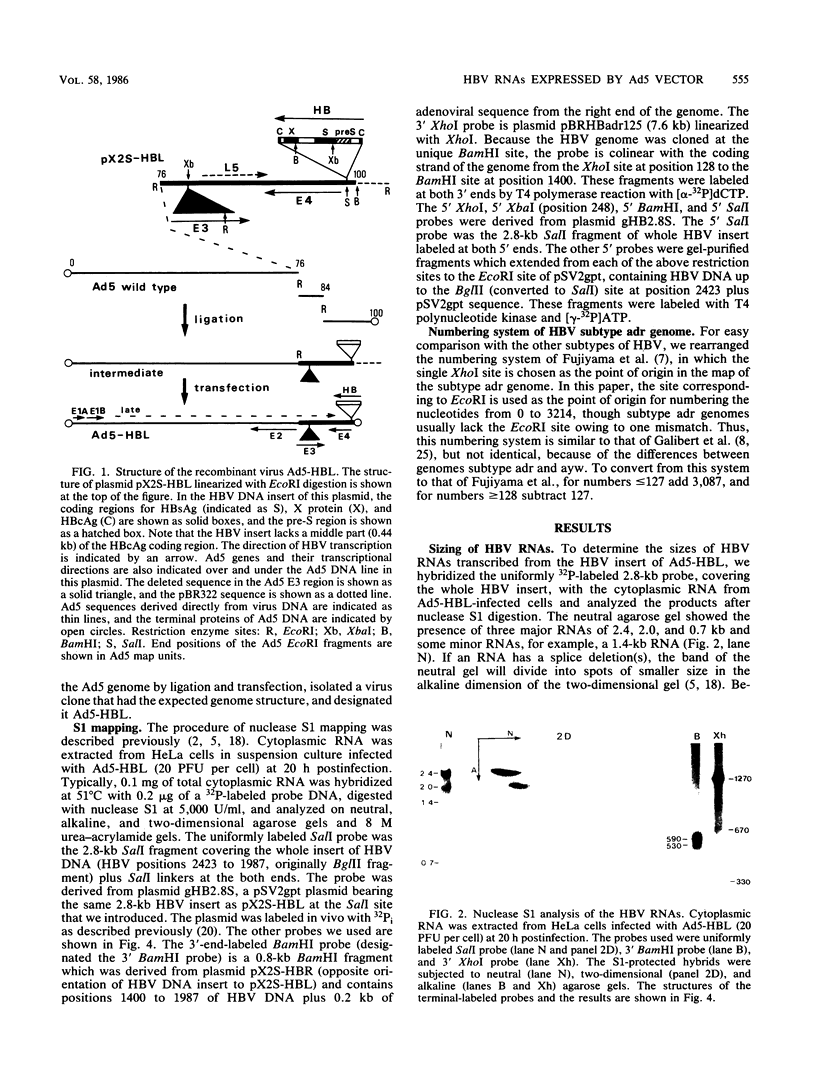
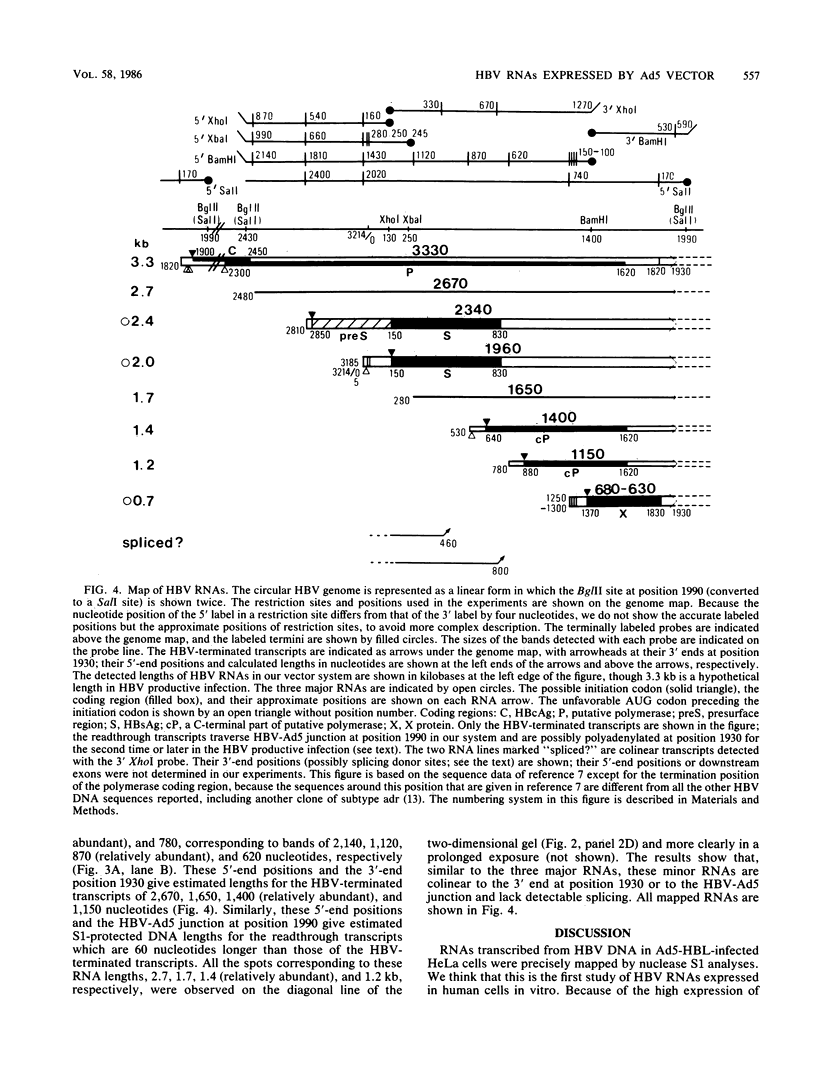
Ad5-HBL is a type 5 adenovirus bearing the large BglII fragment (2.8 kilobases; 87% of the total genome) of hepatitis B virus (HBV), subtype adr. Eight HBV RNAs expressed in HeLa cells infected with Ad5-HBL were mapped by the nuclease S1 technique. Three major RNAs spanning 2.4, 2.0, and 0.7 kilobases of the HBV sequences cover the coding regions of "presurface" plus surface antigen, surface antigen alone, and "X" protein, respectively. The 5' segment of an RNA which could code for core antigen (HBcAg) was also detected. All major HBV RNAs initiate from mutually exclusive 5' ends, terminate at the unique 3' end within the HBcAg coding region (except readthrough species), and have no spliced deletion, forming a novel RNA family structure. No TATA box-like sequences were found near the 5' end of these RNAs, except in the case of the 2.4-kilobase RNA. About two thirds of total HBV RNA does not terminate at the mapped 3'-end position, suggesting the termination signal is functionally inefficient. Since the potential 5' end of HBcAg mRNA was mapped at the same position as the minus-strand nick of HBV DNA previously reported, we propose a model that requires inefficient poly(A) addition to produce an RNA which serves both as HBcAg mRNA and as the putative RNA template of minus-strand DNA synthesis in the HBV life cycle.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Lee F., Harrison T., Williams J., Sharp P. A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979 Aug;17(4):935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Hernandez N., Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983 Sep 22;305(5932):336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Schaller H. Hepatitis B virus transcription in the infected liver. EMBO J. 1984 Sep;3(9):2191–2196. doi: 10.1002/j.1460-2075.1984.tb02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fujiyama A., Miyanohara A., Nozaki C., Yoneyama T., Ohtomo N., Matsubara K. Cloning and structural analyses of hepatitis B virus DNAs, subtype adr. Nucleic Acids Res. 1983 Jul 11;11(13):4601–4610. doi: 10.1093/nar/11.13.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F., Mandart E., Fitoussi F., Tiollais P., Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979 Oct 25;281(5733):646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Gough N. M. Core and E antigen synthesis in rodent cells transformed with hepatitis B virus DNA is associated with greater than genome length viral messenger RNAs. J Mol Biol. 1983 Apr 25;165(4):683–699. doi: 10.1016/s0022-2836(83)80274-5. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub O., Rall L. B., Truett M., Shaul Y., Standring D. N., Valenzuela P., Rutter W. J. Synthesis of hepatitis B surface antigen in mammalian cells: expression of the entire gene and the coding region. J Virol. 1983 Oct;48(1):271–280. doi: 10.1128/jvi.48.1.271-280.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Onda H., Sasada R., Igarashi K., Sugino Y., Nishioka K. The complete nucleotide sequences of the cloned hepatitis B virus DNA; subtype adr and adw. Nucleic Acids Res. 1983 Mar 25;11(6):1747–1757. doi: 10.1093/nar/11.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasek M., Goto T., Gilbert W., Zink B., Schaller H., MacKay P., Leadbetter G., Murray K. Hepatitis B virus genes and their expression in E. coli. Nature. 1979 Dec 6;282(5739):575–579. doi: 10.1038/282575a0. [DOI] [PubMed] [Google Scholar]
- Pourcel C., Louise A., Gervais M., Chenciner N., Dubois M. F., Tiollais P. Transcription of the hepatitis B surface antigen gene in mouse cells transformed with cloned viral DNA. J Virol. 1982 Apr;42(1):100–105. doi: 10.1128/jvi.42.1.100-105.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M., McGeoch D. J. Identification and mapping of two polypeptides encoded within the herpes simplex virus type 1 thymidine kinase gene sequences. J Virol. 1981 May;38(2):593–605. doi: 10.1128/jvi.38.2.593-605.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall L. B., Standring D. N., Laub O., Rutter W. J. Transcription of hepatitis B virus by RNA polymerase II. Mol Cell Biol. 1983 Oct;3(10):1766–1773. doi: 10.1128/mcb.3.10.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Oya Y., Yamamoto K., Yuasa T., Shimojo H. Construction of nondefective adenovirus type 5 bearing a 2.8-kilobase hepatitis B virus DNA near the right end of its genome. J Virol. 1985 Jun;54(3):711–719. doi: 10.1128/jvi.54.3.711-719.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Sato J., Handa H., Shiroki K., Shimojo H. Mapping of RNAs transcribed from adenovirus type 12 early and VA RNA regions. Virology. 1981 Oct 30;114(2):379–398. doi: 10.1016/0042-6822(81)90219-1. [DOI] [PubMed] [Google Scholar]
- Saito I., Shiroki K., Shimojo H. mRNA species and proteins of adenovirus type 12 transforming regions: identification of proteins translated from multiple coding stretches in 2.2 kb region 1B mRNA in vitro and in vivo. Virology. 1983 Jun;127(2):272–289. doi: 10.1016/0042-6822(83)90143-5. [DOI] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring D. N., Rutter W. J., Varmus H. E., Ganem D. Transcription of the hepatitis B surface antigen gene in cultured murine cells initiates within the presurface region. J Virol. 1984 May;50(2):563–571. doi: 10.1128/jvi.50.2.563-571.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenlund A., Lamy D., Moreno-Lopez J., Ahola H., Pettersson U., Tiollais P. Secretion of the hepatitis B virus surface antigen from mouse cells using an extra-chromosomal eucaryotic vector. EMBO J. 1983;2(5):669–673. doi: 10.1002/j.1460-2075.1983.tb01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Tiollais P., Pourcel C., Dejean A. The hepatitis B virus. Nature. 1985 Oct 10;317(6037):489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- Will H., Cattaneo R., Pfaff E., Kuhn C., Roggendorf M., Schaller H. Expression of hepatitis B antigens with a simian virus 40 vector. J Virol. 1984 May;50(2):335–342. doi: 10.1128/jvi.50.2.335-342.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]