Abstract
A new bis-nitrilotriacetic acid (NTA) chelate with catechol anchor was synthesized and immobilized on superparamagnetic iron oxide nanoparticles. When loaded with Ni(II), these bis-NTA-immobilized nanoparticles were shown to bind polyhistidine (His×6-tagged) fusion proteins in their native, folded conformations that commercial microbeads failed to bind under identical conditions. Control experiments with a mono-NTA chelate immobilized on iron oxide nanoparticles indicate a similarly high affinity for His×6-tagged native proteins, suggesting that the high density of the mono-NTA chelate presented by the nanoparticles allows the binding of the His×6-tag to more than one Ni-NTA moiety on the surface. This study shows that the multivalency strategy can be utilized to enhance the binding of His×6-tagged proteins in their native, folded conformations. We further demonstrated the selective purification of His×6-tagged proteins from crude cell lysates by using the Ni(II)-loaded iron oxide nanoparticles. The present platform is capable of efficient purification of His×6-tagged proteins that are expressed at low levels in mammalian cells. This work thus presents a novel nanoparticle-based high-capacity protein purification system with shorter incubation times, proportionally large washes, and significantly smaller elution volumes compared to commercially available microbeads.
INTRODUCTION
As we enter the proteomic era, there is an ever-increasing need for efficient protein purification techniques that allow for direct isolation of proteins from cell lysates.(1) Among many currently used protein purification strategies,(2) immobilized metal affinity chromatography (IMAC) has emerged as one of the most powerful techniques for the purification of recombinant proteins.(3) In the most common IMAC implementation, a His×6 tag that comprises of six consecutively placed histidine residues is incorporated into the C- or N-terminus of a recombinant protein. The His×6 tag binds strongly to a divalent metal chelate such as the Ni(II) nitrilotriacetate complex (Ni-NTA) which is in turn immobilized on a resin. It is believed that four of the six coordination sites on the octahedral Ni(II) center are occupied by the NTA ligand and the remaining coordination sites are occupied by two of the six imidazole moieties in the His×6 tag.(4)
The IMAC strategy allows the purification of His×6-tagged proteins in one or two steps to achieve a moderate degree of purity. Moreover, the His×6 tag is relatively small and generally does not interfere with the native structure and function of the tagged protein. Since its discovery in late 1980s, Ni-NTA based IMAC has been widely used for purifying recombinant proteins, and many His×6-tagged proteins are now commercially available. While this strategy has proven to be largely successful, a significant percentage of recombinant proteins remain difficult to purify by IMAC.(5) Low protein expression is a key contributor to such difficulties as the target protein concentration may be less than 0.1% of the cleared crude lysate when overexpressed in E. coli, and much lower when overexpressed in mammalian cells. Numerous functionally active proteins can only be expressed in mammalian cells, which poses new challenges to protein purification techniques. The target protein structure may also add to purification difficulties by blocking the His×6-tag from the Ni-NTA moiety on the surface of microbeads. Consequently, commercial Ni-NTA resins cannot capture a large number of His×6-tagged proteins under native conditions. To overcome this problem, purification under denaturing conditions has been used to gain full access to the His×6-tag, but overall protein purification efficacy significantly decreases because of the unreliability and inefficiency of in vitro protein re-folding to their native functional conformations.
Significant efforts have been devoted to gaining a better understanding of the Ni-NTA/His×6-tag interactions and thus improving the IMAC protein purification efficiency over the past few years. The strength of the Ni-NTA/His×6-tag interaction was, for example, recently assessed using scanning force microscopic techniques.(6) Ebright et al. elegantly demonstrated enhanced binding of the His×6-tag to a bivalent Ni-NTA system over a monovalent Ni-NTA control using fluorescence anisotropy and fluorescence resonance energy transfer measurements.(7) Piehler et al. has further studied the enhanced affinity of multivalent Ni-NTA-derived molecules toward the histidine tag in great detail.(8,9,10) In the present work, we wish to design a superparamagnetic iron oxide immobilized bivalent Ni-NTA chelate system with the aim of improving IMAC purification of His×6-tagged proteins by strengthening the interactions between the His×6-tag and the bivalent Ni-NTA chelate. The bis-Ni-NTA-immobilized nanoparticles were shown to be capable of binding His×6-tagged proteins in their native, folded conformations that failed to bind commercial microbeads under identical conditions. We have demonstrated that the present system is superior to commercial magnetic beads in binding to His×6-tagged proteins and is useful for isolating target proteins that are overexpressed at low levels in the mammalian cells. Control experiments with a mono-NTA chelate immobilized on iron oxide nanoparticles indicated a similarly high affinity for His×6-tagged proteins, suggesting that the very high density of the mono-NTA chelate presented by the nanoparticles allows the binding of the His×6-tag to more than one NTA moiety on the surface. Thus, this work demonstrates that the multivalency strategy can be utilized to enhance the binding of His×6-tagged proteins in their native, folded conformations.
MATERIALS AND METHODS
General
Nα,Nα-bis(carboxymethyl)-L-lysine was purchased from Fluka. 2-(3,4-Bis-benzyloxyphenyl)-ethylamine-trifluoroacetate-salt (Bn-DA-TFA) and Nα,Nα-bis(carboxymethyl)-L-lysine tribenzyl ester-trifluoroacetate-salt (Bn-NTA-lys-TFA) were prepared from literature procedures.(1,11) All other chemicals were purchased from Sigma-Aldrich and used as received unless otherwise noted. Carboxy-terminated magnetic microparticles were purchased from Bangs Laboratories, Inc. Purified recombinant His×6-endostatin (His×6-rmES) was purchased from Calbiochem. Recombinat human His×6-ubiquitin (His×6-Ub) and mouse His×6-ubiquitin C-terminal hydrolase L-1 (His×6-UCH-L1) were from Boston Biochem. 1H and 13C{1H} NMR spectra were obtained on a Bruker 400MHz AVANCE or Bruker 400MHz DRX. Mass spectra were obtained from Voyager DE-Pro MALDI-MS or HP/Agilent LC/Ion-trap MS in the Department of Chemistry at Duke University. Liquid scintillation counting experiments are performed on a Tri-Carb 2900TR Liquid Scintillation Analyzer. SDS-PAGE gels were quantified using the AlphaEaseFC program. Transmission Electron Microscopy (TEM) images were taken with a JEM-100CXII transmission electron microscope at 100 KV.
Synthesis of dimethyl 5-(3,4-bis(benzyloxy)phenethylcarbamoyl)benzene-1,3-dioate (1)
To a stirring solution of benzene-1,3,5-tricarbonyltrichloride (1.7 g, 6.40 mmol) in dry CH2Cl2 (170 mL) at 0°C was added Bn-DA-TFA (0.536 g, 1.20 mmol) in dry CH2Cl2 (100 mL) dropwise over 5 hours. The solution was allowed to warm to room temperature. The solvent was removed under reduced pressure to produce a white residue. Methanol was added and stirred for a few hours. The excess methanol was removed under reduced pressure to produce a white solid. The product was isolated using flash column chromatography (ethyl acetate/hexanes, 2:3 v/v; Rf = 0.4). The solvents were removed under reduced pressure to yield a white solid of 1 (0.225 g, 0.408 mmol, 34.0% yield). 1H NMR (400MHz, CDCl3): 8.75 (s, 1H), 8.52 (s, 2H), 7.40 (d, 4H), 7.29 (m, 6H), 6.90 (d, 1H), 6.81 (s, 1H), 6.72 (d, 1H), 5.11 (s, 2H), 5.01 (s, 2H), 3.92 (s, 6H), 3.64 (q, 2H), 2.83 (t, 2H); 13C NMR (CDCl3, 100 MHz) δ 165.4, 165.3, 149.0, 147.6, 137.2, 137.0, 135.4, 132.9, 132.0, 131.9, 130.9, 128.3, 127.7, 127.2, 121.5, 115.5, 115.3, 71.3, 71.2, 52.4, 41.4, 34.9, 31.5; MALDI-MS: [M+H]+ m/z 554.0 (calcd 554.6); [M+Na]+, m/z 577.2 (calcd 576.6); [M+K]+ m/z 592.8 (calcd 592.7).
Synthesis of 5-(3,4-bis(benzyloxy)phenethylcarbamoyl)benzene-1,3-dioic acid (2)
To a stirring solution of 1 (0.160, 0.289 mmol) in acetone (6 mL) was added 1N NaOH (4 mL). After 3 hours, complete deprotection was confirmed by TLC. The solution was acidified to pH 3 using 1N HCl. The solvent was removed and the product was stirred in H2O. The precipitate of 2 was collected by suction filtration and recrystallized in acetone (0.145 g, 0.276 mmol, 95.5% yield). 1H NMR (400MHz, MeOD): 8.42 (s, 1H), 8.21 (s, 2H), 7.15−6.90 (m, 10H), 6.67 (s, 1H), 6.62 (d, 1H), 6.49 (d, 1H), 4.73 (s, 4H), 3.28 (t, 2H), 2.54 (t, 2H); 13C NMR (DMSO, 100 MHz) δ 166.4, 164.6, 148.3, 146.8, 137.5, 137.4, 135.5, 132.7, 132.2, 132.0, 131.8, 128.4, 127.8, 127.7, 127.6, 127.5, 121.2, 115.2, 114.8, 70.3, 70.2, 41.1, 34.4; MALDI-MS: [M+H]+ m/z 525.9 (calcd 526.6).
Synthesis of 5-(3,4-bis(benzyloxy)phenethylcarbamoyl)benzene-1,3-benzyl-NTA-lysine (3)
The diacid 2 (0.140 g, 0.266 mmol) was stirred in dry CH2Cl2 (13 mL) to form a cloudy suspension. 0.40 mL of SOCl2 was slowly added and stirring continued at 50 °C for 6 hours to form a clear solution. The solvent was removed under reduced pressure to give the 5-(3,4-bis(benzyloxy)phenethylcarbamoyl)-benzene-1,3-dioyl dichloride intermediate which was used without further purification.
To a stirring mixture of Bn-NTA-lys-TFA (0.408 g, 0.632 mmol), TEA (0.5 mL), and CHCl3 (5 mL) was added a solution of 5-(3,4-bis(benzyloxy)phenethylcarbamoyl)benzene-1,3-dioyl dichloride in CHCl3 (10 mL) dropwise at 0 °C. The reaction was allowed to warm to room temperature and stir overnight. After removal of solvents under reduced pressure, column chromatography (ethyl acetate/hexanes, 3:2 v/v; Rf = 0.5) was used to isolate a pure white solid of 3 (0.281 g, 0.181 mmol, 67.9% yield). 1H NMR (CDCl3, 400 MHz) δ 8.33 (s, 3H), 7.41−7.25 (m, 35H), 6.95 (t), 6.86 (d, 2H), 6.80 (s, 2H), 6.69 (d, 2H), 5.10 (s, 8H), 5.06 (s, 2H), 4.98 (s, 4H), 3.68 (s, 4H), 3.59 (t, 4H), 3.46 (t, 1H), 3.32 (t, 2H) 2.75 (t, 4H), 1,69−1.24 (m, 6H); 13C NMR (CDCl3, 100 MHz) δ 172.8, 171.7, 166.3, 149.3, 148.0, 137.6, 137.5, 135.9, 135.8, 135.6, 135.4, 132.4, 128.8, 128.7, 128.6, 128.5, 128.0, 127.7, 121.7, 116.2, 115.9, 71.6, 66.8, 66.7, 64.5, 53.1, 40.2, 35.4, 29.9, 28.5, 23.1; MALDI-MS: [M+H]+ m/z 1556.3 (calcd 1555.8); [M+Na]+, m/z 1577.5 (calcd 1577.8); [M+K]+, m/z 1593.1 (calcd 1593.9).
Synthesis of 5-(3,4-bis(hydroxy)phenethylcarbamoyl)benzene-1,3-NTA-lysine (4)
3 (0.043 g, 0.052 mmol) was dissolved in 1.5 mL of CHCl3 and 6 mL of MeOH and placed in a Parr flask with 15 mg Pd/C (10%). The mixture was shaken in a Parr reactor for 24 hours under H2 (60 psi). Upon complete deprotection, the palladium catalyst was filtered off using Celite and solvent removed under reduced pressure. The product was purified by recrystallization in MeOH/CH2Cl2 to give 4 in a quantitative yield. 1H NMR (MeOD, 400 MHz) δ; 8.29 (s, 3H), 6.60 (s, 1H), 6.59 (s, 1H), 6.47 (d, 2H), 3.71−3.21 (m, 16H), 2.68 (t, 2H), 1.63−1.45 (m, 8H), 1.18 (m, 4H); 13C NMR (CDCl3, 100 MHz) δ; 174.1, 173.0, 168.6, 146.7, 144.7, 136.6, 132.0, 130.0, 121.1, 117.0, 116.4, 66.9, 54.3, 43.1, 40.7, 35.9, 29.8, 29.8, 24.4; MALDI-MS: [M]+, m/z 833 (calcd 833.8); [M+Na]+, m/z 856 (calcd 856.8).
Synthesis of methyl 3,5-bis(3,4-bis(benzyloxy)phenethylcarbamoyl)benzoate (5)
To a stirring solution of benzene-1,3,5-tricarbonyltrichloride (0.320 g, 1.20 mmol) in dry CH2Cl2 (170 mL) at 0 °C was added Bn-DA-TFA (0.536 g, 1.20 mmol) in dry CH2Cl2 (100 mL) dropwise over 5 hours. The solution was allowed to warm to room temperature. The solvent was removed under reduced pressure to produce a white residue. Methanol was added and allowed to stir for a few hours. The excess methanol was removed under reduced pressure to produce a white solid. The product was isolated using silica flash column chromatography (ethyl acetate/hexanes, 1:1 v/v; Rf = 0.4). The solvents were removed under reduced pressure to yield a white solid of 5 (0.298 g, 0.349 mmol, 29.0% yield). 1H NMR (CDCl3, 400 MHz) δ 8.44 (s, 2H), 8.28 (s, 1H), 7.45−7.25 (m, 20H), 6.88 (d, 2H), 6.81 (s, 2H), 6.71 (d, 2H), 6.56 (t, 2H), 5.10, (s, 8H), 3.88 (s, 3H), 3.60 (q, 4H), 2.80 (t, 4H); 13C NMR (CDCl3, 100 MHz) δ 165.6, 165.3, 148.7, 147.4, 136.9, 136.8, 135.1, 131.9, 130.5, 130.3, 129.6, 128.2, 127.6, 127.2, 127.1, 121.4, 115.4, 115.0, 71.1, 52.2, 41.3, 34.7; MALDI-MS: [M+Na]+, m/z 877.0 (calcd 878.0); [M+K]+, m/z 893.0 (calcd 894.1).
Synthesis of 3,5-bis(3,4-bis(benzyloxy)phenethylcarbamoyl)benzoic acid (6)
5 (0.250 g, 0.292 mmol) was stirred in a solution of 3 mL of acetone and 1 mL of 1M NaOH overnight. The solution was acidified to pH 3 using 1N HCl, added dropwise. The solvent was removed and the product was stirred in H2O. The precipitate was collected by vacuum filtration to give 6 in quantitative yield. 1H NMR (MeOD, 400 MHz) δ 8.53 (s, 2H), 8.37 (s, 1H), 7.40−7.20 (m, 20H), 6.93 (s, 2H), 6.91 (s, 2H), 6.76 (d, 2H), 5.03 (s, 8H), 3.54 (t, 4H), 2.81 (t, 4H); 13C NMR (CDCl3, 100 MHz) δ 169.2, 164.9, 150.4, 148.9, 138.9, 138.8, 136.3, 134.3, 131.9, 129.8, 129.5, 128.9 128.8, 123.1, 117.2, 116.9, 72.7, 72.5, 42.8, 37.0; MALDI-MS: [M+Na]+, m/z 863.4 (calcd 864.0).
Synthesis of benzyl 6-(N3,N5-bis(3,4-bis(benzyloxy)phenethyl)benzene-1,3,5-tricarboxamido)-2-(bis(((benzyloxy)carbonyl)methyl)amino)hexanoate (7)
A mixture of Bn-NTA-lys-TFA (0.060 g, 0.071 mmol), 6 (0.075 g, 0.142 mmol), and HOBt (0.010 g, 0.074 mmol) was stirred in 3 mL anhydrous DMF for 5 minutes. Dicyclohexylcarbodiimide (0.015 g, 0.073 mmol) was added and the mixture was allowed to stir overnight. The solvents were removed under reduced pressure. The product 7 was isolated with silica gel flash column chromatography (ethyl acetate/hexanes, 3/2 v/v; Rf = 0.4). Yield: 0.094 g (97.7%). 1H NMR (CDCl3, 400 MHz) δ 8.43 (s, 1H), 8.41 (s, 2H), 7.50−7.26 (m, 40H), 7.04 (s, 3H), 6.87 (s, 1H), 6.85 (d, 1H), 6.72 (d, 1H), 5.12 (s, 8H), 5.07 (s, 8H), 3.74 (s, 8H), 3.59 (t, 2H), 3.51 (t, 2H), 3.35 (t, 4H), 2.79 (t, 2H), 1.75−1.28 (m, 12H); 13C NMR (CDCl3, 100 MHz) δ 172.5, 171.5, 165.9, 148.9, 147.6, 137.2, 137.1, 135.3, 132.0, 128.5, 128.4, 128.3, 128.2, 128.1, 127.7, 127.4, 127.3, 121.5, 115.7, 115.2, 71.3, 66.5, 66.4, 64.0, 52.8, 41.5, 39.9, 35.0, 27.9, 25.5, 24.8; MALDI-MS: [M+H]+, m/z 1356.4 (calcd 1355.6); [M+Na]+, m/z 1378.0 (calcd 1378.6); [M+K]+, m/z 1393.9 (calcd 1394.7).
Synthesis of 6-(N3,N5-bis(3,4-dihydroxyphenethyl)benzene-1,3,5-tricarboxamido)-2-(bis(carboxymethyl)amino)hexanoic acid (8)
To a solution 7 (0.094 g, 0.069 mmol) in of 5 mL of MeOH and 10 mL of CHCl3 was added 20 mg Pd/C (10%). The mixture was placed in a Parr reactor bottle and rocked for 24 hours at 60 psi H2. Upon complete deprotection, the catalyst was filtered off using Celite and the solvents were removed under reduced pressure. 8 was purified by recrystallization in MeOH/CH2Cl2 in a quantitative yield. 1H NMR (MeOD, 400 MHz) δ 8.34 (s, 3H), 6.69 (s, 2H), 6.67 (s, 2H), 6.56 (d, 2H), 3.64 (t, 4H), 3.60 (t, 1H), 3.33 (s, 4H), 3.29 (t, 2H), 2.77 (t, 4H), 1.90−1.50 (m, 4H), 1.27 (p, 2H); 13C NMR (CDCl3, 100 MHz) δ; 175.3, 174.1, 168.5, 146.1, 144.7, 136.6, 132.0, 129.4, 121.1, 117.0, 116.4, 68.2, 55.1, 43.1, 40.3, 35.9, 26.4, 25.9, 23.6. MALDI-MS: [M+H]+, m/z 724.8 (calcd 725.7); [M+Na]+, m/z 746.8 (calcd 747.7).
Surface modification of iron oxide nanoparticles with bis-NTA-derived Ni(II) chelate and mono-NTA-derived Ni(II) chelate
. The ligand 4 (0.5 mL of a 10 mg/mL MeOH solution) was placed in 1.25 mL distilled water. To this aqueous solution was added 1 mL of iron oxide nanoparticles in n-hexanes (16.3 mg/mL). The two layers were sonicated for 1 hour, and repeatedly washed with n-hexanes, then methanol until the nanoparticles were readily dispersed in distilled water. Using a magnet, the nanoparticles were collected and placed in 2 mL of distilled water. The bis-NTA-pendant ligand-immobilized nanoparticles (0.5 mL) were placed in 2M NiCl2 solution and sonicated for 1 hour. The Ni(II)-loaded nanoparticles were stored in the NiCl2 solution before use. Iron oxide nanoparticle modified with mono-NTA Ni(II)-loaded nanoparticles were similarly prepared.
Preparation of Ni(II)-loaded magnetic microparticles
An aliquot of 0.5 mL (2.4 μmol carboxyl) BioMag Carboxyl magnetic iron oxide microparticles (∼1.5 μm mean diameter, ∼20 mg/mL particle concentration, ∼4.8 μmol/mL carboxy-surface titration) was washed with distilled water (3×500 μL), then with acetone (3×500 μL). Aliquots of N-hydroxysuccinimide (NHS) (25 μL, 30 mg/mL acetone) and dicyclohexylcarbodiimide (50 μL, 50 mg/mL acetone) were added and incubated overnight. The NHS-activated particles were washed with acetone and used without further purification.
A 240 μM solution of Nα,Nα-bis(carboxymethyl)-L-lysine was prepared by dissolving 0.629 mg Nα,Nα-bis(carboxymethyl)-L-lysine in 35 μL of triethylamine, 500 μL of acetone, and 500 μL of distilled water. The NHS-activated microparticles were washed with distilled water, then placed in 100 μL of the Nα,Nα-bis(carboxymethyl)-L-lysine solution. After overnight incubation, the particles were washed with distilled water. The particles were placed in a 1M NiCl2 solution for 1 hour before use.
Isolation of recombinant mouse His×6-Endostatin (His×6-rmES) with Ni(II)-loaded nanoparticles
The Ni(II)-loaded nanoparticles (7−10 μL; 16.3 mg/mL) were washed in distilled water (3×50 μL) and a binding buffer (3×50 μL; 50 mM phosphate buffer system, 300 mM NaCl, 10 mM imidazole, pH 8.0). The washed nanoparticles were incubated with 10 μg of His×6-rmES in 40 μL of binding buffer for 5−60 minutes at 4 °C. Using a centrifuge and magnet, the supernatant was collected and nanoparticles washed in 50 μL aliquots of binding buffer (FT: Flow Through, W1: Wash 1, W2: Wash 2, W3: Wash 3). After the third wash, 50 μL of the elution buffer (50 mM phosphate buffer system, 300 mM NaCl, 250 mM imidazole, pH 8.0) was added and the mixture tumbled for 10 minutes at 4 °C. The eluents were similarly collected by centrifugation and magnetic immobilization (E1: Elution 1, E2: Elution 2). Non-specifically bound protein was released from the nanoparticles by boiling at 95 °C for 4 minutes in a Laemmli SDS-PAGE gel loading buffer. The volumes of the aliquots were reduced in a Speedvac before loading to an SDS-PAGE (12.5%, 180V, 50 min). After separation, the SDS-PAGE gel was stained in a Coomassie Blue solution (0.1% Coomassie blue, 10% acetic acid, 40% methanol) overnight. Recombinant human His×6-ubiquitin (His×6-Ub), and recombinant mouse His×6-ubiquitin C-terminal hydrolase L-1 (His×6-UCH-L1) were isolated with Ni(II)-loaded nanoparticles in the same fashion. Protein isolation by commercial magnetic agarose beads was performed according to the manufacturer's instructions (Qiagen).
Isolation of His×6-rmES by Ni(II)-loaded microparticles
The Ni(II)-loaded microparticles (10 μL, 20mg/mL) were washed in distilled water (3×50 μL) and binding buffer (3 × 50 μL). The washed microparticles were incubated with 10 μg of His×6-rmES in 40 μL of binding buffer for 60 minutes at 4 °C. The beads were washed and the bound protein eluted similarly. All the fractions were loaded to an SDS-PAGE for analysis as previously described.
Expression of [35S]-methionine-labeled His×6-GMAP-210
The gene coding a 70 kDa fragment of GMAP-210 (Golgi-associated microtubule-binding protein) was PCR-amplified from a human total RNA library (Stratagene). After introducing the T7 promoter and TMV 5’-UTR by second PCR, the cDNA was used as template for a coupled in vitro transcription/translation (TNT) reaction in the presence of 10 μCi [35S]-methionine (Perkin-Elmer) in a total volume of 25 μL for 90 minutes at 30 °C. Protein synthesis was confirmed by SDS-PAGE and autoradiography. The protein was stored at −80 °C until used.
Isolation of [35S]-methionine-labeled His×6-GMAP-210 with Ni(II)-loaded nanoparticles
The Ni(II)-loaded nanoparticles (10 μL; 16.3 mg/mL) were washed in distilled water (3×50 μL) and binding buffer (3×50 μL). The washed nanoparticles were incubated with 5 μL of TNT reaction mixture in 100 μL of binding buffer for 60 minutes at 4 °C. The nanoparticles were washed and the bound protein eluted according to the procedures detailed above. All the fractions including the remaining nanoparticles were transferred to scintillation vials for radioactivity measurements using a liquid scintillation counter. Protein isolation using commercial magnetic agarose beads (Qiagen) was performed according to the manufacturer's instructions, and scintillation counts were measured as mentioned above.
Expression and purification of His×6-UGT from 293T cells using nanoparticles
The cDNA corresponding to the open reading frame of C. elegans UGT was amplified from a C. elegans cDNA library (Invitrogen) using high fidelity Platinum Pfx DNA polymerase (Invitrogen) and sequence-specific primers with CACC at the 5’ end of the forward primer. The full-length cDNA was gel purified and subcloned into the pcDNA3.1D/V5-His TOPO vector (Invitrogen) according to the manufacturer's instructions. The resulting gene-containing plasmid was confirmed by PCR and sequence analysis.
293T cells were cultured in Dulbecco's Minimal Essential Media (DMEM, Gibco) supplemented with 10% Fetal Bovine Serum (FBS, Sigma), 1% L-glutamate (Gibco), and 1% Penicillin/Streptomycin. Cell culture was maintained at 37 °C with 5% CO2. On the day of transfection, the pre-formed UGT plasmid DNA-Lipofectamine complex (500 μL) was added directly to each well. Cells were incubated at 37 °C in 5% CO2 for 4 hours. Afterwards, the media was changed and 2 mL of fresh DMEM (with 10% FBS and 1% L-glutamate) was added. After incubation at 37 °C in a 5% CO2 incubator for a total of 24 hours to allow protein expression, cells were harvested and lysed using a commercial extraction buffer (Biovision). Intact cells were removed by centrifugation and the supernatants were cleared at 21,000×g for 15 minutes at 4 °C. Levels of protein expression were assessed by separation on SDS-PAGE gels, followed by Western blotting onto nitrocellulose membranes (Amersham Pharmacia), and probing with anti-V5 antibody (Invitrogen). The lysates that were prepared were used for binding studies.
To purify the His×6-UGT expressed in 293T cells, the pre-washed, Ni(II)-loaded nanoparticles (10 μL; 16.3 mg/mL) were incubated with 50 μL of cleared crude cell lysate (∼4 μg/μL total protein) in 50 μL of binding buffer for 15 minutes at 4 °C. After washing nanoparticles three times using 50 μL of binding buffer, the bound proteins were eluted using 15 μL of elution buffer. Aliquots of each fraction were loaded to an SDS-PAGE for separation. The protein was detected by Coomassie blue staining.
RESULTS AND DISCUSSION
Surface Modification of Iron Oxide Nanoparticles with Bis-NTA and Mono-NTA Ni(II) Chelates
The iron oxide magnetic nanoparticles were synthesized using protocols previously described.(12,13,14) The nanoparticles were washed in ethanol and transferred into n-hexanes and used without further purification. The concentration of the nanoparticles was estimated based on the dry mass after the removal of the solvents. Iron oxide nanoparticles prepared by different routes gave similar results for protein binding and purification experiments.
Although a number of functional groups have been immobilized on iron oxide nanoparticles, their binding strength and their ease of surface immobilization vary significantly. Since Xu and co-workers recently demonstrated the efficient immobilization of dopamine derivatives on iron oxide nanoparticles,(15) we have decided to use this strategy to immobilize bis-NTA Ni(II) chelate for protein purifications. The new bis-NTA ligand with a catechol group (4) was synthesized according to Scheme 1. Compound 4 was synthesized in 22% overall yield in 4 steps starting from known benzyl-protected dopamine trifluoroacetate salt and benzene-1,3,5-tricarbonyltrichloride. New compounds 1−4 were characterized by 1H and 13C{1H} NMR spectroscopy and mass spectrometry.
Scheme 1.
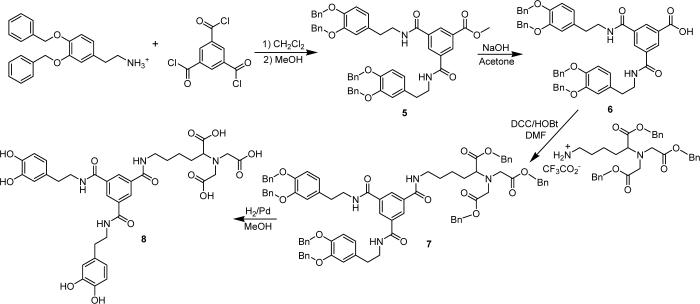
In order to determine the effects of multivalency on protein binding, we have also prepared a new mono-NTA ligand with two catechol groups (8) by using a 1:1 ratio of dopamine hydrochloride and benzene-1,3,5-tricarbonyltrichloride as shown in Scheme 2. Compound 8 was synthesized in an overall yield of 28% in 4 steps. New compounds 5−8 were characterized by 1H and 13C{1H} NMR spectroscopy and mass spectrometry.
Scheme 2.
Surface modification of iron oxide nanoparticles with new NTA-derived ligands was accomplished by simply stirring a biphasic mixture of the ligand in water and iron oxide nanoparticles in hexanes. The NTA-ligand-immobilized nanoparticles were charged with Ni(II) ions by ultrasonic mixing in 2M NiCl2. The iron oxide nanoparticles with different coatings were characterized by transmission electron microscopy (TEM). TEM images of ∼10 nm monodisperse iron oxide nanoparticles (prepared by thermal decomposition) before and after surface modifications illustrate the retention of nanoparticle size and shape, and monodispersity throughout monovalent and bivalent NTA-ligand surface modification and Ni(II)-loading. Slight aggregation of Ni(II)-loaded nanoparticles was however observed in the TEM images (Figure 1).
Figure 1.
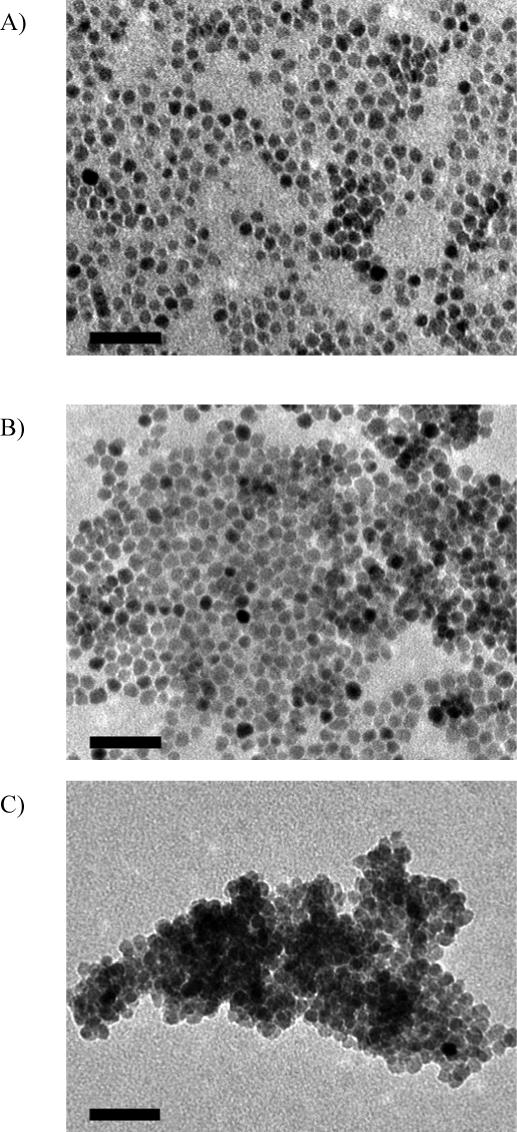
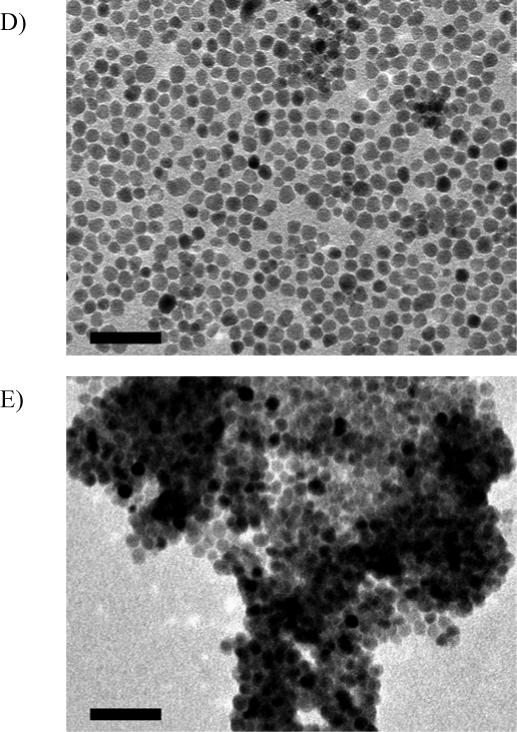
TEM images of monodisperse iron oxide nanoparticles (∼10 nm in diameter): A) Prepared by thermal decomposition and stabilized with oleic acid; B) Modified with bis-NTA ligand; C) Ni(II)-loaded bis-NTA nanoparticle; D) Modified with mono-NTA ligand; E) Ni(II)-loaded mono-NTA nanoparticle. The scale bar is 50 nm.
Binding of His×6-tagged Proteins Labeled with [35S]-Methionine
Immobilization of proteins onto the nanoparticles and subsequent elution were initially demonstrated with His×6-tagged, [35S]-methionine-labeled GMAP-210 protein synthesized via coupled in vitro transcription and translation (Scheme 3). In a typical binding-elution experiment, the direct lysate (DL) signal is equal to the sum of the flow through (FT), washes (W1, W2, W3), elutions (E1, E2), and residual resin signals. The signal from each fraction was measured using liquid scintillation counting.
Scheme 3.

Since only a small percentage of [35S]-methionine was incorporated into the recombinant protein in the TNT reaction, a majority of the radioactivity in both the commercial beads and nanoparticle systems came from unbound free [35S]-methionine in the FT (Figure 2 and Table S1). Following the removal of FT, the resin was washed three times until the background was low, before elution by imidazole. This scintillation counting assay demonstrates that the NTA-modified nanoparticles can specifically capture His×6-tagged proteins followed by efficient elution with imidazole. We also examined the relative protein binding capacity of nanoparticles as compared to the commercial beads. It was found that the 10 μL (0.32% v/v suspension, assuming Fe3O4 density of 5.15 g/cm3) of nanoparticle (16.3 mg/mL) had a similar binding capacity to 10 μL (5% v/v suspension) of commercial magnetic agarose beads. It appears that the unoptimized nanoparticles used in this work have higher residual radioactivity after elutions when compared to the commercial beads, presumably due to relatively strong nonspecific binding. This result was within expectation, since the commercial beads were coated with agarose, which has low non-specific binding to proteins. We think that the non-specific binding of the nanoparticle could be minimized if its surface is modified with hydrophilic molecules such as PEG that are known to resist nonspecific protein binding.(16)
Figure 2.
Graphical representation of [35S]-labeled His×6-GMAP-210 binding scintillation counting assay for bis-Ni-NTA nanoparticles (solid bars) and commercial Ni-NTA agarose beads (open bars). Fractions collected and radioactivity quantified in counts per minute (CPM), DL = Direct Lysate, FT = Flow Through, W1 = Wash 1, W2 = Wash 2, W3 = Wash 3, E1 = Elution 1, E2 = Elution 2.
Binding of Overexpressed His×6-tagged Proteins
We next carried out protein binding studies of several commercially available recombinant His×6-tagged proteins. One of the major problems in using Ni-NTA in protein purification is that numerous His×6-tagged proteins can only be purified under denaturing conditions. Consequently, a complicated refolding process is needed that often results in low yield of the active protein. In this work, we focus on examining whether the NTA-modified nanoparticles can be used under native conditions to purify His×6-tagged proteins that require denaturing conditions when commercial NTA beads are used. We used recombinant murine His×6-endostatin as an example. Endostatin is a potent angiogenesis inhibitor that has shown therapeutic suppression of tumor-induced angiogenesis in mice implanted with tumors.(17) O'Reilly et al. reported that recombinant murine endostatin with His×6 tag could be expressed in E. coli as insoluble inclusion bodies and purified using Ni-NTA resin under denaturing conditions. Endostatin thus purified underwent an inefficient refolding process before it could be used in xenograft mouse studies. We found that the already folded endostatin could not be captured when commercial Ni(II)-NTA beads were used. As shown in Figure 3, approximately 90% of His×6-rmES was present in the FT, and almost no protein was detected in the elutions, indicating that commercial beads failed to capture His×6-rmES under native conditions. Significantly, Ni(II)-loaded nanoparticles readily bound His×6-rmES without the use of denaturing conditions. A 10 μL volume of Ni(II)-loaded nanoparticles (163 μg) is found to have a binding capacity of 5.6 μg under unoptimized conditions. The His×6-rmES was readily eluted by imidazole and was shown to be biologically active in inducing apoptosis in cow pulmonary artery endothelial cells (Figure S1)(18). Confocal microscopic images showed characteristic apoptotic rings for the cow pulmonary artery endothelial cells after annexin-V PE staining. This result indicates that milligrams of a biologically active native protein can be purified using approximately 30 mg of Ni(II)-loaded nanoparticles. We have also successfully recycled and reused the modified nanoparticles in His×6-rmES binding without Ni(II) reloading. The recycled nanoparticle appears to have a similar binding capacity to the pristine sample based on SDS-PAGE analysis (Figure S2).
Figure 3.
Binding assay of His×6-rmES: (A) Commercial magnetic agarose beads (Qiagen): no protein was bound to the bead or eluted by imidazole. Most of the protein was washed away unbound in the FT. (B) Bis-Ni-NTA NPs: His×6-tag protein was bound and a majority was eluted from the nanoparticles by imidazole. (C) Graphical representation of densitometry quantification of SDS-PAGE gels.
We also compared the binding of other His×6-tagged proteins that do not require denaturing conditions for Ni-NTA based purifications. Ubiquitin is a protein found throughout all eukaryotic cells and plays key roles in highly specific protein degradation. The small His×6-tagged ubiquitin (9.3 kDa) is expected to have an affinity for both the commercial beads as well as the surface modified nanoparticles. SDS-PAGE analysis confirms the affinity of the commercial beads and the Ni(II)-loaded nanoparticle (Table 1 and Figure S3). Densitometry analysis shows the binding capacity of 20 μL (5% v/v suspension) of commercial beads is 1.1 μg His×6-Ub, while that of 10 μL (0.32% v/v suspension) of bis-NTA-Ni(II)-loaded nanoparticle is 2.3 μg. In comparing the binding of 10 μL (0.32% v/v suspension) of nanoparticle for His×6-rmES (21.3 kDa, 5.6 μg) and 6×His-Ub (9.3 kDa, 2.3 μg), it is estimated that the binding capacity of the Ni(II)-loaded nanoparticles is approximate 25 pmol of proteins/μL.
Table 1.
Intensity Density Values (% IDV) compiled from densitometry quantification of SDS-PAGE results for collected fractions comparing bis-Ni-NTA-modified nanoparticles and commercial Ni-NTA agarose beads in His×6-tagged protein binding experiment (10 μg total protein).
| FT | W1 | W2 | W3 | E1 | E2 | NP/resin | |
|---|---|---|---|---|---|---|---|
| 6×His-rmES + 2NiNTA-NP | 30.48 | 1.05 | 1.70 | 0.67 | 56.20 | 1.21 | 8.70 |
| 6×His-rmES + Com. Beads | 89.58 | 6.06 | 0.44 | 0.44 | 0.54 | 0.18 | 2.74 |
| 6×His-Ub + 2NiNTA-NP | 26.30 | 18.22 | 9.80 | 3.07 | 23.21 | 7.85 | 11.54 |
| 6×His-Ub + Com. Beads | 39.35 | 23.76 | 8.31 | 5.10 | 10.70 | 6.24 | 6.54 |
| 6×His-UCH-L1 + 2NiNTA NP | 31.80 | 12.38 | 1.25 | 1.16 | 34.78 | 3.30 | 15.33 |
| 6×His-UCH-L1 + Com. Beads | 83.16 | 6.19 | 0.89 | 0.89 | 0.58 | 0.03 | 8.27 |
Binding studies on His×6-UCH-L1 (∼25 kDa) were also used to demonstrate the nanoparticle's ability to capture His×6-enzymes (Table 1 and Figure S4). Binding capacities of 40 μL (5% v/v suspension) of commercial beads and 10 μL (0.32% v/v suspension) of nanoparticle are 0.06 μg and 3.48 μg His×6-UCH-L1, respectively. One major concern in NTA microbeads based protein purification is the large elution volume that requires additional concentration steps before being used in the subsequent experiments. While lyophilization can be used to concentrate most proteins, the process reduces the biological activity of many other proteins. Ultra-filtration is another widely used method in volume reduction. However, the loss of purified proteins could be significant, presumably due to nonspecific interactions with the membrane. Our Ni(II)-loaded nanoparticles disperse well in aqueous solution, and therefore significantly smaller elution volume can be used. This advantage eliminates the need of volume reduction and makes it possible to directly use the proteins after purification. Our studies clearly demonstrated that the present magnetic, recyclable nanoparticles are able to bind important His×6-proteins in native, active conformations with a high capacity and thus show significant advantages over the commercial microbeads. Since the chosen His×6-proteins have a wide range of molecular weights (9387.6 − 74406.8 Da) and isoelectric points (5.43 − 9.33), we do not believe that their binding to the present Ni(II)-loaded nanoparticles are due to simple electrostatic interactions.
Purification of Recombinant His×6-tagged Proteins from Crude Mammalian Cell Lysate
Proteins expressed in mammalian cells are usually soluble and active, and therefore extremely useful in studying their physiological significances. The biggest drawback is that their expression levels are very low, making the purification challenging. The nanoparticle platform was previously used for protein purification from E. Coli cell lysate,(15) in which proteins are much more easily overexpressed. We investigated whether our NTA-loaded nanoparticles could be used to purify recombinant proteins expressed in mammalian cells. 293T cells were used to express a recombinant His×6-UGT protein. The lysate was prepared for purification of 6×His-UGT protein by the bis-Ni(II)-NTA modified nanoparticles. Figure 4 shows the staining of DL, FT, W3, E1, E2, and nanoparticle fractions and clearly indicates the purification of desired His×6-tagged protein from the mammalian crude lysate. The elution E1 is significantly cleaner than the direct lysate, with few impurities. These nonspecifically bound impurities are probably due to proteins with many amino acid residues that can coordinate to the immobilized metal ion. Nonetheless, the present nanoparticle system has shown great promise in the purification of His×6-tagged proteins from crude mammalian cell lysates.
Figure 4.
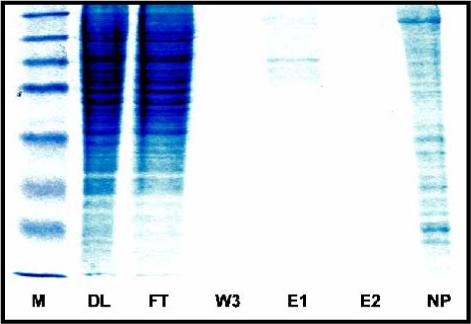
Mammalian Cell Lysate Fractionation. SDS-PAGE (12.5% separation gel) results of flag-His×6-UGT expressed in 293T cells showing marker (M), direct lysate (DL, 10% dilution), flow through (FT, 10%), wash 3 (W3), elution 1 (E1), elution 2 (E2), and protein residue on nanoparticle (NP).
Nonspecific Interactions
Nonspecific interactions between the protein and nanoparticles do not appear to be significant when the NTA nanoparticles are chelated with Ni(II). Table 1 shows approximately 8.7% of His×6-rmES remained on the Ni(II) loaded bis-NTA-modified nanoparticles after elution, slightly higher than that on commercial microbeads. Without the chelation of Ni(II), His×6-rmES was found to have a high affinity for the bis-NTA-modified nanoparticle (Figure S5). Densitometry measurements show that more than 45% of the 6×His-rmES was present in the sum of the washes and elutions, while 39% of the protein remained on the bis-NTA-modified nanoparticles.
On the other hand, freshly prepared iron oxide nanoparticles (without surface modifications) had little interaction with His×6-rmES. Over 70% of the 10 μg of loaded protein was detected in the FT and over 90% was detected in the sum of the FT and washes. The nanoparticles retained approximately 7% of the His×6-rmES after elution, a significant reduction from the 39% retention of Ni(II)-free bis-NTA-modified nanoparticles. The nonspecific interaction of the rmES to the nanoparticle is thus a result of the surface modifications. To rule out the potential role of residual oleic acid on the nanoparticle surface, we carried out a binding experiment using particles without oleic acid stabilization. Nonspecific interaction was observed for the oleic acid-free nanoparticles as well.
As the ratio of His×6-rmES (μg) to bis-Ni(II)-loaded NTA nanoparticle (mg) is increased from 10.2 to 61.3, the amount of non-specifically bound protein appears to remain constant (Figure 5). The specific his-tag binding to Ni(II)-NTA occurs after the full capacity of the nonspecific interaction is reached at a 6×His-rmES (μg) to bis-Ni(II)-loaded NTA nanoparticle (mg) ratio between 10.2 and 15.3. Maximum binding capacity of the nanoparticle via the His×6-tag appears to have been reached with a His×6-rmES (μg) to bis-Ni(II)-loaded NTA nanoparticle (mg) ratio of 61.3 since some 6×His-rmES is detectable in the FT.
Figure 5.
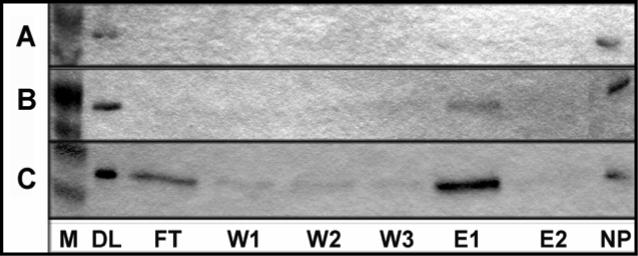
Preference toward nonspecific interaction before his-tag—Ni-NTA interaction demonstrated through increasing ratios of 6×His-rmES protein (μg) to bis-Ni-NTA nanoparticle (mg) A: 10.2, B: 15.3, C: 61.3. (M = Marker, FT = Flow Through, W1 = Wash 1, W2= Wash 2, Wash 3 = W3, E1 = Elution 1, E2 = Elution 2, NP = protein bound nonspecifically to nanoparticle.)
Various attempts were made to block the nonspecific interaction of the target protein (Figure S6). We first attempted to block the nonspecific binding with BSA. The surface modified nanoparticles were pre-incubated with BSA before the His×6-rmES binding experiment was performed. Unfortunately, BSA did not effectively block the His×6-rmES nonspecific binding. An attempt of blocking the His×6-rmES nonspecific interaction of the nanoparticle with a smaller protein (His×6-Ub) did not seem to result in any noticeable improvement by SDS-PAGE analysis.
Surface Density of the Ni-NTA Chelates
The mono-NTA ligand with two dopamine anchors was synthesized to present monovalency of the Ni(II)-NTA moiety onto the nanoparticle surface. Interestingly, an SDS-PAGE binding assay of the mono-Ni(II)-NTA-modified nanoparticle showed a similarly effective binding with His×6-rmES (Figure S7). Furthermore, we compared the mono- and bis-Ni(II)-NTA ligand modified nanoparticles by elution with variable imidazole concentrations, and no significant difference was observed. Unlike commercial Ni-NTA beads, the His×6-rmES could not be eluted from the mono- and bis-Ni(II)-NTA-modified nanoparticles at a pH as low as 4.0 (without the use of imidazole, Figure S8). These results are suggestive of similar binding affinity of mono- and bis-Ni(II)-NTA-modified nanoparticles, probably a result of high surface coverage of mono-Ni(II)-NTA moieties.
To demonstrate the importance of high surface coverage, we conjugated Ni(II)-NTA to micrometer-sized, carboxy-terminated commercial beads (∼1.5 μm diameter). NTA-lysine was conjugated to the beads via the amide bond using the DCC/NHS coupling protocol. Upon loading with Ni(II), these beads were found to bind His×6-rmES as effectively as mono- and bis-Ni(II)-NTA modified nanoparticles (Figure S9). This experiment unambiguously supports the notion that enhanced binding of His×6-tagged protein can be achieved by taking advantage of the multivalency effect.
CONCLUSIONS
New bis-nitrilotriacetic acid (NTA) chelates with catechol anchors were synthesized and immobilized on superparamagnetic iron oxide nanoparticles. When loaded with Ni(II), these NTA-immobilized nanoparticles were shown to bind His×6-tagged proteins in their native, folded conformations with retained activity that commercial microbeads fail to bind under identical conditions. A series of control experiments suggest that the multivalency strategy can be utilized to enhance the binding of His×6-tagged proteins in their native, folded conformations. We further demonstrated the selective purification of His×6-tagged proteins from direct cell lysates by using the Ni(II)-loaded magnetic nanoparticles. The present platform is capable of efficient purification of His×6-tagged proteins that are expressed at low level in mammalian cells. This work thus presents a novel nanoparticle-based high-capacity protein purification system with shorter incubation times, proportionally large washes, and significantly smaller elution volumes compared to currently available microbeads.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge NIH (U54-CA119343) for funding this research. We thank Mr. William J. Reiter for taking the TEM images.
Footnotes
Supporting Information Available. Detailed experimental procedures, data table, and figures areavailable free of charge via the Internet at http://pubs.acs.org.
References
- 1.Porath J, Olin B. Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry. 1983;22:1621–1630. doi: 10.1021/bi00276a015. [DOI] [PubMed] [Google Scholar]
- 2.Garrett RH, Grisham CM. Biochemistry. 2nd Edition Saunders College Publishing; Fort Worth: 1999. [Google Scholar]
- 3.Porath J, Kristiansen T. Biospecific affinity chromatography and related methods. In: Neurath H, Hill RL, editors. The Proteins. 3rd edition Vol. 1. Academic Press; New York: 1975. pp. 95–178. [Google Scholar]
- 4.Hochuli E, Dobeli H, Schacher A. New metal chelate absorbent selective for proteins and peptides containing neighboring histidine residues. J. Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 5.Braun P, Hu Y, Shen B, Halleck A, Koundinya M, Harlow E, LaBaer J. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2654–2659. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conti M, Falini G, Samorì B. How Strong Is the Coordination Bond between a Histidine Tag and Ni - Nitrilotriacetate? An Experiment of Mechanochemistry on Single Molecules. Angew. Chem. Int. Ed. 2000;39:215–218. [PubMed] [Google Scholar]
- 7.Kapanidis AN, Ebright YW, Ebright RH. Site-specific incorporation of fluorescent probes into protein: hexahistidine-tag-mediated fluorescent labeling using (Ni2+:nitrilotriacetic acid)n-fluorochrome conjugates. J. Am. Chem. Soc. 2001;123:12123–12125. doi: 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- 8.Lata S, Gavutis M, Tampé R, Piehler J. Specific and Stable Fluorescence Labeling of Histidine-Tagged Proteins for Dissecting Multi-Protein Complex Formation. J. Am. Chem. Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 9.Lata S, Reichel A, Brock R, Tampé R, Piehler J. High-Affinity Adaptors for Switchable Recognition of Histidine-Tagged Proteins. J. Am. Chem. Soc. 2005;127:10205–10215. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- 10.Hutshenreiter S, Neumann L, Rädler U, Schmitt L, Tampé R. Metal-Chelating Amino Acids As Building Blocks For Synthetic Receptors Sensing Metal Ions And Histidine-Tagged Proteins. ChemBioChem. 2003;4:1340–1344. doi: 10.1002/cbic.200200455. [DOI] [PubMed] [Google Scholar]
- 11.Brook MA, Stan RS. Can, Pat. Appl. CA. 2004 02309486. [Google Scholar]
- 12.Sahoo Y, Pizem H, Fried T, Golodnitsky D, Burstein L, Sukenik CN, Markovich G. Alkyl Phosphonate/Phosphate Coating on Magnetite Nanoparticles: A Comparison with Fatty Acids. Langmuir. 2001;17:7907–7911. [Google Scholar]
- 13.Sun SH, Murray CB, Weller D, Folks L, Moser A. Monodisperse FePt Nanoparticles and Ferromagnetic FePt Nanocrystal Superlattices. Science. 2000;287:1989–1992. doi: 10.1126/science.287.5460.1989. [DOI] [PubMed] [Google Scholar]
- 14.Park J, An K, Hwang Y, Park J-G, Noh H-J, Kim J-Y, Park J-H, Hwang N-M, Hyeon T. Ultra-large-scale syntheses of monodisperse nanocrystals. Nature Mat. 2004;3:891–895. doi: 10.1038/nmat1251. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, Xu K, Gu H, Zheng R, Liu H, Zhang X, Guo Z, Xu B. Dopamine as A Robust Anchor to Immobilize Functional Molecules on the Iron Oxide Shell of Magnetic Nanoparticles. J. Am. Chem. Soc. 2004;126:9938–9939. doi: 10.1021/ja0464802. [DOI] [PubMed] [Google Scholar]
- 16.Bentzen EL, Tomlinson ID, Mason J, Gresch P, Warnement MR, Wright D, Sanders-Bush E, Blakely R, Rosenthal SJ. Surface Modification To Reduce Nonspecific Binding of Quantum Dots in Live Cell Assays. Bioconj. Chem. 2005;16:1488–1494. doi: 10.1021/bc0502006. [DOI] [PubMed] [Google Scholar]
- 17.O'Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 18.Dhanabal M, Ramchandran R, Waterman MJ, Lu H, Knebelmann B, Segal M, Sukhatme VP. Endostatin induces endothelial cell apoptosis. J. Biol. Chem. 1999;274:11721–11726. doi: 10.1074/jbc.274.17.11721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.