Abstract
The translocator protein (18kDa; TSPO), previously known as peripheral-type benzodiazepine receptor, is a high affinity cholesterol- and drug-binding mitochondrial protein involved in various cell functions including steroidogenesis, apoptosis, and proliferation. TSPO is highly expressed in secretory and glandular tissues, especially in steroidogenic cells, and its expression is altered in certain pathological conditions such as cancer and neurological diseases. In this study, we characterized the regulatory elements present in the region of the TPSO promoter extending from 515 to 805-bp upstream of the transcription start site, an area previously identified as being important for transcription. Promoter fragments extending 2.7-kb and 805-bp upstream of the transcription start site were able to direct enhanced green fluorescent protein expression to Leydig cells of the testis, theca cells of the ovary, and cells of the adrenal cortex in transgenic animals. This expression pattern perfectly mimicked endogenous TSPO expression. Functional characterization of the 515 to 805-bp region revealed the presence of one Specificity protein 1/Specificity protein 3 (Sp1/Sp3) and two v-ets erythroblastosis virus E26 oncogene homolog (Ets) binding sites that are important for transcriptional activity in both MA-10 mouse Leydig tumor cells and NIH/3T3 whole mouse embryo fibroblasts. GA-binding protein α (GABPα) – a member of the Ets family of transcription factors – was found to be associated with the endogenous TSPO promoter. We conclude that Sp1/Sp3 and members of the Ets family of transcription factors bind to specific binding sites in the TSPO promoter to drive basal TSPO gene transcription.
Keywords: TSPO, peripheral benzodiazepine receptor, promoter, Sp1, Sp3, Ets, transcription regulation
Introduction
The translocator protein (18kDa) (TSPO), previously known as the peripheral-type benzodiazepine receptor (PBR), has been implicated in many important cellular functions including proliferation and apoptosis, immunomodulation, porphyrin transport, and anion transport (1). TSPO has been most widely studied for its role in the transfer of cholesterol from the cytoplasm to the inner mitochondrial membrane, the rate-determining step of steroid hormone production, where it cooperates with the steroidogenic acute regulatory protein (StAR) (2-7). Consistent with its ability to bind cholesterol and facilitate its transport during steroidogenesis, TSPO is primarily localized in the outer mitochondrial membrane (8), and is a member of a multimeric protein complex whose other constituents include the voltage-dependent anion channel and adenine nucleotide carrier (9). TSPO is also localized to the nucleus and the perinuclear region in breast cancer cells (10) as well as to the plasma membrane of erythrocytes (11).
Variable amounts of TSPO have been detected in all adult tissues examined to date as well as in embryonic tissues (12, 13). Adrenal glands, pineal glands, salivary glands, olfactory epithelium, ependyma, and gonads are particularly rich in TSPO (1, 12, 13). Renal and myocardial tissue exhibit intermediate levels of expression, and the liver and brain exhibit low levels of expression (12, 14). Aside from its abundance in adrenal gland and the gonads, selective localization of TSPO to steroid-producing cells within these tissues supports a role for TSPO in steroid hormone production (8, 15).
TSPO expression is physiologically and pharmacologically modulated. The pituitary gland appears to regulate its expression since hypophysectomy leads to a dramatic decrease in TSPO in the adrenal glands, testis, and ovaries (16, 17). Similarly, adrenalectomy is associated with decreased renal TSPO levels, which can be restored after administration of aldosterone but not dexamethasone (18, 19). Other molecules affecting TSPO expression include interleukin-1 (20), dopamine, serotonin and norepinephrine (21), ginkgolide B (22), TNF-α (23), and several peroxisome proliferators (24).
Altered TSPO expression has been observed in several pathological conditions. For example, several cancers, including those of the breast (10, 25), colon (26), ovary (27), and liver (28), have been associated with increased levels of TSPO, suggesting a role for this protein in tumorigenesis. Changes in the levels of the receptor have also been observed in neurological and psychiatric disorders including Alzheimer's and Huntington's diseases (29-31) and panic disorders (32, reviewed in 33). Finally, hypo- and hyper-thyroidism is associated with altered TSPO expression in the heart, kidney, liver and testis (34, 35).
TSPO cDNA has been cloned for a number of species. In human it encodes a 169 amino acid protein (36-38). The gene is composed of four exons, and exon 1 and half of exon 4 are untranslated (39, 40). In human, mouse, and rat, the short first exon is separated from exon 2 by a large intron containing several areas of repetitive sequence. Transcription of the gene initiates at 56 and 25 nucleotides upstream of the exon1-intron1 junction in rats and humans, respectively (39, 40). In mice, initiation of transcription primarily occurs at an adenine nucleotide situated 61 nucleotides upstream of the translation start site on the cDNA, but internal initiation is also observed (13). The mouse TSPO promoter has been recently cloned, and the proximal area has been functionally characterized in steroidogenic and non-steroidogenic cells (13). Sequence analysis revealed the presence of a series of GC boxes and the absence of TATA or CCAAT boxes. In addition, two overlapping Sp1/Sp3 sites in the proximal promoter were found to be crucial for basal transcription in all cell lines tested.
Despite the abundance of data regarding expression, relatively little is known about the transcriptional regulation of TSPO. In the present study, we set out to identify and characterize the promoter elements present in the 515 to 805 bp region upstream of the TSPO transcription start site. The 805 bp mouse TSPO promoter fragment (as well as the 2.7 kb fragment) was capable of driving expression of enhanced green fluorescent protein (EGFP) to the Leydig cells of the testis, theca cells of the ovary, and cells of the adrenal cortex of transgenic mice, mimicking endogenous TSPO expression. Binding sites for v-ets erythroblastosis virus E26 oncogene homolog (Ets), and specificity protein (Sp) families of transcription factors identified within this region were found to be important for basal promoter activity. These findings appear to be biologically relevant, as members of the Ets family of transcription factors were found to be associated with the endogenous TSPO promoter in MA-10 and NIH/3T3 cells.
Materials And Methods
Transgenic mice
For generation of transgenic mice, the 2700 bp, 805 bp, and 123 bp fragments of the mouse TSPO promoter were freed from plasmids pGL3-2700, pGL3-805, and pGL3-123, respectively (13), with restriction enzyme digestion. The fragments were subcloned in plasmid pd2EGFP (BD Biosciences, San Jose, CA) immediately upstream of the open reading frame (ORF) for EGFP. The resulting plasmids were digested with restriction enzymes and fragments containing only the promoter, EGFP ORF and a polyA signal (devoid of any sequences of bacterial origin) were gel purified and injected into the pronucleus of one cell stage FVB inbred mouse embryos (Taconic, Germantown, NY). Embryos were re-implanted into the oviducts of pseudo-pregnant foster mothers and allowed to develop to term. To identify founder animals, tail DNA was screened by PCR and Southern blot analysis for incorporation of the transgene. The mice were housed and bred in a specific pathogen-free facility. Animals were maintained on a 12-h light, 12-h dark schedule, and food and water were provided ad libitum. All procedures involving animals were performed in accordance with current federal and university guidelines and were reviewed and approved by the Georgetown University Institutional Animal Use and Care Committee.
Transgenic animals were euthanized by carbon dioxide inhalation in accordance with the Animal Welfare Act and institutional guidelines. Tissues to be analyzed were immediately excised and frozen in OCT compound. Seven micron-thick sections were obtained using a Leica CM1900 cryostat (Leica Microsystems Inc, Bannockburn, IL), mounted onto charged slides, counterstained with DAPI (Invitrogen Corp, Carlsbad, CA), and coverslipped in an antifade solution (Invitrogen, Cat # P-7481). Samples were observed and photographed using an Olympus fluoview-FV300 laser scanning confocal microscope (Olympus America Inc, Melville, NY). Background subtraction was carried out using the Olympus Fluoview V. 3C software, with baseline fluorescence being set equal to that emitted from equivalent non-transgenic tissue.
In vitro DNAse I footprinting
A fragment of the TSPO promoter extending from −740 bp to −420 bp was amplified by PCR and cloned in pCR4TOPO vector (Invitrogen Corp.). After sequence verification, the fragment was released by restriction enzyme digestion, isolated, and labeled on the noncoding strand using Klenow enzyme and 32P dATP. Unincorporated nucleotides were removed using a Biospin 6 column (Bio-Rad, Hercules, CA). For DNAse I analysis, approximately 2 ng (40,000 cpm) of the probe were incubated with 20 μg of nuclear extracts from MA-10 cells in the presence of 20 mM Tris pH 7.9, 60 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 0.05% NP-40, 5% glycerol, and 1 μg poly(dI-dC).poly(dI-dC). After allowing complexes to form for 20 min, increasing amounts of DNAse I (Amersham, Piscataway, NJ) were added to parallel reactions and reactions were allowed to proceed for 2 min at room temperature. Reactions were terminated by adding stop solution (200 mM NaOAc, 30 mM EDTA, 0.1% SDS, and 60 μg/ml yeast tRNA). DNA was then phenol/chloroform extracted, ethanol precipitated, and resuspended in loading buffer. DNA fragments were separated on 8% polyacrylamide sequencing gels alongside GC sequencing reactions as size markers. Images were acquired using a phosphor imager.
Plasmids and in vitro site-directed mutagenesis
Plasmids pGL3-805, -585, and -515, which contain the 805, 585, and 515 bp fragments of the mouse TSPO promoter, respectively, cloned upstream of the firefly luciferase gene have been previously described (13). Plasmids pGL3-805m1-m9, each one carrying a 12 bp substitution mutation, as well as plasmid pGL3-805del lacking the −636 to −684 bp region of the TSPO promoter, were constructed by PCR using modified primers containing copies of the NsiI and NheI restriction enzyme recognition sequences at the 5′end, with which the original sequence was substituted. Plasmids pGL3-805mEts, -805mA/P, -805mE/A/P, -805mEts.1, -805mEts.2, -805mEts.1/2 contained two bp substitution mutations (Table 1) and were also constructed by PCR using modified primers and pGL3-805 as template. Plasmids pGL3-585mEts.2 and pGL3-585mN containing substitution mutations (Table 1) were constructed by PCR using modified primers and pGL3-585 as template. All constructs were fully sequenced for verification.
Table 1.
Mutations introduced into plasmids and oligonucleotides used in EMSA
| Mutations introduced into plasmids | ||
|
| ||
| WT sequence | -773-GGTTAAGAACAGGAAGCCC-755- | -568-TGACTCACAGGAAGAGGTT-550- |
| pGL3-805m8 | GGTGCTAGCATGCATGCCC | no mutation |
| pGL3-805mEts/-805mEts.1 | GGTTAAGAACATTAAGCCC | no mutation |
| pGL3-805mA/P | GGTTACTAACAGGAAGCCC | no mutation |
| pGL3-805mE/A/P | GGTTACTAACATTAAGCCC | no mutation |
| pGL3-805mEts.2/pGL3-585mEts.2 | no mutation | TGACTCACATTAAGAGGTT |
| pGL3-805mEts.1/2 | GGTTAAGAACATTAAGCCC | TGACTCACATTAAGAGGTT |
|
| ||
| WT sequence | -543-TGCCAGCAAGTGGGTGT-527 | -570-ATTGACTCACAGG-558 |
| pGL3-585mN | -543-TGCCAGGCTAGCGGTGT-527 | no mutation |
|
| ||
|
Oligonucleotides used in EMSA
| ||
| Ets.1 | GCTGGTTAAGAACAGGAAGCCCGGGA | |
| Ets.1-m1 | GCTGGTTAAGAACATTAAGCCCGGGA | |
| Ets.1-m2 | GCTGGTTAAGAACCGTAAGCCCGGGA | |
| AR/PR-m1 | GCTGGTTACTAACAGGAAGCCCGGGA | |
| AR/PR-m2 | GCTGGTTACGCACAGGAAGCCCGGGA | |
| FP1 | GGGCTGCCAGCAAGTGGGTGTGGCCA | |
| FP1 mutant | GGGCTGCCAGGCTAGCGGTGTGGCCA | |
|
| ||
Cell culture, transfections, and luciferase reporter assays
MA-10 cells were a generous gift of Dr. Mario Ascoli (University of Iowa, IA) and were maintained in DMEM/F12 50:50 medium supplemented with 5% horse serum and 2.5% fetal bovine serum (FBS) at 37°C in a 3.7% CO2 atmosphere. NIH/3T3 cells were maintained in DMEM supplemented with 10% FBS.
For transfection, cells were plated in six-well plates at a density of 200,000 cells/well. Transfection was performed 24 h after plating, using Fugene 6 reagent (Roche Applied Science, Indianapolis, IN) for MA-10 cells and Polyfect reagent (Qiagen, Valencia, CA) for NIH/3T3 cells, according to the manufacturer's instructions. All reporter plasmids were used at equimolar amounts and pUC19 DNA was employed to keep the total amount of DNA constant. Plasmid pRL-TK (Promega Corp., Madison, WI) expressing the Renilla luciferase gene under the control of the thymidine kinase promoter was used to normalize transfection efficiency. Twenty-four hours after transfection, the cells were washed, and total lysates were obtained using passive lysis buffer (Promega Corp). Samples were processed with the Dual-Luciferase Reporter assay (Promega Corp.), and activity was measured using an automated plate reader.
Preparation of nuclear extracts and electromobility shift assay (EMSA)
Nuclear extracts were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Chemical Co., Rockford, IL). For EMSA, double-stranded 5′ biotinylated oligonucleotides Ets.1, Ets.2, and FP1 (Table 1) were synthesized and incubated with 10 μg of nuclear extracts either from MA-10, Y-1, or NIH/3T3 cells. Unlabeled double-stranded Ets.1 and Ets.2 oligonucleotides were used for competition experiments. The following mutated oligonucleotides were also used, where indicated (Table 1): Ets.1-m1, Ets.1-m2, AR/PR-m1, AR/PR-m2, Ets.2-m, Ets.2-mPEA, Ets.2-mAP1, Ets.2-mcMyb, and FP1 mutant. EMSA was carried out using the LightShift Chemiluminescent EMSA kit (Pierce Chemical Co.), according to the supplied protocol, in the presence of EDTA and Mg++. Complexes were separated on 6% nondenaturing polyacrylamide gels, transferred to nylon membranes, and processed for visualization. All antibodies used were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Real time quantitative reverse transcription PCR (qRT-PCR)
Total RNA (500 ng) extracted with the RNeasy mini kit (Qiagen) was reverse transcribed using the Superscript III first-strand cDNA synthesis system for RT-PCR (Invitrogen Corp.) according to the manufacturer's instructions. After digestion with RNase H, first strand cDNA was subjected to 40 cycles of PCR, and amplimers were detected with a DNA engine Opticon 2 continuous fluorescence detection system (MJResearch Inc., Waltham, MA). Amplimers of mouse TSPO mRNA and 18S were detected using SYBR green (Applied Biosystems, Foster City, CA) and specific primers. Amplimers of Ets1, Ets2, GABPα, GABPβ1, and Elk1 were detected using the corresponding Taqman gene expression assays and Taqman chemistry (Applied Biosystems). All data were normalized against amplification of 18S and quantified using the standard curve method.
siRNA transfections
Pooled siRNAs for mouse Ets1, Ets2, GABPα, and GABPβ1 were purchased from Dharmacon Inc. (Chicago, IL). For transfection, MA-10 and NIH/3T3 cells were plated in six-well plates at a density of 150,000 cells/well. Twenty-four hours later, cells were transfected using Xtreme Gene reagent (Roche Applied Science) following the manufacturer's instructions. Forty-eight hours after transfection, cells were lysed, and total RNA was extracted using the RNeasy mini kit (Qiagen). Mock transfections (no siRNA) and transfections using a scrambled siRNA (Applied Biosystems) served as controls.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT kit (Active Motif, Carlsbad, CA) according to a slightly modified version of the manufacturer's protocol. Briefly, cells were grown in three 150 mm dishes to 80% confluency and were crosslinked with media containing 1% formaldehyde for 10 min at room temperature. After crosslinking, cells were collected and homogenized using a dounce homogenizer. Nuclei were pelleted and resuspended. Using a Vibracell VC 130 sonicator (Sonics & Materials Inc., Newtown, CT) fitted with a 3 mm stepped microtip, nuclear DNA was then sheared to an average size of 500 bp with seven 20 sec pulses at 25% power. After shearing, chromatin was immunoprecipitated with 3 μg of antibody overnight at 4°C, and complexes were captured using protein-G beads. After washing and reversal of crosslinks, DNA was subjected to PCR using primers specific for the mouse TSPO distal promoter. Immunoprecipitates obtained with normal mouse IgG served as negative controls.
Sequence and statistical analysis
Sequence analysis was performed with Vector NTi (Invitrogen Corp.), and identification of putative transcription factor binding sites was done with MatInspector V2.2 software (Genomatix Inc., Munchen, Germany). Statistical analyses were performed with Excel (Microsoft Corp., Redmont, WA). Group comparisons were performed using the student's t test and ANOVA as appropriate. Values of p < 0.05 were accepted as significant. All data are expressed as mean ± SD.
Results
The mouse TSPO promoter directs expression of EGFP to testicular Leydig, ovarian theca, and adrenal cortical cells in transgenic mice
We have previously reported that a fragment of the TSPO promoter extending 123 bp upstream of the most commonly used transcription start site possesses approximately 75% of the maximum activity observed with any TSPO promoter fragment in MA-10 Leydig and Y-1 adrenal cortical cells but only 50% of the maximum activity in NIH/3T3 fibroblasts (13). The shortest promoter fragment supporting full activity in all three cell lines extends 805 bp upstream of transcription start site and exhibits similar activity when compared to a fragment extending 2.7 kb (13). To identify the fragment of the promoter that is able to mimic the expression pattern of endogenous TSPO, we generated transgenic mice expressing EGFP under the control the above-mentioned fragments of the mouse TSPO promoter. The gonads and adrenal glands were chosen for analysis, since TSPO expression is highly specific and very well documented in these tissues. All three transgenic lines containing the 2.7 kb construct demonstrated specific expression of EGFP in the Leydig cells of the testis, the theca cells of the ovary, and adrenal cortical cells (Fig. 1). Of the two transgenic lines containing the 805 bp construct, both also demonstrated specific expression of EGFP in the Leydig cells of the testis, the theca cells of the ovary, and adrenal cortical cells (Fig. 1). In contrast, a more widespread expression in steroidogenic and non-steroidogenic cells was observed in these three tissues in animals harboring the 123 bp construct. Expression of EGFP under the control of the 805 and 123 bp promoter fragments was also observed in almost all other tissues examined (data not shown), with various intensities. These data suggest that the promoter fragment extending 805 bp upstream of the transcription start site may possess all the necessary elements for specific expression of TSPO.
Figure 1.
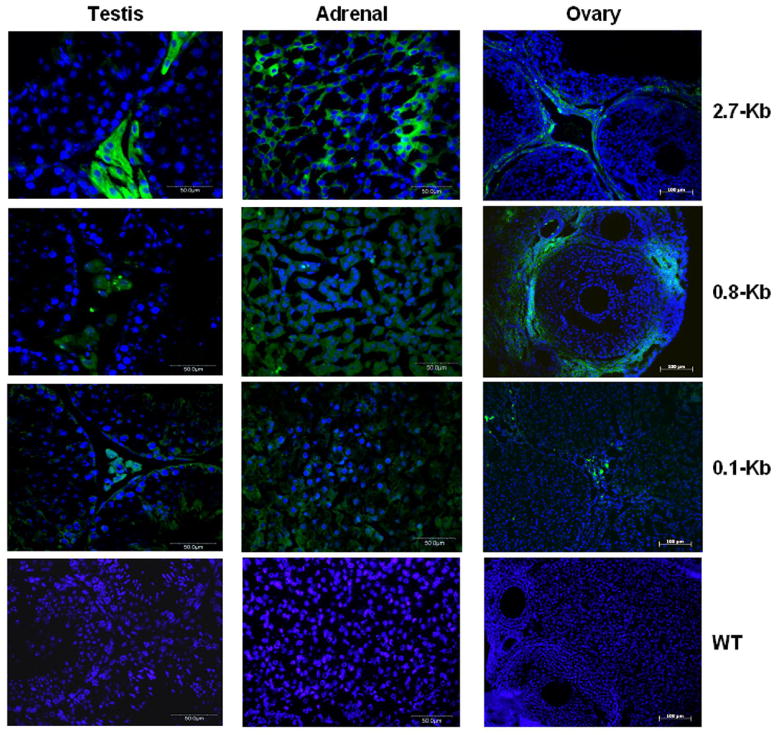
2.7 kb and 805 bp fragments of the TSPO promoter direct expression of EGFP to testicular Leydig, ovarian theca, and adrenal cortical cells in transgenic mice. Fragments of the mouse TSPO promoter extending approximately 2700 bp, 805 bp, and 123 bp upstream of the transcription start site were cloned upstream of the ORF of enhanced green fluorescence protein (EGFP). Transgenic animals generated from these constructs were examined for tissue-specific expression of EGFP (green). Histological sections were counterstained with DAPI (blue). Scale bars represent 50 μm for testis and adrenal and 100 μm for ovary.
Functional characterization of −515 to −585 bp of the mouse TSPO promoter and identification of a Sp1/Sp3 site important for basal promoter activity
We have previously found that the area of the promoter between -515 and -123 bp does not contribute significantly to basal activity in our model systems; whereas, the sequence between -805 and -515 does (13; Fig. 2). This stretch of nucleotides also imparts cell-specific expression of TSPO in adrenal, testis and ovary (Fig. 1). Thus, we analyzed this region for important promoter elements using a combined DNAse I in vitro footprinting, sequence, and site-specific mutant analysis approach. For these studies, we chose to use the MA-10 mouse Leydig tumor cells and NIH/3T3 whole mouse embryo fibroblasts since they express high and low levels of TSPO, respectively.
Figure 2.

Mouse TSPO promoter sequence extending 486 to 812 bp upstream of the transcriptional start site. Underlined sequences denote the core of putative transcription factor binding sites.
Incubation of MA-10 nuclear extract with a radiolabeled fragment of the TSPO promoter extending from -740 to -420 bp, revealed that one area is protected from digestion (FP1, Fig. 3A). Sequence analysis revealed that this protected area coincides with a putative element for transcription factor Nkx3.1 and also comprised an atypical Sp1/Sp3 binding site (Fig. 2).
Figure 3.
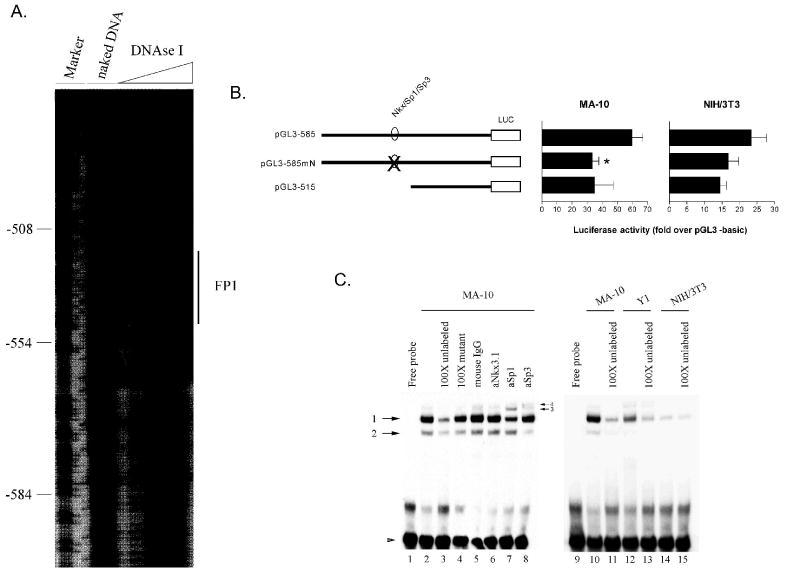
An Sp1/Sp3 site situated between −515 and −585 bp is important for TSPO promoter activity. A. In vitro DNAse I footprinting analysis of the area between −585 to −515 bp of the TSPO promoter. Analysis was performed with MA-10 nuclear extract. The area protected from digestion is indicated (FP1). B. Reporter assays were performed in MA-10 and NIH/3T3 cells with wild type luciferase reporter constructs or constructs carrying a substitution mutation on the Sp1/Sp3 binding site (pGL3-585mN; see Table 1). Luciferase activity is expressed as a fold increase over background, which was measured using the promoterless plasmid pGL3-basic. *p < 0.05. C. Electrophoretic mobility shift assay of nuclear extracts from MA-10, Y-1, or NIH/3T3 cells and a biotinylated double-stranded oligonucleotide spanning protected area FP1. For competition experiments, a 100-fold molar excess of unlabeled probe (lane 3), or a mutant oligonucleotide (lane 4) was used. For super-shift analysis, antibodies specific for Nkx3.1, Sp1 and Sp3 were used (lanes 6-8). Arrows and arrow heads indicate specific complexes and free probe, respectively.
A reporter plasmid carrying a mutation in the core of the putative Nkx3.1/Sp1/Sp3 site was constructed (pGL3-585mN; Table 1) and used to examine the contribution of this site to TSPO transcription. As shown in Figure 3B, mutation of the putative Nkx3.1/Sp1/Sp3 site resulted in a marked decrease in promoter activity in MA-10 cells. This decrease was similar to that seen after deletion of the -585 to -515 bp sequence. Mutation of this site led to only a modest decrease in activity in NIH/3T3 cells.
To determine whether Sp1, Sp3, and/or Nkx3.1 are able to bind this site, EMSA was performed using a biotinylated double-stranded oligonucleotide spanning protected area (FP1, Table 1). Incubation of FP1 with nuclear proteins from MA-10 cells resulted in two shifted complexes (complexes 1 and 2, Fig. 3C, lane 2). Addition of 100-fold molar excess of unlabeled FP1 resulted in reduced density of both of these complexes (lane 3), indicating that binding is specific. Addition of 100-fold molar excess of an unlabeled oligonucleotide carrying a mutation (FP1m, Table 1) did not have any effect (lane 4), indicating that this mutation prevents proteins from binding. Inclusion of an antibody specific for Nkx3.1 transcription factor neither had an effect on the density of these two complexes nor resulted in the formation of a super-shifted complex (lane 6). On the other hand, addition of an antibody specific for Sp1 resulted in super-shifted complex 3 and a reduction in intensity of complex 1 (lane 7) indicating that Sp1 is part of the latter complex. Addition of an anti-Sp3 antibody led to super-shifted complex 4 and depletion of complex 2, indicating that Sp3 is the main component of the latter complex. Indeed, nuclear extracts from MA-10 cells were found to be highly enriched in Sp1 and Sp3 when compared with nuclear extracts from Y-1 and NIH/3T3 cells (lanes 12-15), in agreement with previous results (13).
Functional characterization of−585 to −805 bp of the mouse TSPO promoter in NIH/3T3 cells
To identify strong positive elements in the −585 to −805 bp region, sequence analysis and comprehensive mutagenesis were employed. Sequence analysis of this region revealed the presence of several putative transcription factor binding sites, including elements for NF-Y, STAT3, steroidogenic factor 1 (SF-1), Ets (Ets.1), as well as a half site for the androgen and progesterone receptors (AR/PR) (Fig. 2). For our initial screening we used NIH/3T3 fibroblasts, as this area appears to be more important for basal transcription in this cell line (13). Using pGL3-805 as a template, nine reporter constructs (pGL3-805m1-m9, Fig. 4) were generated, each one carrying a 12 bp substitution mutation. The effect of each mutation was then assessed after transfection. As shown in Figure 4, mutation of the area between −759 and −770 bp (pGL3-805m8), which corresponded to a putative binding site for Ets family transcription factors and a half site for androgen and progesterone receptors (AR and PR respectively), led to an approximate 50% decrease in activity. This decrease was comparable to that observed after deletion of the whole area between −805 and −585 bp (pGL3-585, Fig. 4). Interestingly, this 12 bp substitution resulted in a similar decrease in activity of the promoter construct in MA-10 cells (Fig.5A), although deletion of the area between −805 and −585 bp in this cell line results in only a 15% decrease (13). The remaining 12 bp substitution mutations as well as a deletion between −636 and −684 bp (pGL3-805del, Fig. 4), did not alter the activity of the promoter construct significantly in NIH/3T3 cells.
Figure 4.
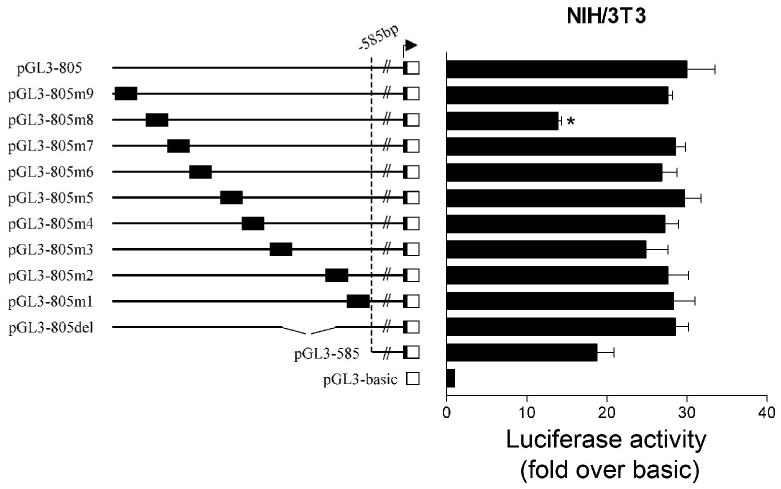
Functional characterization of −585 to −805 bp of the mouse TSPO promoter in NIH/3T3 cells. Luciferase reporter assays were performed using a series of nine constructs (pGL3-805m1-m9) that carried 12 bp substitution mutations (pGL3m1: -596-607 bp, -m2: -630-641 bp, -m3: -679-690 bp, -m4: -693-704 bp, -m5: -706-717 bp, -m6: -732-743 bp, -m7: -746-757 bp, -m8: -759-770 bp, -m9: -791-802 bp) spanning almost the entire −585 to −805 bp region. A reporter construct lacking the sequence between −636 and −684 bp (pGL3-805del) was also tested. Luciferase activity is expressed as a fold increase over background, obtained after transfection with pGL3-basic. Solid boxes indicate the position of the mutated sequence. The arrow indicates the transcription start site. *p< 0.05.
Figure 5.
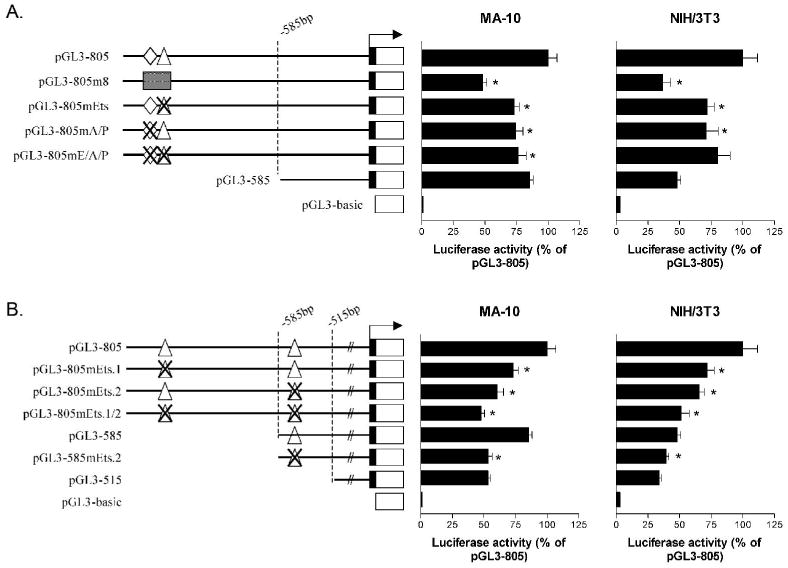
Putative Ets sites located in the−515 to −805 bp region of the mouse TSPO promoter are strong positive elements regulating transcriptional activity. A. Luciferase reporter assays were performed in MA-10 and NIH/3T3 cells using constructs carrying 2 bp substitution mutations for Ets (pGL3-805mEts), AR/PR (pGL3-805mA/P), and both Ets and AR/PR (pGL3–805mE/A/P). B. Luciferase reported assays were performed in MA-10 and NIH/3T3 cells using constructs carrying identical 2 bp substitution mutations in the context of the upstream 805 bp fragment (pGL3-805mEts.2) and the upstream 585 bp fragment (pGL3-585mEts.2). A reporter construct carrying 2 bp substitution mutations on both Ets sites (pGL3-805mEts.1/2) was also examined. Results are expressed as a percentage of the activity of the WT promoter construct (pGL3-805). *p <0.05. The transcription initiation site is indicated by an arrow.
A putative binding site for Ets and an AR/PR half site act as positive elements in the regulation of TSPO promoter activity
To assess the potential contribution of the Ets family of transcription factors and AR/PR on basal transcription of TSPO, reporter plasmids carrying 2 bp substitution mutations for the Ets binding site (pGL3-805mEts), the AR/PR binding site (pGL3-805mA/P), or both sites (pGL3-805mE/A/P) were constructed (Table 1). Mutation of either site resulted in an approximate 25% decrease in promoter activity in both MA-10 and NIH/3T3 cells (Fig. 5A). Mutation of both sites had no additive effect (Fig. 5A), suggesting that only one of the two sites may be utilized at any time.
Identification of a putative Ets binding site located between −515 and −585 bp that is important for basal TSPO promoter activity
In a search for additional putative Ets and/or AR/PR binding sites in the TSPO promoter, we identified a second Ets site (Ets.2, Fig. 2) situated between −515 and −585 bp. As shown in Figure 5B, a 2 bp substitution mutation of Ets.2 in the context of the 805 bp promoter (pGL3-805mEts.2) significantly (p<0.05) decreased promoter activity in both MA-10 and NIH/3T3 cells when compared to WT (pGL3-805, Fig. 5B). An identical mutation of this site in the context of the 585 bp promoter (pGL3-585mEts.2) resulted in a significant (p<0.05) decrease of activity in MA-10 cells, similar to that seen after deletion of −585 to −515 bp. However, mutation of this site in the context of the 585 bp promoter in NIH/3T3 cells did not result in a comparable decrease in activity. Finally, mutation of both Ets sites (pGL3-805mEts.1/2) resulted in an approximate 50% decrease in activity in both cell types (versus wild type; Fig. 5B). Thus, mutation of both Ets sites had a greater effect on promoter activity than mutation of either Ets.1 or Ets.2 alone. These results suggest that both putative sites are important cis-regulatory elements for the activity of the mouse TSPO promoter.
GABPα, a member of the Ets family of transcription factors, binds its corresponding putative elements efficiently and specifically in vitro
To test whether putative Ets.1 and Ets.2 sites were viable binding sites, EMSA analysis was performed using MA-10, NIH/3T3, and Y-1 nuclear proteins. Incubation of NIH/3T3 nuclear extracts with biotinylated, double-stranded oligonucleotide containing the Ets.1 site (Table 1) resulted in one shifted complex (Fig. 6, lane 2). The specificity of this binding was confirmed by the addition of 100-fold excess of unlabeled Ets.1 or an unrelated oligonucleotide (Epstein Barr virus nuclear antigen, EBNA) (lanes 3 and 8, respectively). The addition of 100-fold excess unlabeled oligonucleotides carrying two different mutations on the putative Ets site (Ets.1-m1 and Ets.1-m2, Table 1; Fig. 6, lanes 4 and 5, respectively) did not compete with the specific complex, indicating that an intact Ets binding site is required for the interaction. In contrast, addition of AR/PR-m1 and AR/PR-m2 oligonucleotides (Table 1) carrying two different mutations on the AR/PR putative binding site competed with the specific complex, indicating that the AR/PR half site is not required for the interaction. Addition of mouse a-Ets1, a-Ets2, and a-Elk1 antibodies did not super-shift or immunodeplete the specific complex (lanes 15-17). However, addition of a-GABPα antibody blocked the formation of the specific complex (lane 18), suggesting that GABPα takes part in the interaction. Surprisingly, incubation of Ets.1 oligonucleotide with either MA-10 or Y-1 nuclear proteins did not result in specific shifted complexes (Fig. 6, lanes 10, 11 and 20, 21) under a wide range of binding conditions (not all data shown). The results obtained for Ets.2 were similar to those for Ets.1 (data not shown).
Figure 6.
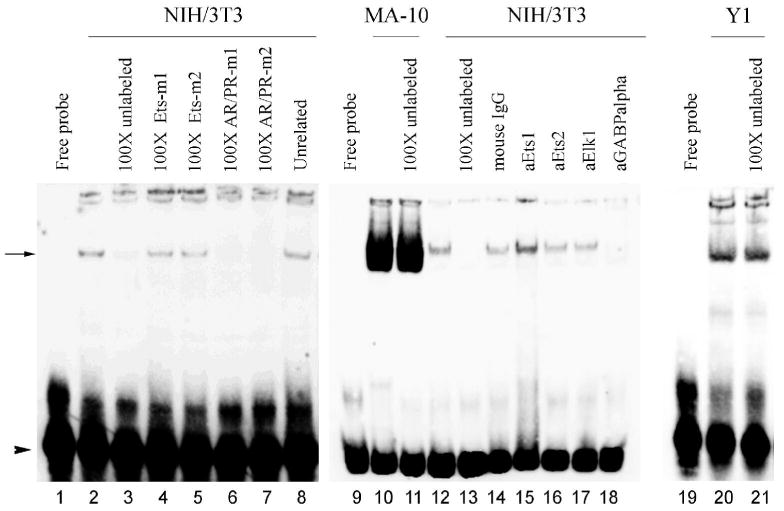
GABPα binds to its corresponding putative binding site in vitro. Electrophoretic mobility shift assay was performed using double-stranded, biotinylated oligonucleotide Ets. and nuclear extract from NIH/3T3, MA-10, or Y-1 cells. Competition experiments with unlabeled probe were performed using Ets.1 oligonucleotide (lanes 3, 11, 13, and 21), mutant oligonucleotides Ets-m1 and Ets-m2 (lanes 4 and 5), mutant oligonucleotides AR/PR-m1 and AR/PR-m2 (lanes 6 and 7), and an unrelated oligonucleotide (lane 8). Super-shift analysis was performed using antibodies specific for Ets1 Ets2, Elk1 and GABPα (lanes 15-18). An arrow indicates specific complex, and the arrow head indicates free, unbound probe.
Members of the Ets family of transcription factors are differentially expressed in NIH/3T3 and MA-10 cells
The preceding data suggest that Ets transcription factors and, more specifically, GABP may play an important role in TSPO transcription regulation. It is possible that different levels of this factor in cells can lead to different transcription rates for TSPO. To test this hypothesis, levels of Ets in NIH/3T3 and MA-10 cells were examined using real time quantitative reverse transcription PCR. Five members of the Ets family of were investigated based on their ability to bind TSPO promoter sequences and their widespread expression. As shown in Figure 7, Ets1 mRNA was undetectable in MA-10 cells after 40 cycles of PCR but was present at low levels in NIH/3T3 cells. Ets2 mRNA was present at low levels in MA-10 cells but at high levels in NIH/3T3 cells. GABPα levels, which were relatively low in both cell lines, were higher in MA-10 cells than in NIH/3T3 cells. Finally, GABPβ1 and Elk1 mRNA were found to be present at low and moderate levels, respectively and were similar in both cell lines. For comparison, the levels of steady state TSPO mRNA in both cell lines is also shown.
Figure 7.
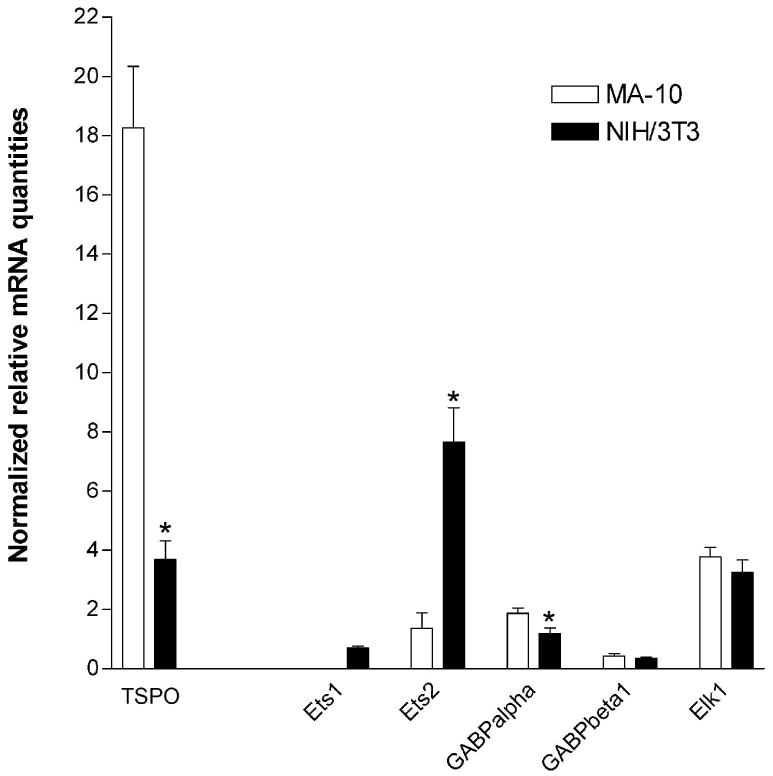
Ets1, Ets2, GABPα, GABPβ1, Elk1, and TSPO mRNA expression in MA-10 and NIH/3T3 cells. Total RNA isolated from MA-10 and NIH/3T3 cells was reverse transcribed and subjected to 40 cycles of real time quantitative PCR. All data were normalized against 18S rRNA levels. Data are derived from two independent experiments performed in triplicate. *p <0.05.
Reduction of Ets1, Ets2, GABPα, and GABPβ1 mRNA leads to changes in TSPO steady state mRNA levels
To investigate the functional role of GABP, Ets1, and Ets2 in the regulation of TSPO transcription, siRNAs were employed. In MA-10 cells, transfection of GABPα siRNA effectively knocked down GABPα mRNA levels and resulted in an approximate 25% decrease in TSPO mRNA levels (p < 0.05; Fig. 8A). GABPβ1 siRNA also resulted in a statistically significant decrease in TSPO mRNA (∼35%, p < 0.05; Fig. 8A). Although transfection of Ets2 siRNA in these cells led to an approximate 75% reduction in Ets2, this did not have any effect on TSPO mRNA levels (Fig. 8A).
Figure 8.
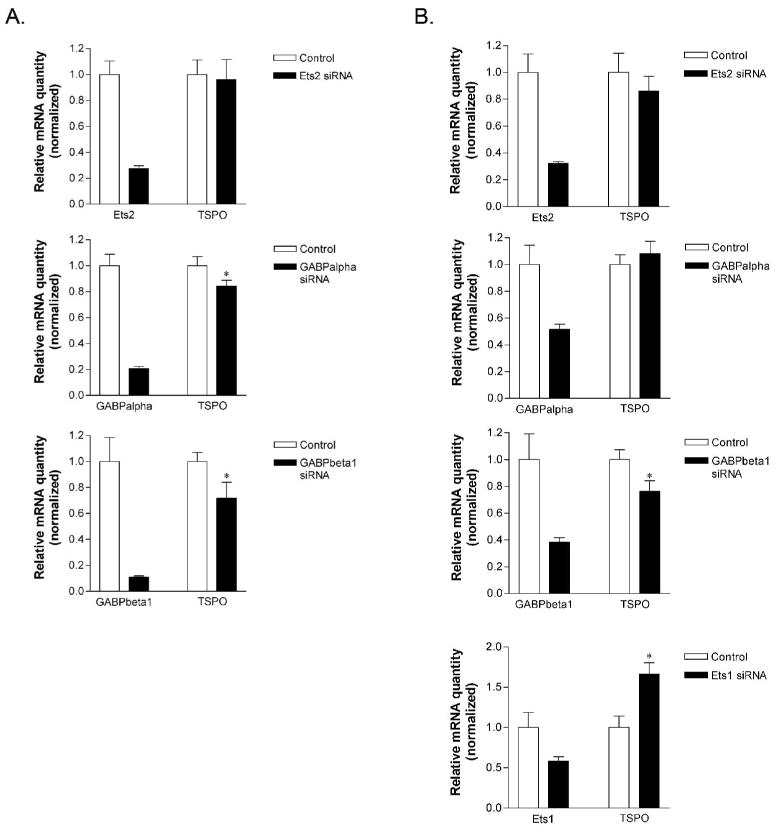
Reduction of GABPα and GABPβ1 mRNA leads to reduced steady-state TSPO mRNA levels. A. MA-10 cells or B. NIH/3T3 cells were transfected with the indicated siRNAs for 48 h, and levels of TSPO, Ets2, GABPα, and GABPβ1 mRNA were quantified using real time PCR. Data are derived from two independent experiments performed in triplicate. *p <0.05.
In NIH/3T3 cells, transfection of GABPα siRNA reduced GABPα mRNA by 50% but had no effect on TSPO mRNA levels (Fig. 8B). Similar to MA-10 cells, a 60% reduction of GABPβ1 mRNA led to a 25% reduction in TSPO mRNA (p < 0.05). Reduction of Ets2 mRNA did not alter TSPO mRNA levels. Interestingly, a 50% reduction in Ets1 mRNA led to a 65% increase in TSPO mRNA (p<0.05).
GABPα and Ets1 transcription factors are bound to the endogenous TSPO promoter in MA-10 and NIH/3T3 cells, respectively
To determine whether members of the Ets family of transcription factors are bound to the endogenous TSPO promoter in intact cells, the ChIP assay was utilized. MA-10 and NIH/3T3 cells were crosslinked using formaldehyde, and chromatin was immunoprecipitated with antibodies specific for Ets1, Ets2, GABPα and Elk1 after shearing. DNA was then subjected to PCR with primers specific for the distal part of the mouse TSPO promoter (arrows, Fig. 9A). As shown in Figure 9B, GABPα was found to be associated with the distal part of TSPO promoter in intact MA-10 cells. However, in NIH/3T3 cells, Ets1, Ets2, and GABPα were found to be associated with the TSPO promoter, with the former being the most prominent (Fig. 9B).
Figure 9.
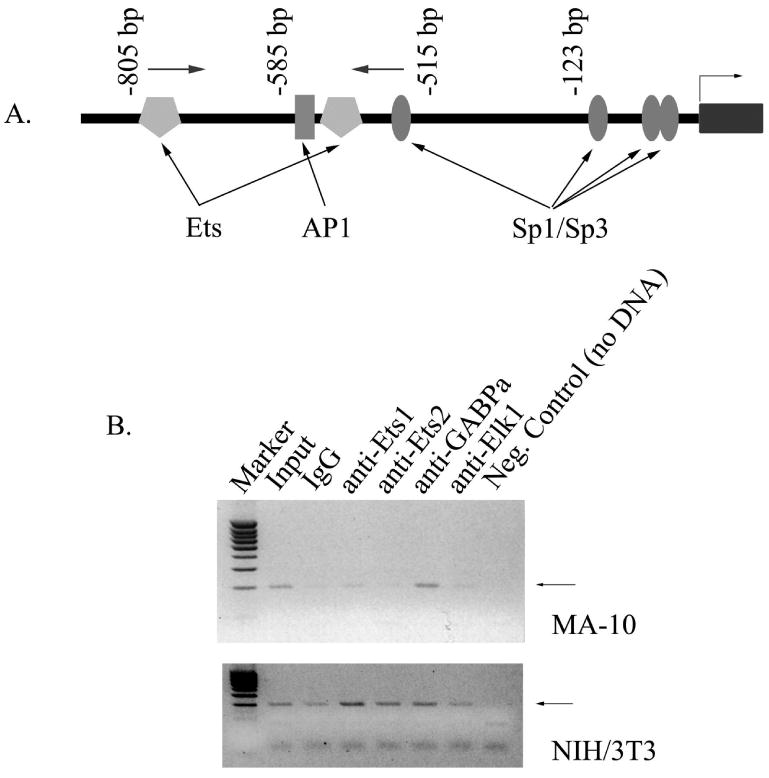
GABPα and Ets1 are bound to the endogenous TSPO promoter in intact MA-10 and NIH/3T3 cells, respectively. A. Schematic representation of the mouse TSPO promoter showing transcription factor binding sites important for activity. Dark rectangular indicates exon 1, while arrows indicate the position of primers used for chromatin immunoprecipitation assay. B. Proteins and DNA from MA-10 and NIH/3T3 cells were crosslinked using formaldehyde. Chromatin shearing, immunoprecipitation with antibodies specific for Ets1, Ets2, GABPα and Elk1, reversal of crosslinks, and isolation of DNA were performed. DNA was then amplified by PCR using primers specific for the distal area of TSPO promoter. Input represents amplification before immunoprecipitation. Normal mouse IgG and a reaction without template served as negative controls. Black arrows indicate specific amplimers.
Discussion
Extensive research on TSPO has suggested a role in many important cellular functions including steroidogenesis, mitochondrial respiration, cellular proliferation, and apoptosis. At the behavioral level, TSPO may also function in coping with stress and anxiety disorders. The involvement of TSPO in such important functions has made it a potential target for treatment of several diseases including cancer, which is often associated with overexpression of the receptor, several psychiatric disorders, which have been associated with reduced levels of TSPO, as well as brain injury. Therefore, understanding the transcriptional regulation of TSPO is of foremost importance.
Previous cloning and functional characterization of the mouse TSPO promoter has indicated that, in addition to two overlapping Sp1/Sp3 sites in the proximal area, the −515 −805 bp region appears to be important for basal transcriptional regulation. It has also been shown that a fragment of the promoter extending 805 bp upstream of the transcription start site possesses maximum transcriptional activity in several cell lines (13). In the current study we show that the promoter fragment extending 805 bp upstream of the transcription start site is able to direct expression of EGFP in Leydig cells of the testis, theca cells of the ovary, and cells of the adrenal cortex, in a fashion that mimics the expression of endogenous TSPO in those tissues. This finding, together with the observation that the sequences between −515 and −123 bp do not appear to be important for transcriptional regulation (13), have led us to focus our search for important cis-regulatory elements in the area between −515 and −805 bp.
Within this region, a DNase I digestion protected area binds to both Sp1 and Sp3 and is important for basal transcription. Sp1 and Sp3 are ubiquitously expressed and bind virtually identical DNA sequences with comparable affinity. Sp1 has been found to act as a transcriptional activator for a large number of genes including structural proteins, metabolic enzymes, cell cycle regulators, transcription factors, and growth factors (41-44). Sp3, on the other hand, can function either as an activator or as a repressor. It achieves the latter through competition with Sp1 for the same sites (45, 46). This has led to the proposal that the function of Sp3 under any given set of conditions is determined by its ratio to Sp1 (41). It is possible that the TSPO transcription rate is regulated by tissue-specific abundance of Sp1 and/or Sp3. In our model systems (MA-10 and NIH/3T3 cells), higher levels of Sp1 and Sp3 are associated with higher levels of TSPO.
Our data also reveal that two Ets sites situated within the −515 − 805 bp region of the promoter are able to bind GABP in vitro and that mutation of these elements leads to decreased promoter activity. Moreover, GABP is associated with the distal area of the TSPO promoter in intact cells. In accordance with these findings, down-regulation of GABP components leads to a reduction in TSPO steady state mRNA. The Ets family of transcription factors consists of at least thirty members. All members share a highly conserved domain that mediates binding to purine-rich sequences containing a core GGAA/T sequence found in many gene promoters (47). Ets transcription factors can activate or repress transcription in cooperation with other transcription factors/ co-factors, and several of them are known to be nuclear targets of signaling pathways (48). Most of their properties, including DNA binding, transcriptional activity, protein-protein interactions, subcellular localization, and stability, are regulated via post-translational modification (47, 48). GABP is an unusual member of the Ets family, in that it is composed of two subunits, one of which confers DNA binding properties (α subunit) and another that confers transactivating activity (β subunit). The abundance and specific expression of Ets factors in different tissues may be, at least in part, responsible for the differences in expression of TSPO. When comparing our model systems, higher levels of GABP mRNA are associated with higher levels of steady state TSPO mRNA. GABP is also a target of c-Raf and JNK (49), and it physically interacts and cooperates with Sp1 and Sp3 for the activation of at least one promoter (50). Interestingly, absence of Ets1 mRNA is also associated with high levels of TSPO mRNA, an observation that is corroborated by the fact that induced downregulation of Ets1 in NIH/3T3 cells results in increased TSPO mRNA. Despite the presence of Ets2 and Ets1/ Ets2 mRNA, in MA-10 and NIH/3T3 cells, respectively, none of these factors were able to bind the TSPO Ets sites in vitro. This may be due to intra molecular interactions among autoinhibitory domains present in these proteins, which prevent them from binding DNA in vitro by an allosteric mechanism (51).
Interestingly, mutation of a half site for AR/PR found adjacent to the first Ets site led to a decrease in activity of the 805 bp promoter fragment. Simultaneous mutation of both the AR/PR and Ets sites did not have any additive effect, suggesting that only one of the two sites is used at any time. In support of this possibility is the close proximity of the two sites. That is, binding of two different transcription factors may be mutually exclusive due to allosteric hindrance. Given that neither androgen nor progesterone receptors are expressed in NIH/3T3 cells (unpublished observations), it is likely that the observed effect is attributable solely to the Ets site and that decrease in activity observed after mutating the AR/PR element are probably due to alteration of the sequences surrounding the core of the Ets binding site. Such alterations have been found to prevent binding of Ets factors or to favor binding of other members of the family present in the cells (52). In MA-10 cells, where the presence of androgen and progesterone receptors has only been studied indirectly and androgen/progesterone regulation via the classical nuclear receptors is unclear (53-55), the observed effects on TSPO promoter activity are also likely attributable to the Ets site.
It is important to note that the DNAse footprinting studies also revealed a partially protected area upstream of FP1 which coincides with the second Ets site characterized in the present study, as well as with a putative AP1 site located in the immediate vicinity. In the future, it will be of great interest to characterize this AP1 site, since AP1 proteins have been found to functionally cooperate with both Ets and Sp transcription factors in the regulation of several promoters (56, 57).
In conclusion, modulation and binding of different Ets transcription factors as well as abundance of Sp1 and Sp3 and possible cooperation with GABP may be critical for tissue- and cell-specific regulation of the TSPO promoter. These findings may provide insight into the aberrant expression of TSPO in several pathological conditions and may aid in the identification of targets for intervention.
Acknowledgments
This work was supported by Grant R01 ES07747 from the National Institutes of Health.
Abbreviations
- TSPO
translocator protein 18kDa
- PBR
peripheral-type benzodiazepine receptor
- StAR
steroidogenic acute regulatory protein
- Sp1
specificity protein 1/Sp1 transcription factor
- Sp3
specificity protein 3/Sp3 transcription factor
- Ets
v-ets erythroblastosis virus E26 oncogene homolog
- GABP
GA binding protein transcription factor
- TNF
tumor necrosis factor
- EGFP
enhanced green fluorescent protein
- ORF
open reading frame
- EMSA
electromobility shift assay
- ChIP
chromatin immunoprecipitation
- Nkx3.1
NK3 transcription factor related, locus 1 (Drosophila)
- AR
androgen receptor
- PR
progesterone receptor
- EBNA
Epstein Barr virus nuclear antigen
- SF-1
steroidogenic factor 1
- NF-Y
nuclear transcription factor Y
- STAT3
signal transducer and activator of transcription 3
- c-Raf
v-raf-1 murine leukemia viral oncogene homolog 1
- JNK
c-Jun N-terminal kinase
- AP1
activator protein 1
References
- 1.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacaperre JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Papadopoulos V. In search of the function of the peripheral-type benzodiazepine receptor. Endocr Res. 2004;30:677–684. doi: 10.1081/erc-200043971. [DOI] [PubMed] [Google Scholar]
- 3.Papadopoulos V, Widmaier EP, Amri H, Zilz A, Li H, Culty M, Castello R, Philip GH, Sridaran R, Drieu K. In vivo studies on the role of the peripheral benzodiazepine receptor (PBR) in steroidogenesis. Endocr Res. 1998;24:479–487. doi: 10.3109/07435809809032636. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Yao Z, Degenhardt B, Teper G, Papadopoulos V. Cholesterol binding at the cholesterol recognition/ interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc Natl Acad Sci USA. 2001;98:1267–1272. doi: 10.1073/pnas.031461598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacapere JJ, Papadopoulos V. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids. 2003;68:569–585. doi: 10.1016/s0039-128x(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 7.Hauet T, Yao ZX, Bose HS, Wall CT, Han Z, Li W, Hales DB, Miller WL, Culty M, Papadopoulos V. Peripheral-type benzodiazepine receptor-mediated action of steroidogenic acute regulatory protein on cholesterol entry into leydig cell mitochondria. Mol Endocrinol. 2005;19:540–554. doi: 10.1210/me.2004-0307. [DOI] [PubMed] [Google Scholar]
- 8.Anholt RR, Pedersen PL, De Souza EB, Snyder SH. The peripheral-type benzodiazepine receptor: Localization to the mitochondrial outer membrane. J Biol Chem. 1986;261:576–583. [PubMed] [Google Scholar]
- 9.McEnery MW, Snowman AM, Trifiletti RR, Snyder SH. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci USA. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999;59:831–842. [PubMed] [Google Scholar]
- 11.Olson JM, Ciliax BJ, Mancini WR, Young AB. Presence of peripheral-type benzodiazepine binding sites on human erythrocyte membranes. Eur J Pharmacol. 1988;152:47–53. doi: 10.1016/0014-2999(88)90834-5. [DOI] [PubMed] [Google Scholar]
- 12.Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A. Enigma of the peripheral benzodiazepine receptor. Pharmacol Rev. 1999;51:629–650. [PubMed] [Google Scholar]
- 13.Giatzakis C, Papadopoulos V. Differential Utilization of the Promoter of Peripheral-Type Benzodiazepine Receptor by Steroidogenic Versus Non-steroidogenic Cell Lines and the Role of Sp1 and Sp3 in the Regulation of Basal Activity. Endocrinology. 2004;145:1113–1123. doi: 10.1210/en.2003-1330. [DOI] [PubMed] [Google Scholar]
- 14.Anholt RR, De Souza EB, Oster-Granite ML, Snyder SH. Peripheral-type benzodiazepine receptors: autoradiographic localization in whole-body sections of neonatal rats. J Pharmacol Exp Ther. 1985;233:517–526. [PubMed] [Google Scholar]
- 15.De Souza EB, Anholt RR, Murphy KM, Snyder SH, Kuhar MJ. Peripheral-type benzodiazepine receptors in endocrine organs: autoradiographic localization in rat pituitary, adrenal, and testis. Endocrinology. 1985;116:567–573. doi: 10.1210/endo-116-2-567. [DOI] [PubMed] [Google Scholar]
- 16.Anholt RR, De Souza EB, Kuhar MJ, Snyder SH. Depletion of peripheral-type benzodiazepine receptors after hypophysectomy in rat adrenal gland and testis. Eur J Pharmacol. 1985;110:41–46. doi: 10.1016/0014-2999(85)90026-3. [DOI] [PubMed] [Google Scholar]
- 17.Bar-Ami S, Fares F, Gavish M. Effect of hypophysectomy and hormone treatment on the induction of peripheral-type benzodiazepine binding sites in female rat genital axis. Horm Metab Res. 1989;21:106–107. doi: 10.1055/s-2007-1009164. [DOI] [PubMed] [Google Scholar]
- 18.Basile AS, Weissman BA, Skolnick P. Maximal electroshock increases the density of [3H]Ro 5-4864 binding to mouse cerebral cortex. Brain Res Bull. 1987;19:1–7. doi: 10.1016/0361-9230(87)90158-4. [DOI] [PubMed] [Google Scholar]
- 19.Basile AS, Ostrowski NL, Skolnick P. Aldosterone-reversible decrease in the density of renal peripheral benzodiazepine receptors in the rat after adrenalectomy. J Pharmacol Exp Ther. 1987;240:1006–1013. [PubMed] [Google Scholar]
- 20.Moynagh PN, O'Neill LA, Williams DC. Interleukin-1 and phorbol myristate acetate modulate the peripheral-type benzodiazepine receptor in lymphocytes and glial cells. Biochem Pharmacol. 1993;46:821–827. doi: 10.1016/0006-2952(93)90490-n. [DOI] [PubMed] [Google Scholar]
- 21.Itzhak Y, Norenberg MD. Regulation of peripheral-type benzodiazepine receptors in cultured astrocytes by monoamine and amino acid neurotransmitters. Brain Res. 1994;660:346–348. doi: 10.1016/0006-8993(94)91311-0. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulos V, Kapsis A, Li H, Amri H, Hardwick M, Culty M, Kasprzyk PG, Carlson M, Moreau JP, Drieu K. Drug-induced inhibition of the peripheral-type benzodiazepine receptor expression and cell proliferation in human breast cancer cells. Anticancer Res. 2000;20:2835–2847. [PubMed] [Google Scholar]
- 23.Rey C, Mauduit C, Naureils O, Benahmed M, Louisot P, Gasnier F. Up-regulation of mitochondrial peripheral benzodiazepine receptor expression by tumor necrosis factor alpha in testicular leydig cells. Possible involvement in cell survival. Biochem Pharmacol. 2000;60:1639–1646. doi: 10.1016/s0006-2952(00)00500-1. [DOI] [PubMed] [Google Scholar]
- 24.Gazouli M, Yao ZX, Boujrad N, Corton JC, Culty M, Papadopoulos V. Effect of peroxisome proliferators on Leydig cell peripheral-type benzodiazepine receptor gene expression, hormone-stimulated cholesterol transport, and steroidogenesis: role of the peroxisome proliferator-activator receptor alpha. Endocrinology. 2002;143:2571–2583. doi: 10.1210/endo.143.7.8895. [DOI] [PubMed] [Google Scholar]
- 25.Han Z, Slack RS, Li W, Papadopoulos V. Expression of peripheral benzodiazepine receptor (PBR) in human tumors relationship to breast, colorectal and prostate tumor progression. Journal of Receptors and Signal Transduction. 2003;23:225–238. doi: 10.1081/rrs-120025210. [DOI] [PubMed] [Google Scholar]
- 26.Katz Y, Eitan A, Amiri Z, Gavish M. Dramatic increase in peripheral benzodiazepine sites in human colonic adenocarcinoma as compared to normal colon. Eur J Pharmacol. 1988;148:483–484. doi: 10.1016/0014-2999(88)90135-5. [DOI] [PubMed] [Google Scholar]
- 27.Katz Y, Ben-Baruch G, Kloog Y, Menczer J, Gavish M. Increased density of peripheral benzodiazepine-binding sites in ovarian carcinomas as compared with benign ovarian tumours and normal ovaries. Clin Sci (Lond) 1990;78:155–158. doi: 10.1042/cs0780155. [DOI] [PubMed] [Google Scholar]
- 28.Venturini I, Zeneroli ML, Corsi L, Avallone R, Farina F, Alho H, Baraldi C, Ferrarese C, Pecora N, Frigo M, Ardizzone G, Arrigo A, Pellicci R, Baraldi M. Up-regulation of peripheral benzodiazepine receptor system in hepatocellular carcinoma. Life Sci. 1998;63:1269–1280. doi: 10.1016/s0024-3205(98)00388-9. [DOI] [PubMed] [Google Scholar]
- 29.Bidder M, Ratzoni G, Weizman A, Blumensohn R, Norymberg M, Tyano S, Gavish M. Platelet benzodiazepine binding in Alzheimer's disease. Biol Psychiatry. 1990;28:641–643. doi: 10.1016/0006-3223(90)90403-o. [DOI] [PubMed] [Google Scholar]
- 30.Owen F, Poulter M, Waddington JL, Mashal RD, Crow TJ. [3H]R05-4864 and [3H]flunitrazepam binding in kainate-lesioned rat striatum and in temporal cortex of brains from patients with senile dementia of the Alzheimer type. Brain Res. 1983;278:373–375. doi: 10.1016/0006-8993(83)90276-7. [DOI] [PubMed] [Google Scholar]
- 31.Messmer K, Reynolds GP. Increased peripheral benzodiazepine binding sites in the brain of patients with Huntington's disease. Neurosci Lett. 1998;241:53–56. doi: 10.1016/s0304-3940(97)00967-1. [DOI] [PubMed] [Google Scholar]
- 32.Pini S, Martini C, Abelli M, Muti M, Gesi C, Montali M, Chelli B, Lucacchini A, Cassano GB. Peripheral-type benzodiazepine receptor binding sites in platelets of patients with panic disorder associated to separation anxiety symptoms. Psychopharmacology (Berl) 2005;181:407–411. doi: 10.1007/s00213-005-2247-x. [DOI] [PubMed] [Google Scholar]
- 33.Veenman M, Gavish M. The peripheral-type benzodiazepine receptor and the cardiovascular system. Implications for drug development. Pharmacol Ther. 2006;110:503–524. doi: 10.1016/j.pharmthera.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Gavish M, Weizman A, Okun F, Youdim MB. Modulatory effects of thyroxine treatment on central and peripheral benzodiazepine receptors in the rat. J Neurochem. 1986;47:1106–1110. doi: 10.1111/j.1471-4159.1986.tb00727.x. [DOI] [PubMed] [Google Scholar]
- 35.Kragie L, Smiehorowski R. Altered peripheral benzodiazepine receptor binding in cardiac and liver tissues from thyroidectomized rats. Life Sci. 1994;55:1911–1918. doi: 10.1016/0024-3205(94)00523-0. [DOI] [PubMed] [Google Scholar]
- 36.Sprengel R, Werner P, Seeburg PH, Mukhin AG, Santi MR, Grayson DR, Guidotti A, Krueger KE. Molecular cloning and expression of cDNA encoding a peripheral-type benzodiazepine receptor. J Biol Chem. 1989;264:20415–20421. [PubMed] [Google Scholar]
- 37.Riond J, Mattei MG, Kaghad M, Dumont X, Guillemot JC, Le Fur G, Caput D, Ferrara P. Molecular cloning and chromosomal localization of a human peripheral-type benzodiazepine receptor. Eur J Biochem. 1991;195:305–311. doi: 10.1111/j.1432-1033.1991.tb15707.x. [DOI] [PubMed] [Google Scholar]
- 38.Parola AL, Stump DG, Pepperl DJ, Krueger KE, Regan JW, Laird HE., 2nd Cloning and expression of a pharmacologically unique bovine peripheral-type benzodiazepine receptor isoquinoline binding protein. J Biol Chem. 1991;266:14082–14087. [PubMed] [Google Scholar]
- 39.Lin D, Chang YJ, Strauss JF, 3rd, Miller WL. The human peripheral benzodiazepine receptor gene: cloning and characterization of alternative splicing in normal tissues and in a patient with congenital lipoid adrenal hyperplasia. Genomics. 1993;18:643–650. doi: 10.1016/s0888-7543(05)80367-2. [DOI] [PubMed] [Google Scholar]
- 40.Casalotti SO, Pelaia G, Yakovlev AG, Csikos T, Grayson DR, Krueger KE. Structure of the rat gene encoding the mitochondrial benzodiazepine receptor. Gene. 1992;121:377–382. doi: 10.1016/0378-1119(92)90147-h. [DOI] [PubMed] [Google Scholar]
- 41.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 42.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 43.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J Biol Chem. 2002;276:29116–29125. doi: 10.1074/jbc.M104130200. [DOI] [PubMed] [Google Scholar]
- 45.Ihn H, LeRoy EC, Trojanowska M. Oncostatin M stimulates transcription of the human alpha2(I) collagen gene via the Sp1/Sp3-binding site. J Biol Chem. 1997;272:24666–24672. doi: 10.1074/jbc.272.39.24666. [DOI] [PubMed] [Google Scholar]
- 46.Majello B, De Luca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 47.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;11:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 48.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 49.Flory E, Hoffmeyer A, Smola U, Rapp UR, Bruder JT. Raf-1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galvagni F, Capo S, Oliviero S. Sp1 and Sp3 physically interact and cooperate with GABP for the activation of the utrophin promoter. J Mol Biol. 2001;306:985–996. doi: 10.1006/jmbi.2000.4335. [DOI] [PubMed] [Google Scholar]
- 51.Jonsen MD, Petersen JM, Xu QP, Graves BJ. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang CY, Petryniak B, Ho IC, Thompson CB, Leiden JM. Evolutionarily conserved Ets family members display distinct DNA binding specificities. J Exp Med. 1992;175:1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]; J Exp Med. 1993;178:1133. Erratum in. [Google Scholar]
- 53.Houk CP, Pearson EJ, Martinelle N, Donahoe PK, Teixeira J. Feedback inhibition of steroidogenic acute regulatory protein expression in vitro and in vivo by androgens. Endocrinology. 2004;145:1269–1275. doi: 10.1210/en.2003-1046. [DOI] [PubMed] [Google Scholar]
- 54.Burgos-Trinidad M, Youngblood GL, Maroto MR, Scheller A, Robins DM, Payne AH. Repression of cAMP-induced expression of the mouse P450 17 alpha-hydroxylase/C17-20 lyase gene (Cyp17) by androgens. Mol Endocrinol. 1997;11:87–96. doi: 10.1210/mend.11.1.9871. [DOI] [PubMed] [Google Scholar]
- 55.Schwarzenbach H, Chakrabarti G, Paust HJ, Mukhopadhyay AK. Gonadotropin-mediated regulation of the murine VEGF expression in MA-10 Leydig cells. J Androl. 2004;25:128–139. doi: 10.1002/j.1939-4640.2004.tb02768.x. [DOI] [PubMed] [Google Scholar]
- 56.Reddy SP, Vuong H, Adiseshaiah P. Interplay between proximal and distal promoter elements is required for squamous differentiation marker induction in the bronchial epithelium: role for ESE-1, Sp1, and AP-1 proteins. J Biol Chem. 2003;278:21378–21387. doi: 10.1074/jbc.M212258200. [DOI] [PubMed] [Google Scholar]
- 57.Hao H, Qi H, Ratnam M. Modulation of the folate receptor type beta gene by coordinate actions of retinoic acid receptors at activator Sp1/ets and repressor AP-1 sites. Blood. 2003;101:4551–4560. doi: 10.1182/blood-2002-10-3174. [DOI] [PubMed] [Google Scholar]


