Abstract
Phage Mu transposes promiscuously, employing MuB protein for target capture. MuB forms stable filaments on A/T-rich DNA, and a correlation between preferred MuB binding and Mu integration has been observed. We have investigated the relationship between MuB-binding and Mu insertion into ‘hot’ and ‘cold’ Mu targets within the E. coli genome. Although higher binding of MuB to select hot versus cold genes was seen in vivo, the hot genes had an average A/T content and were less preferred targets in vitro, whereas cold genes had higher A/T values and were more efficient targets in vitro. These data suggest that A/T-rich regions are unavailable for MuB binding, and that A/T content is not a good predictor of Mu behavior in vivo. Insertion patterns within two hot genes in vivo could be superimposed on those obtained in vitro in reactions employing purified MuA transposase and MuB, ruling out the contribution of a special DNA structure or additional host factors to the hot behavior of these genes. While A/T-rich DNA is a preferred target in vitro, a fragment made up exclusively of A/T was an extremely poor target. A continuous MuB filament assembled along the A/T region likely protects it against the action of MuA. Our results suggest that MuB binds E. coli DNA in an interspersed manner utilizing local A/T richness, and facilitates capture of these bound regions by the transpososome. Actual integration events are then directed to sites that are in proximity to MuB filaments but are themselves free of MuB.
Keywords: Mu DNA transposition, target site selection, MuA transposase, MuB filament
Introduction
Transposable elements employ a variety of strategies for selecting target sites, and display a wide spectrum of target specificities.1, 2 The transposases of some elements choose target sites directly, while others use accessory proteins to mediate this choice. The sequence, structure, as well as transcription and replication status of DNA can influence insertion preference of different elements. The study of target site selectivity provides insights not only into transposition mechanisms, but genome structure and function as well.
Phage Mu is an extremely efficient transposon which gets its name ‘mutator’ from its ability to insert essentially randomly within the E. coli chromosome,3 a randomness confirmed in early studies by fine-mapping of Mu insertions within a single gene.4 Later studies, however, showed preferential regions for Mu insertions within a plasmid,5 near the control region of some genes,6,7 as well as in the whole E. coli genome.8, 9 The preferential integration observed within a plasmid was correlated to binding of the accessory protein MuB.10 DNaseI footprinting identified a MuB-protected region on the plasmid, and Mu insertions were seen to occur on either side of this protected region. A target sequence consensus of 5'-NY(G/C)RN-3' was identified, which was shown to be independent of the presence of MuB i.e reflected the preference of the transposase.10 A more detailed in vitro analysis of this target site consensus was carried out,11 and a bias of CGG at the central three positions was identified in vivo.9
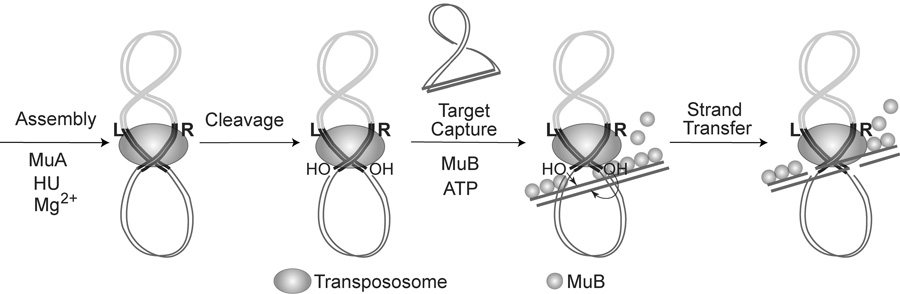
Mu transposition in vitro requires assembly of the transposase MuA on the ends of the Mu genome with assistance from DNA supercoiling and accessory proteins.12 Within the assembled transpososome, Mu ends first undergo single-strand cleavage, followed by strand transfer to target DNA (Fig. 1). The MuB protein plays a critical role in capturing target DNA and promoting intermolecular transposition. MuB polymerizes cooperatively and non-specifically on double-stranded DNA, exhibiting a tendency to form larger polymers or filaments on A/T-rich DNA.13, 14 Formation of MuB polymers requires ATP or ATPγS.15 A catalytic cycle of ATP-binding and hydrolysis leads to conformational changes that are coupled to polymer formation and dissolution.14, 16, 17 MuB dissociation is stimulated by MuA, which promotes integration into the MuB-bound DNA.18–20
Figure 1.
Steps in Mu DNA transposition. In the presence of the accessory host factor HU and divalent metal ions, the transposase MuA assembles into a transpososome on L and R ends of Mu present on supercoiled DNA. Mu ends undergo single stranded cleavages to generate 3’OHs, which act as nucleophiles in the subsequent chemical step of strand transfer into target DNA in the presence of MuB.
Consistent with early genetic studies, a recent microarray analysis of Mu transposition targets in E. coli showed that the majority of transposition sites were distributed throughout the genome.8 However, 4% of the genes were hot spots and 1% cold spots. Highly transcribed genes appeared to be protected from Mu integration. Similar results were obtained in a study of Mu transposition in Salmonella.21 This study showed in addition that a plasmid partitioning protein ParB, which forms a stable filament at the par locus, is responsible for the cold spot for Mu transposition at this locus. It appears therefore that Mu transposition is occluded by a transcribing polymerase, or by a high density of proteins strongly bound to DNA in the cold spots.
What features are responsible for the occurrence of hot spots? Are these regions A/T rich and hence attract the transpososome by binding MuB? Do they have an altered DNA structure/topology that facilitates target capture? Are there host proteins that direct the transpososome to these regions? In this study we show that A/T content is not a good predictor of Mu behavior in vivo, and that MuA and MuB proteins are the primary determinants of target site selection into hot genes. We show that MuB promotes target capture while blocking integration within MuB-bound DNA. We infer that MuB forms short unstable filaments on DNA in vivo, and discuss the implications of our results for target immunity, a process that prevents Mu from self-integration.
Results
A/T content of genes exhibiting high and low target site preference in vivo
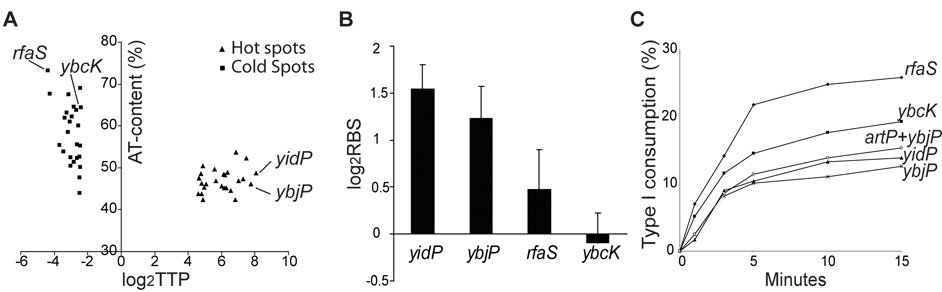
Manna et al. measured the relative abundance of all gene sequences covalently linked to the right end of Mu DNA within phage propagated in E. coli, and derived transposition target preference or TTP values.8 To do so, they amplified host DNA attached to the Mu right end and hybridized it to DNA microarrays printed with 4,290 E. coli genes. Although the array lacks intergenic sequences, these comprise a very small fraction of the genome in E. coli. TTP values showed that 95% of transposition events were distributed throughout the genome without gross bias toward any part of the chromosome; the TTP values for these genes were within 4-fold of the median value of 1.0. However, 4% of the genes had TTP values >4 (hot spots), while 1% had TTP values <4 (cold spots). Given the preference of MuB for A/T rich DNA, we wished to examine the relationship of targets with high and low TTP values with the A/T-content of their DNA.
When the A/T content of 25 hot and cold Mu target genes in E.coli was plotted against their log2TTP values, we observed that the average A/T content of the hot targets was around 50%, while that of the cold targets ranged from 44% to 73%, with half of the latter targets having an A/T value above 60% (Fig. 2A). To investigate this phenomenon further, two of the hottest genes, yidP and ybjP, and two of the coldest genes, rfaS and ybcK (see Table 1 in Ref. 8), were chosen for more detailed studies.
Figure 2.
A. A/T content of 25 hot and cold Mu target genes in E. coli. The log2TTP data is taken from Table 1 in Ref. 8, and A/T content of each gene is based on the E. coli K12 genome sequence (NC_000913). Indicated genes were used for further study. B. The relative binding strength (RBS) of MuB on hot and cold genes in vivo. MuB binding was measured by ChIP assays using MuB antibody (see Methods). Bound DNA was amplified by PCR and quantitated. RBS or relative binding strength is the ratio of specific to non-specific binding. The binding data are an average of three repeats performed for the following regions showing the strongest binding within each gene: yidP (49% A/T), 351–717bp; ybjP (46% A/T), 210–516 bp; rfaS (73% A/T), 601–936 bp; ybcK (64% A/T), 351–700bp. Student-t tests were performed, and the p-values (p<0.05) suggest that MuB binding to hot targets genes was significantly stronger than that to cold target genes. C. In vitro target efficiency of hot and cold in vivo targets. Strand transfer was initiated by adding equal amounts of the cleaved Mu complex assembled on pSP104 to 2-fold molar excess of PCR-amplified linear target DNA derived from the indicated genes. Reaction aliquots taken at various times were run on an agarose gel, and donor or target consumption measured by quantifying the intensity of the appropriate DNA bands. Target sizes are as follows: yidP (717 bp), ybjP (516 bp), rfaS (936 bp), ybcK (1527 bp), artP + ybjP (1452 bp).
MuB binding to hot sites in vivo is significantly stronger than to cold sites
To determine the relationship between in vivo target preference and MuB-binding, ChIP experiments were conducted for the four chosen genes yidP (717 bp), ybjP (516 bp), rfaS (936 bp) and ybcK (1527 bp), using MuB antibody bound to protein-A beads to immunoprecipitate MuB-bound DNA. Controls without antibody were used to assess the contribution of non-specific binding. The ratio of specific to non-specific binding is referred to as relative binding strength or RBS. PCR reactions using primer pairs designed to amplify 300–400 bp segments of DNA spanning the entirety of each gene showed the RBS values for all the segments within an individual gene to be comparable. One segment from each gene showing the best binding was therefore chosen as a representative for comparison of binding among the four genes (Fig. 2B). MuB binding to the two hot genes yidP and ybjP was seen to be significantly higher than to the two cold genes rfaS and ybcK. This implies that high Mu integration frequency is correlated with a higher MuB-binding preference, but that MuB-binding preference is not correlated with the A/T content of the target DNA, at least for the genes examined here (see Fig. 2A).
Target preference of in vivo sites reverses in vitro
To determine if the hot and cold behavior of in vivo targets was maintained in vitro, the four representative genes were used as target in transposition reactions in vitro. Efficiency of target use was determined in a two-step transposition reaction, where the Mu transpososome assembled on a mini-Mu plasmid was first allowed to undergo cleavage at Mu ends, followed by addition of the cleaved complex to the four linear target DNAs (see Fig. 1). Strand transfer efficiency, measured as consumption of the cleaved donor DNA, is shown in Fig. 2C. The results were similar when target consumption was followed (not shown). We note that the donor was not completely consumed in these experiments because linear targets are not used as efficiently as supercoiled targets. Fig. 2C shows that the cold in vivo target rfaS with the highest A/T content (73%) was used most efficiently in vitro, followed by second cold in vivo target ybcK (64% A/T). The two hot in vivo targets ybjP and yidP (46% and 49% A/T, respectively) were used at slower but similar rates. Only half the donor was consumed at the end of the reaction in the latter reactions when compared to the rfaS reaction. Thus, the in vivo target preference was reversed in vitro, with hot in vivo targets consumed less efficiently than cold targets. To rule out the influence of target size on the outcome of the results, the size of the smallest target ybjP was increased to 1452 bp by including its upstream gene artP. This gene has a similar A/T content (47%) to ybjP and is the fourth favored hot spot.8 The transposition result for the combined larger target was similar to that seen for ybjP alone (Fig. 2C).
The higher in vitro target efficiency of rfaS and ybcK, which have a higher A/T content than ybjP and yidP (Fig. 2A), is consistent with the reported preferential use of A/T-rich targets in vitro.14, 17 It follows from the data presented in Figs. 2A–C, that although transposition target preference in vivo is apparently correlated with MuB distribution, this distribution must be determined by factors other than A/T-content alone.
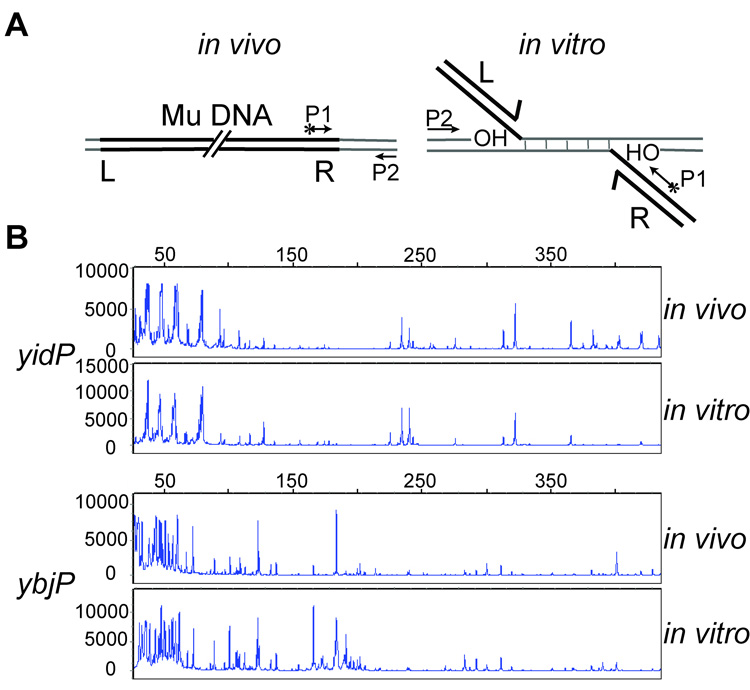
Pattern of Mu target site selection is similar in vivo and in vitro
Since the efficiency of target use in vivo was apparently reversed in vitro, the pattern of Mu transposition into these targets might also be different under the two conditions. To obtain in vivo profiles, we amplifed yidP, ybjP, rfaS and ybcK sequences linked to the Mu R end in virions obtained from induction of a Mu lysogen (Fig. 3A, left). To obtain in vitro Mu insertion profiles, the strand transfer products generated in the presence of MuA and MuB in vitro were used as templates in PCR reactions employing the same primer pairs as those used to obtain in vivo patterns (Fig. 3A, right). Analysis of the size and abundance of the products for both sets of reactions is shown in Fig. 3B.
Figure 3. In vivo and in vitro patterns of Mu integration on two hot targets.
A. PCR strategy for amplification of in vivo insertion sites packaged in Mu phage (left), or in vitro insertion sites from strand transfer products generated as described in Fig. 2C. (right). Fluorescently labeled (*) P1 primer hybridizes within the Mu R end, and P2 hybridizes to one end of the gene being analyzed. B. PCR products from yidP and ybjP reactions were subjected to Fragment Length Analysis as described in Methods. Numbers on the X-axis refer to nucleotides. The intensity of the fluorescent signal is represented by arbitrary numbers on the Y axis.
The hot genes yidP and ybjP amplified well from the induced phage population, whereas the cold genes rfaS and ybcK were barely detectable in the PCR reactions. The in vivo and in vitro profiles for the two hot genes are shown in Fig. 3B. These profiles are remarkably similar not only for the sites of insertion, but also for the relative frequency of their usage as judged by the relative heights of the individual insertion peaks. This shows that MuA and MuB are the primary, if not the sole, determinants of the spectrum of Mu insertions observed within these genes in vivo.
Mu insertion patterns are robust, and are primarily a property of MuA
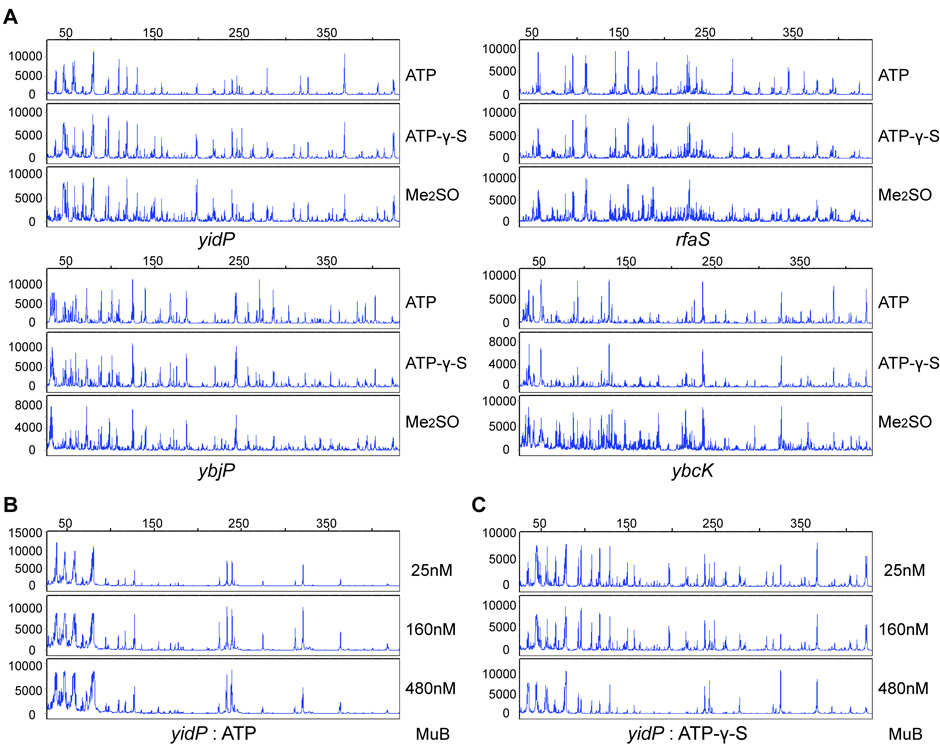
To determine if in vitro insertion patterns are influenced by the pattern of MuB bound to the DNA, we compared these in the presence of ATP or ATPγS. In the presence of ATP, Mu insertion patterns must reflect the steady state distribution of MuB on DNA, which is determined by the dynamics of MuB–DNA association/dissociation. In the presence of ATPγS, where MuB does not dissociate from DNA, the insertion patterns should reflect the initial distribution of MuB. We also set up strand transfer reactions in the presence of Me2SO, where the MuB requirement can be bypassed; MuB is therefore not included in these reactions. Insertions obtained under these conditions should report on the contribution of MuA alone to the profile. The results are shown in Fig. 4.
Figure 4. Contribution of MuB to Mu insertion within hot and cold targets in vitro.
A. Strand transfer reactions carried out in the presence of MuB included either ATP or ATPγS, and employed min-Mu plasmids as donor and linear DNA as target. MuB concentration was 160nM. Reactions in the absence of MuB employed precleaved oligonucleotide Mu R ends as donors. Insertions were analysed as in Fig. 3. B, C. Reactions using the yidP target with increasing MuB concentrations in the presence of ATP or ATPγS.
Surprisingly, the insertion profiles were independent of ATP or ATPγS for all four targets (Fig. 4A). Reactions performed in the absence of MuB (Me2SO) showed a broader distribution of Mu insertions, particularly evident in the rfaS and ybcK targets. However, the signature peaks of insertions seen in the presence of MuB were distinctly recognizable even in the absence of MuB. Indeed, the new insertions in the absence of MuB were clustered around the insertion peaks seen in the presence of MuB, and could be the result of now-vacant MuB binding regions. While it is known that the target site consensus is determined by MuA,10 the results in Fig. 4A show that the frequency of usage of specific insertion sites is also determined largely by MuA.
To understand why the Mu insertion profile was similar in MuB reactions employing either ATP or ATPγS, MuB concentration was varied over a 20-fold range for the yidP target (standard reactions contain 160nM MuB). If MuB binds to higher affinity sites first, then in the presence of ATPγS the pattern of MuB distribution on DNA should vary with increasing MuB concentrations. If however, the assembly of MuB polymers on DNA involves independent stochastic events as concluded by a recent study, 22 then the pattern of Mu insertions should not be significantly different at the different MuB concentrations, unless Mu integration is inhibited by excess MuB that cannot dissociate from DNA. The results support the latter scenario (Fig. 4B,C). The insertion pattern was similar at all the MuB concentrations tested, with some suppression observable at saturating MuB concentrations (480nM) in the presence of ATPγS (note reduction of the relative heights of insertion peaks, particularly between the 100–250 region).
We conclude that the pattern of Mu insertion within a target is robust i.e. unperturbed over a wide range of MuB concentrations, whether in the presence of ATP or ATPγS. While MuB assists target DNA capture, it does not influence target site selection.
MuB suppresses integration within a synthetic A/T-rich DNA fragment
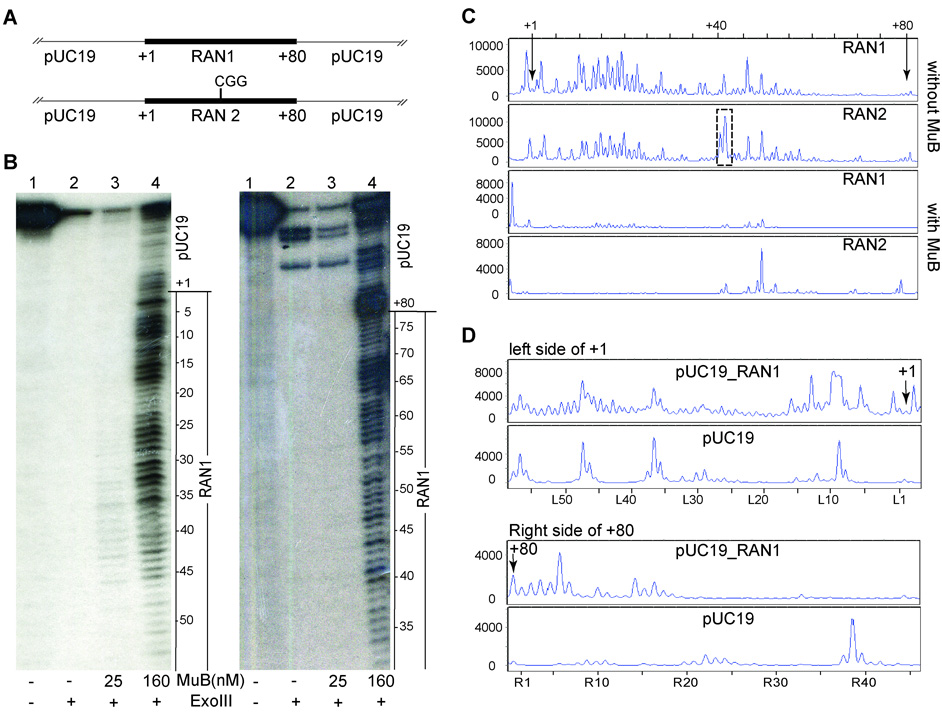
Data presented in Fig. 4A show that the Mu insertion profile broadened in the absence of MuB. In other words, the presence of MuB suppressed the use of several potential target sites. Is suppression caused by MuB binding to these sites? In order to address the relationship between the bound MuB and choice of insertion sites, we followed Mu integration within a target containing a continuous stretch of A/T base pairs. We synthesized 80bp A/T-only DNA fragments without or with the 5 bp Mu target consensus at its center (Fig. 5A; RAN1 and RAN2, respectively). These were cloned into pUC19 and analyzed for their MuB-binding (Fig. 5B) and Mu integration patterns (Fig. 5C, D).
Figure 5. MuB binding and Mu integration within A/T-rich DNA.
A. 80bp A/T-containing RAN1 and RAN2 oligonucleotides were cloned in pUC19 (see Suppl Table 1). RNA2 has an additional CGG starting at position 40. B. PCR-amplified RAN1 DNA 32P-labeled at 5’ end of the top (left panel) or bottom strand (right panel) of flanking pUC19 DNA, was used for ExoIII protection assays in the presence of increasing MuB. C. Comparison of Mu insertion profiles within RAN1 and RAN2 in pUC19, in the presence and absence of MuB. Position of the central target consensus CGG in RAN2 is marked with a dotted box. Nucleotide windows spanning + 1 to +80 nucleotides of the RAN sequence are marked on the X-axis. Other descriptions as in Fig. 3. D. Comparison of insertion sites within pUC19 DNA with those in the same region flanking either side of the RAN1 fragment in the presence of MuB. The X-axis shows a 60 nucleotide region either to the left (L) or right (R) of RAN1.
ExoIII protection was used to demonstrate MuB binding to RAN substrates labeled at the 5’ end of the top strand or the complementary bottom strand in flanking pUC19 DNA. ExoIII, which catalyzes the stepwise release of nucleotides from the 3’end of DNA, displayed specific digestion patterns with increasing MuB concentration, indicative most likely of MuB-bound regions on the DNA which slow its travel (Fig. 5B). More DNA was bound at 160 nM than at 25 nM MuB, indicated by a larger series of undigested substrate bands. At 480 nm MuB, the DNA was completely protected (not shown). The protection shown is for RAN1; identical protection patterns were observed with RAN2 (not shown). On the top strand, the region marked +1 corresponds to the start of the A/T region at the left end of the DNA fragment (Fig. 5B, left panel). Thus, the start of this region coincides with the start of the MuB-bound region. On the bottom strand, the region marked +80 corresponds to the end of the A/T region at the right end of the DNA fragment (Fig. 5B, right panel). Here, the MuB-protected region extended into flanking pUC19 sequence.
The Mu insertion pattern obtained on the RAN1 and RAN2 plasmid substrates in the absence of MuB shows that insertions are distributed through most of the AT-rich segment, although they were highest around the centrally placed Mu target consensus on the RAN2 substrate (Fig. 5C, dotted box around CGG). In the presence of 160 nM MuB, insertions are almost eliminated on the RAN1 substrate. On the RAN2 substrate, an insertion spike was observed near, but not at the internal CGG site, suggesting perhaps that interruption of the A/T sequence destabilized the MuB filament around this region.
To examine the insertion pattern outside the A/T region, we determined insertion profiles in the pUC19 DNA flanking the +1 and +80 regions of the RAN1 substrate (Fig. 5D). Insertions occurred preferentially near, and were concentrated within, a 15–20 bp window at both the left and right junctions of the A/T and pUC19 region. Insertions in DNA beyond this region were not influenced by MuB bound to the A/T segment, as determined by comparison of the insertion profile to control pUC19 DNA.
We conclude from these data that a MuB filament was formed on the A/T DNA, and that this filament inhibited Mu integration within its interior. The increased frequency of Mu insertions near the junction of the A/T and pUC19 DNA, and the relative absence of insertions beyond the junction region, suggests that MuB promotes insertions only in its immediate vicinity outside the bound DNA.
DNA sequence surrounding Mu insertion peaks
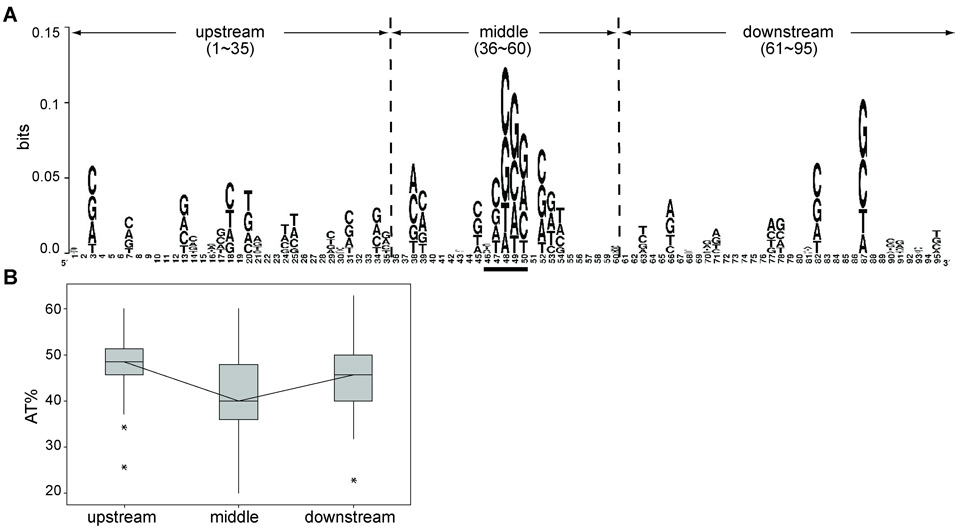
Analysis of 61 insertion peaks in the hot target genes yidP and ybjP, showed a G/C-rich 5 bp target consensus (Fig. 6A). The relatively low conservation value (~0.1 bits) of this consensus is indicative of the generally random nature of Mu transposition. Other than an apparent preference for G/C observed at nucleotide 87, there was little, if any, sequence conservation within the surrounding 95 bp region examined. Assuming that ~25 bp of the target is bound by the transposase active site,29 we compared the A/T content of a 25 bp region with a centrally placed MuA target consensus (Fig. 6A, middle), with that of flanking 35 bp segments (Fig. 6A, upstream and downstream). A significantly higher A/T content was observed in these flanking DNA segments compared to that of the middle segment, as determined by student-t tests (p<0.005), while the A/T difference between upstream and downstream segments was not significant (p>0.05). The median A/T values for the upstream, middle, and downstream segments were 49.6%, 40%, and 45.7%, respectively (Fig. 6B).
Figure 6. Sequence conservation within and around Mu insertion peaks in hot targets.
A. 61 sequences representing transposition peaks with heights greater than 2000, obtained by fragment length analysis of Mu integration into yidP and ybjP in the presence of MuB (see Fig. 4), were aligned with the peaks at the center, and analyzed using the Weblogo program (http://weblogo.berkeley.edu/). The sequence is divided into upstream, middle, and downstream segments as indicated for A/T content analysis shown in B. The 5bp target consensus is underlined in the center. The height of each letter on the x-axis is proportional to the observed frequency of the corresponding nucleotide, and the overall height of each stack is proportional to the sequence conservation, measured in bits on the y-axis. Bits are defined as the difference between the maximum possible distribution and the observed symbol distribution. The maximum conservation for each position is 2 bits for DNA/RNA. B. Boxplots showing the AT percentage of DNA in indicated regions of the 95 nucleotide segment shown in A, constructed using the statistical software Minitab 15. The box represents sequences that fall between 25th and 75th percentile of the data set, the vertical lines above and below the box spanning the upper and lower percentiles. The horizontal line through box is the median value, and lines between boxes connect these values. *, outliers.
These data are consistent with our inference derived from the results presented in Fig. 3–Fig. 5, that MuB likely binds in an interspersed manner to locally A/T-rich segments in natural DNA, preventing integration in the bound regions but directing the transpososome to adjacent DNA sites (insertion peaks) free of MuB.
Discussion
Among well-studied transposons, target choice in vivo can range from high specificity,23 to regional specificity determined either by transcription,24, 25 replication status,26 or structural features in DNA,27–29 to apparently minimal specificity.3, 4, 8 Mu belongs to the last category of elements. During the lytic cycle of Mu growth, a majority of the E. coli and S. enterica genes were observed to receive a uniform distribution of Mu insertions, while a small fraction showed either hot or cold spots for integration.8, 21 A comparison of in vivo and in vitro transposition patterns within the hot and cold spots has provided insights into the target selection process in vivo, as discussed below.
MuB and target preference in vivo
Mu transposition in vitro displays a regional preference coincident with the MuB binding preference for A/T-rich regions.14, 17 We were therefore interested in determining whether there was a direct correlation between the hot and cold Mu targets reported in the in vivo study by Manna et al. 2004, and the A/T content of their DNA. Our analysis of TTP values of the 25 hottest and coldest sites reported by Manna et al. showed that Mu preference for these genes was not correlated to their A/T-content (Fig. 2A). This finding could be extended to the whole genome as well.30 Despite these findings, ChIP experiments showed that MuB binding was an important factor in target selection in vivo, at least for the four genes examined in this study (Fig. 2B).
Although in vivo target preference was not correlated to A/T content, in vitro use of four genes as transposition targets was directly correlated with A/T content (Fig. 2C). These results suggest that the MuB-preferred A/T-rich targets are unavailable for Mu transposition in vivo. The cold behavior of rfaS and ybcK in vivo can be partially explained by their higher TSC values (transcript copy number) compared to those of yidP and ybjP, since transcription appeared to have a negative impact on Mu transposition.8 However, the lac operon has a much higher TSC value (10) compared to that of rfaS (2.46) or ybcK (1.227), yet the lac genes are not as cold a target. Therefore, there must be additional mechanisms occluding Mu transposition into these genes. It is interesting that Mu and HIV, whose integrase proteins have similar structures and share related transposition mechanisms, have opposite regional preferences for targets. While Mu avoids highly transcribed regions, HIV prefers them,25 suggesting a role for accessory proteins in mediating these choices.
If host factors promote or inhibit transposition into the hot and cold genes, the pattern of insertions into these genes would be expected to differ from those obtained in a purified in vitro system. While insertions within the cold genes rfaS and ybcK were barely detectable, the in vivo insertion patterns obtained for the hot genes yidP and ybjP were seen to be superimposable on those obtained for these genes in vitro (Fig. 3). This suggests that the in vivo insertion pattern is generated largely through the activity of MuA and MuB proteins. The data argue against the existence of a special DNA structure or topology that might exist in vivo, or host factors that promote integration within these genes. It appears instead that the Mu proteins have unrestrained access to the hot genes. We suggest that ‘hot’ is a relative term; genes that show up as hot do so because other more transcribed genes fare worse. The conformity of in vivo and in vitro patterns also tells us that knowledge of Mu transposition derived from in vitro studies can be extrapolated in vivo.
MuB and target preference in vitro
The basic underlying pattern of strong integration sites was found to be independent of MuB, and hence a property of MuA (Fig. 4). Thus, MuB only increases the efficiency of finding the target but does not control the profile of Mu insertions obtained. The insertion pattern was robust in that it was reproducible over a 20-fold range of MuB concentrations, and even in the presence of ATPγS where MuB does not dissociate from DNA. The absence of MuB broadened the spectrum of insertion sites chosen, suggesting that MuB-bound regions block integration at the bound sites. Support for this idea came from monitoring insertions into an A/T-rich target, as discussed below; such targets have been observed to polymerize larger MuB filaments.13, 14
Synthetic A/T-only substrates occluded Mu integrations within the A/T sequence in the presence of MuB, Mu insertions being recovered at the immediate junction of A/T and non-A/T regions (Fig. 5). These as well as footprinting results showed that MuB indeed formed a continuous filament on the A/T region and that the interior of the filament was refractory to integration. These results are consistent with earlier data showing Mu integrations occurring on either side of an A/T-rich segment protected by MuB,10 as well as data showing that MuB dissociation occurs mainly through an end-dependent mechanism, dissociation from within the polymer interior being slower.14, 22 Suppression of insertions on the A/T-only substrate with only 160 nM MuB (Fig. 5C), but not on the natural yidP target with even 480 nM MuB (Fig. 4B), shows that the MuB-bound forms on the two substrates have different stabilities.
The frequent distribution of insertions in the four targets analyzed (Fig. 4), suggests that MuB likely binds in valleys between insertion peaks on these natural targets. Analysis of sequences around the insertion peaks showed a G/C-rich 5 bp target consensus as expected, while the DNA flanking this region had a significantly higher A/T content (Fig. 6). That the MuA target consensus favors G/C sequences, while MuB binding is favored at A/T sequences, suggests this arrangement would result in a mutually synergistic rather than competitive activity of MuA and MuB proteins.
Role of the MuB filament
What is the role of the MuB filament in the life of Mu? Unlike a RecA filament which extends and unwinds DNA and promotes homology search and strand exchange within the filament,31 the MuB filament does not appear to change DNA twist (our unpublished data; see also Ref. 22), and is apparently refractory to Mu integration inside the filament. Comparison of the data in Fig. 4 and Fig. 5 would suggest that MuB likely forms short unstable rather than long stable filaments on natural DNA sequences. One role for short unstable filaments might be to increase the target capture efficiency without inhibiting integration. As a corollary, long stable filaments or a complete absence of filaments would prevent integration. The latter explanation has been proposed for ‘target immunity’, a phenomenon where Mu does not utilize its own DNA as an integration substrate.14 According to this proposal, if only the MuB filaments that are assembled on A/T rich regions are acceptable sites of integration, then non-A/T regions will be ignored by the transpososome. From the results reported here, however, it is apparent that A/T content is not a good predictor of Mu behavior in vivo. We are currently testing whether MuB is excluded from the interior of the Mu genome, and if so, is it because it binds poorly per se, or is prevented from binding because of transcription through Mu.
Materials and Methods
DNA and proteins
Procedures for phage growth and DNA isolation from the Mu lysogen HM8305 have been described.32 MuA, MuB and HU proteins were purified as described.33 MuB antibody was obtained by immunization of rabbits with purified protein. Enzymes were from New England Biolabs. Primers used in PCR reactions are listed in Suppl. Table 1.
In vitro transposition and Fragment Length Analysis of PCR products
Cleaved Mu complexes were formed on the mini-Mu plasmid donor pSP10434 as described.35 Strand transfer was initiated by adding equal amounts of the reaction mixture to tubes containing 10µg/ml of various linear target DNAs, 2mM ATP, and 160nM MuB at 30°C. Strand transfer using pre-cleaved R1-R2 oligonucleotide substrates under Me2SO conditions was also performed as described.35 Products were analyzed on 1% agarose gels and DNA band intensities quantified using Bio-Rad Multianalyst software.
For insertion sites analysis, the strand transfer products were excised from gels and used as templates in PCR reactions where the Mu R end primer was labeled at its 5’ end with fluorescent agent 6-FAM (Integrated DNA Technologies). Labeled PCR products were analyzed using Applied Biosystems 3130XL Genetic Analyzer, and interpreted using the analysis software GeneMaker (SoftGenetics LLC, Version 1.5).
Chromatin Immunoprecipitation (ChIP)
HM8305 was grown in LB at 30°C until the culture reached an OD600 of 0.6. Phage replication was induced by inactivating the temperature sensitive repressor at 42°C for 30 minutes. ChIP was performed using anti-MuB antibody as described, but with the following modifications.36 The cell culture was crosslinked with 1% formaldehyde (final) at room temperature for 10 minutes as previously described. Formaldehyde was quenched by 150mM glycine for 5 minutes at room temperature. Cells were harvested by centrifugation and washed twice with Tris-buffered saline (pH7.5). The pellet was resuspended in lysis buffer (10mM Tris-HCl, PH8.0, 10mM EDTA, 50mM NaCl, 20% sucrose, and 4mg/ml lysozyme) containing protease inhibitors (Roche; 1 tablet per 10 ml buffer). The suspension was incubated in 37°C for 30 minutes, followed with the addition of same volume of 2×IP buffer (1×IP = 50mM Hepes-KOH, PH7.5, 150mM NaCl, 1mM EDTA, 1% TritonX-100, 0.1% sodium deoxycholate and 0.1% SDS). Cellular DNA was then sheared by ultrasonication (SONICS Vibra cell®; model VC 505) to an average length of 300–1000 bp. Typically, 2ml of cell extract were sonicated for 5 cycles of 15 seconds each at a duty cycle setting of 40%, keeping the extract on ice for 2 min between each cycle. The cell debris generated was removed by centrifugation, and the supernatant used as input samples for immunoprecipitation. For each reaction, 10µg of purified anti-MuB polyclonal antibody was added to 800 µl of input samples, and no-antibody controls were included as well. After overnight incubation at 4°C, 50µl protein-A agarose beads were added to the samples. After 90 min incubation at room temperature, the agarose beads were centrifuged and washed twice with 1×IP buffer, twice with LiCl buffer (10mM Tris-HCl, pH8.0, 250mM LiCl, 1mM EDTA, 0.5% Nonidet-P40, 0.5% sodium deoxycholate), and once with TE buffer (10mM Tris-HCl, pH8.0, 1mM EDTA) The beads were resuspended in Elution buffer (50mM Tris-HCl, pH8.0, 10mM EDTA and 1% SDS) and heated to 65°C for at least 6 hours to break protein-DNA crosslinks. After full speed centrifugation, the supernatant was carefully transferred to a new tube and treated with 100mg/ml proteinase K at 37°C for 2 hours. The sample was then treated with regular PCR cleanup protocol (Qiaquick PCR purification Kit®).
Immune-precipitated DNA was amplified by PCR, and analyzed by agarose gel electrophoresis. Primers for PCR were designed according to the DNA sequences of yidP, ybjP, rfaS and ybcK, with the length of the amplified fragments being around 300bp. The primers are listed in Suppl Table 1.
Exonuclease III protection assay
32P-labeled DNA was pre-incubated with MuB in the strand transfer buffer at 30°C for 30 min, followed by 10 min of incubation with 100 units of Exo III. The reaction was stopped by addition of equal volume of 2×stop buffer (100mM EDTA, 25mM Tris-HCL, pH7.6, 500 ng/µl yeast tRNA). Proteinase K was added (1 µg/µl, final) at 37°C for 1 hour, the DNA extracted with phenol-chloroform and precipitated with ethanol. Reaction products were analyzed on 8% polyacrylamide denaturing gels.
Supplementary Material
Sequence of oligonucleotides and primers.
Acknowledgements
We thank Makkuni Jayaram for discussions and critical reading of the manuscript. This work was supported by National Institutes of Health grant GM 33247 and in part by the Robert Welch Foundation Grant F-1351.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Craig NL. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 2.Sandmeyer S. Targeting transposition: at home in the genome. Genome Res. 1998;8:416–418. doi: 10.1101/gr.8.5.416. [DOI] [PubMed] [Google Scholar]
- 3.Taylor AL. Bacteriophage-induced mutations in E. coli. Proc Natl Acad Sci. 1963;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukhari AI, Zipser D. Random insertion of Mu-1 DNA within a single gene. Nature New Biology. 1972;236:240–243. [Google Scholar]
- 5.Castilho BA, Casadaban MJ. Specificity of mini-Mu bacteriophage insertions in a small plasmid. J Bacteriol. 1991;173:1339–1343. doi: 10.1128/jb.173.3.1339-1343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Higgins NP. 'Muprints' of the lac operon demonstrate physiological control over the randomness of in vivo transposition. Mol Microbiol. 1994;12:665–677. doi: 10.1111/j.1365-2958.1994.tb01054.x. [DOI] [PubMed] [Google Scholar]
- 7.Manna D, Wang X, Higgins NP. Mu and IS1 transpositions exhibit strong orientation bias at the Escherichia coli bgl locus. J Bacteriol. 2001;183:3328–3335. doi: 10.1128/JB.183.11.3328-3335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manna D, Breier AM, Higgins NP. Microarray analysis of transposition targets in Escherichia coli: the impact of transcription. Proc Natl Acad Sci U S A. 2004;101:9780–9785. doi: 10.1073/pnas.0400745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manna D, Deng S, Breier AM, Higgins NP. Bacteriophage Mu targets the trinucleotide sequence CGG. J Bacteriol. 2005;187:3586–3588. doi: 10.1128/JB.187.10.3586-3588.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuuchi M, Mizuuchi K. Target site selection in transposition of phage Mu. Cold Spring Harb Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- 11.Haapa-Paananen S, Rita H, Savilahti H. DNA transposition of bacteriophage Mu. A quantitative analysis of target site selection in vitro. J Biol Chem. 2002;277:2843–2851. doi: 10.1074/jbc.M108044200. [DOI] [PubMed] [Google Scholar]
- 12.Chaconas G, Harshey RM. Mobile DNA II. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Transposition of phage Mu DNA. Washington DC: ASM Press; 2002. pp. 384–402. [Google Scholar]
- 13.Adzuma K, Mizuuchi K. Steady-state kinetic analysis of ATP hydrolysis by the B protein of bacteriophage Mu. Involvement of protein oligomerization in the ATPase cycle. J Biol Chem. 1991;266:6159–6167. [PubMed] [Google Scholar]
- 14.Greene EC, Mizuuchi K. Visualizing the assembly and disassembly mechanisms of the MuB transposition targeting complex. J Biol Chem. 2004;279:16736–16743. doi: 10.1074/jbc.M311883200. [DOI] [PubMed] [Google Scholar]
- 15.Greene EC, Mizuuchi K. Dynamics of a protein polymer: the assembly and disassembly pathways of the MuB transposition target complex. Embo J. 2002;21:1477–1486. doi: 10.1093/emboj/21.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung PC, Harshey RM. Two mutations of phage Mu transposase that affect strand transfer or interactions with B protein lie in distinct polypeptide domains. J Mol Biol. 1991;219:189–199. doi: 10.1016/0022-2836(91)90561-j. [DOI] [PubMed] [Google Scholar]
- 17.Greene EC, Mizuuchi K. Direct observation of single MuB polymers: evidence for a DNA-dependent conformational change for generating an active target complex. Mol Cell. 2002;9:1079–1089. doi: 10.1016/s1097-2765(02)00514-2. [DOI] [PubMed] [Google Scholar]
- 18.Yamauchi M, Baker TA. An ATP-ADP switch in MuB controls progression of the Mu transposition pathway. EMBO J. 1998;17:5509–5518. doi: 10.1093/emboj/17.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzuma K, Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of Mu B protein. Cell. 1988;53:257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- 20.Greene EC, Mizuuchi K. Target immunity during Mu DNA transposition. Transpososome assembly and DNA looping enhance MuA-mediated disassembly of the MuB target complex. Mol Cell. 2002;10:1367–1378. doi: 10.1016/s1097-2765(02)00733-5. [DOI] [PubMed] [Google Scholar]
- 21.Manna D, Porwollik S, McClelland M, Tan R, Higgins NP. Microarray analysis of Mu transposition in Salmonella enterica, serovar Typhimurium: transposon exclusion by high-density DNA binding proteins. Mol Microbiol. 2007;66:315–328. doi: 10.1111/j.1365-2958.2007.05915.x. [DOI] [PubMed] [Google Scholar]
- 22.Tan X, Mizuuchi M, Mizuuchi K. DNA transposition target immunity and the determinants of the MuB distribution patterns on DNA. Proc Natl Acad Sci U S A. 2007;104:13925–13929. doi: 10.1073/pnas.0706564104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig NL. Tn7: a target site-specific transposon. Mol Microbiol. 1991;5:2569–2573. doi: 10.1111/j.1365-2958.1991.tb01964.x. [DOI] [PubMed] [Google Scholar]
- 24.Boeke JD, Devine SE. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters JE, Craig NL. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol Cell. 2000;6:573–582. doi: 10.1016/s1097-2765(00)00056-3. [DOI] [PubMed] [Google Scholar]
- 27.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bender J, Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci U S A. 1992;89:7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pribil PA, Haniford DB. Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. J Mol Biol. 2000;303:145–159. doi: 10.1006/jmbi.2000.4135. [DOI] [PubMed] [Google Scholar]
- 30.Ge J. A general linear mixed model for bacteriophage Mu transposition target preference. MA in Statistics. University of Texas; 2007. [Google Scholar]
- 31.Bell CE. Structure and mechanism of Escherichia coli RecA ATPase. Mol Microbiol. 2005;58:358–366. doi: 10.1111/j.1365-2958.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 32.Au TK, Agrawal P, Harshey RM. Chromosomal integration mechanism of infecting mu virion DNA. J Bacteriol. 2006;188:1829–1834. doi: 10.1128/JB.188.5.1829-1834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang JY, Kim K, Jayaram M, Harshey RM. Domain sharing model for active site assembly within the Mu a tetramer during transposition: the enhancer may specify domain contributions. Embo J. 1995;14:2374–2384. doi: 10.1002/j.1460-2075.1995.tb07232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathania S, Jayaram M, Harshey RM. Path of DNA within the Mu transpososome. Transposase interactions bridging two Mu ends and the enhancer trap five DNA supercoils. Cell. 2002;109:425–436. doi: 10.1016/s0092-8674(02)00728-6. [DOI] [PubMed] [Google Scholar]
- 35.Lee I, Harshey RM. Importance of the conserved CA dinucleotide at Mu termini. J Mol Biol. 2001;314:433–444. doi: 10.1006/jmbi.2001.5177. [DOI] [PubMed] [Google Scholar]
- 36.Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, Constantinidou C, Struhl K, Church G, Busby SJ. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J Bacteriol. 2004;186:6938–6943. doi: 10.1128/JB.186.20.6938-6943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of oligonucleotides and primers.