Abstract
We evaluated quantitatively the relative cell permeability of peptoids and peptides using a cell-based reporter gene-based assay. Generally, peptoids were much more cell permeable than the corresponding peptides, though the difference decreased with increasing length. These results suggest that peptoids may be useful reagents for manipulating the activities of intracellular proteins.
There is currently considerable interest in developing chemical reagents with which to manipulate protein-protein interactions as tools for chemical biology or as potential drugs. Peptides have the type of protein-binding properties required for this type of application as they are capable of recognizing the relatively large, shallow surfaces typical of interaction domains. However, peptides have well-known limitations as pharmacological agents, the most serious being sensitivity to proteases and limited cell permeability. Therefore, several different types of peptidomimetics have been investigated,1 with the hope of identifying a family of compounds that retain the favorable protein-binding properties of peptides but exhibit superior pharmacokinetic characteristics. In this endeavor, it would be useful to have a general method with which to compare the cell permeability of a given peptidomimetic compound with that of the corresponding peptide. We report here a method that allows such comparisons to be made conveniently and quantitatively using any transfectable cell type. Using peptoids as a model system, we show that these N-alkylated species are up to 20-fold more cell-permeable than analogous peptides. Along with their other favorable properties,2-8 these data suggest that peptoids may be useful reagents for targeting intracellular proteins.
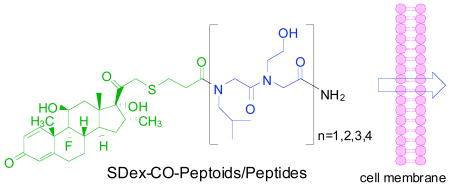
Recently we developed a high-throughput assay for assessing the relative cell permeability of synthetic molecule-dexamethasone (Dex) derivative conjugates.9 A schematic of the system is provided in the Supplementary Material (Fig. S1). Briefly, the molecule of interest is conjugated to a Dex derivative and incubated with HeLa cells that have been transfected with three plasmids. One encodes a fusion protein comprised of the Gal4 DNA-binding domain, the Glucocorticoid Receptor ligand binding domain and the VP16 transactivation domain (Gal4DBD-GRLBD-VP16). The other two carry a Gal4-responsive firefly luciferase reporter gene (pG5B), and a constitutively expressed Renilla reniformis luciferase (pRL-SV40), respectively. Gal4DBD-GRLBD-VP16 is a potentially potent activator of pG5B expression, but is held in an inactive form by tight binding of Hsp90 to the GR LBD. However, binding of the steroid to this domain competes this interaction and allows the activator to drive firefly luciferase expression. Thus, the ratio of firefly luciferase to Renilla luciferase activity in these cells is a reflection of the amount of steroid-GR LBD complex formed, which, in turn, depends on the concentration of steroid conjugate that enters the cell.
For the purposes of this study, we developed a new dexamethasone derivative (that we call SDex-COOH) which retains the high GR LBD binding affinity of the parent steroid and is compatible with solid-phase synthesis.10 Peptoids were synthesized as reported previously11 and the peptides by conventional methods. The N-terminus of each molecule was then capped with the activated ester of SDex-COOH and the conjugates were released from the beads with 95% trifluoroacetic acid. All conjugates were purified by reverse phase HPLC.
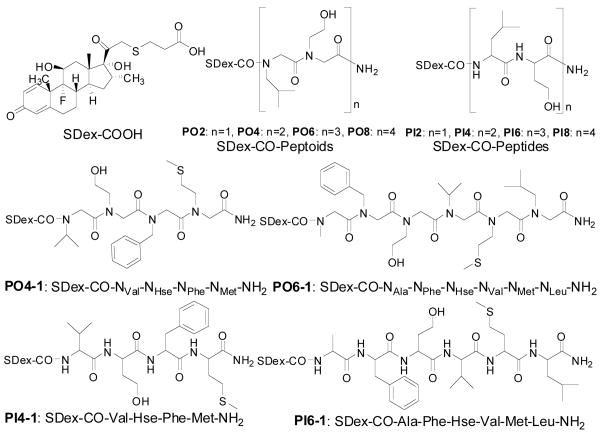
To compare the cell permeability of peptoids and peptides, we employed this chemistry to construct a series of peptide and peptoid conjugates containing between one and four copies of a dimer containing leucine and homoserine side chains (Fig. 1). The analogous peptides and peptoids are isomers and thus provide a fair comparison of the effect on cell permeability of moving the side chain from the α-carbon to the main chain nitrogen.
Figure 1.
Chemical structures of the compounds employed in this study.
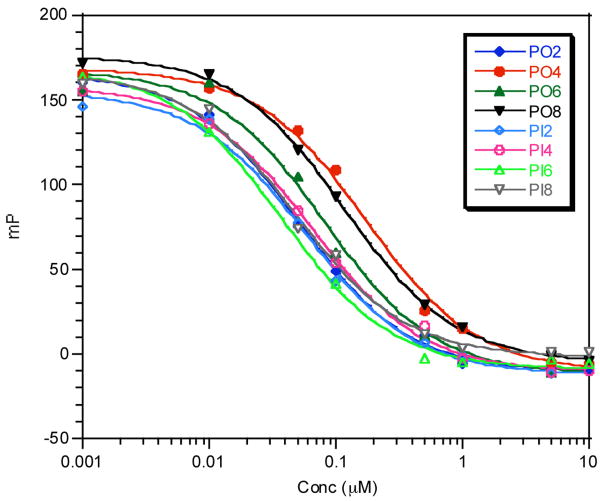
First, we carried out competitive binding assays to measure the in vitro GR binding affinity of each conjugate using fluorescence polarization spectroscopy (Fig. 2) to ensure that the peptide or peptoid fusion had not greatly affected the steroid-GR LBD interaction. This assay monitors displacement of a fluorescein-labeled dexamethasone (Fluormone from Invitrogen) by the conjugate. In most cases, the IC50 values derived from the titration curves were in the nanomolar concentration range, as expected.
Figure 2.
Fluorescence polarization assay for in vitro GR binding affinity. The displacement of a fluorescein-labeled dexamethasone was monitored by fluorescence polarization. The data were plotted as the milipolarization units (mP) versus the concentration of the competitors. IC50 values are shown in Table 1.
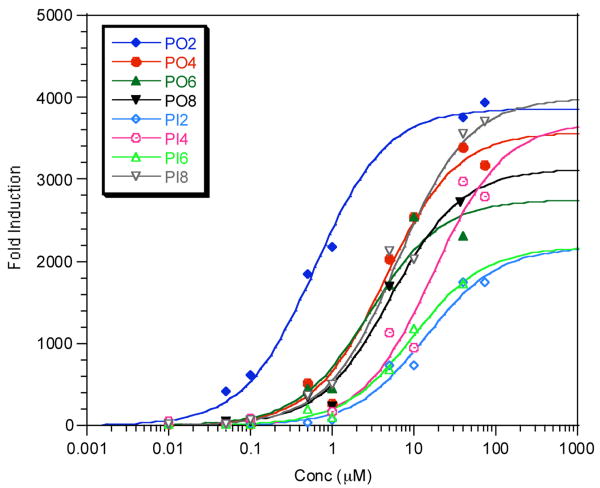
Luciferase assays were then carried out in 96-well plates using HeLa cells transfected with the three plasmids described above. Luminescent intensities were measured 40 hours after treatment with each compound at various concentrations with a dual-luciferase reporter assay system. From the ratio of Gal4-induced luciferase activity to constitutive activity at different concentrations of the conjugates, EC50 values could be determined (Fig. 3). These data were then normalized to correct for the differences in the in vitro GR LBD binding affinities of each compound. Table 1 summarizes these data. The stability of each peptoid or peptide in the serum under these conditions was monitored relative to an internal standard and in all cases the amount of proteolysis was ≤ 10% (data not shown)
Figure 3.
Dose dependent induction of luciferase expression. Luminescent intensities were measured 40 h after treatment of each compound at various concentrations with a dual-luciferase reporter assay system. The data were plotted as the fold induction of luciferase activities versus the concentration of the compounds. EC50 values are summarized in Table 1.
Table 1.
Summary of parameters from in vitro GR binding affinity and luciferase assays.
| Compoundsa | Luciferase induction, EC50 (μM) | In vitro GR binding affinity, IC50 (nM) | Relative permeability factorb |
|---|---|---|---|
| PO2 | 0.62 ± 0.06 | 51 ± 5 | 20.3 |
| PO4 | 4.4 ± 0.9 | 173 ± 20 | 10.2 |
| PO6 | 2.9 ± 0.9 | 82 ± 18 | 7.5 |
| PO8 | 5.2 ± 1.0 | 118 ± 6 | 3.0 |
| PI2 | 14.1 ± 3.3 | 57 ± 9 | - |
| PI4 | 17.1 ± 6.2 | 66 ± 9 | - |
| PI6 | 9.8 ± 1.6 | 37 ± 5 | - |
| PI8 | 6.3 ± 1.2 | 48 ± 6 | - |
| PO4-1 | 6.9 ± 2.5 | 182 ±12 | 26.6 |
| PO6-1 | 1.4 ± 0.2 | 44 ± 4 | 21.0 |
| PI4-1 | 30.3 ± 8.2 | 30 ± 2 | - |
| PI6-1 | 25.4 ± 8.6 | 38 ± 4 | - |
As a reference, isobutyl amide of SDex-COOH: EC50 = 567 nM, IC50 = 14.1 nM.
The relative permeability of peptoids compared to the corresponding peptides, when combined with in vitro GR binding affinity. For example, [EC50(PI2)/EC50(PO2)]/[IC50(PI2)/IC50(PO2)].
The shortest peptoid, PO2 displayed an EC50 value of 0.62 μM, while the corresponding peptide, PI2 had EC50 value of 14.1 μM. Both had nearly identical affinities for the GR LBD in vitro, showing that the peptoid dimer is about 20 times more cell permeable than the isomeric peptide. As can be seen from the data in Table 1, this trend was also observed for the tetrameric, hexameric and octameric isomer pairs, except that the magnitude of the peptoid/peptide permeability ratio decreased with increasing compound size. The tetrameric peptoid was about 10 times more permeable than the isomeric peptide while the hexamers and octamers showed a 7.5- and 3-fold difference, respectively. This appears to be due to the fact that while the permeability of the peptoids decreases, in general, as the size of the compound increases, curiously this is not the case for the peptides. Why peptides in this series do not appear to be less permeable as their size increases from two to eight residues is not clear.
To determine if the conclusion that peptoids are generally more cell permeable than peptides is somehow unique to this series of compounds or a general phenomenon, we synthesized another two peptoids (PO4-1 and PO6-1) with different side chains and the corresponding peptides (PI4-1 and PI6-1) (Fig. 1). The in vitro GR binding affinities (IC50 values) of these compounds were measured and employed to normalize their activities in the cell-based reporter gene assays. As seen in Table 1, the peptoid conjugates each proved to be more than 20 times more cell permeable than the corresponding peptides.
In summary, we evaluated quantitatively the relative cell permeability of analogous peptoids and peptides using a reporter gene-based assay in vivo. Generally, peptoids were much more cell permeable than the corresponding peptides. This suggests that the attachment of side chains to the amide backbone nitrogen atom instead of the α-carbon, which eliminates the polar N-H bond of the peptide, increases the lipophilicity of the molecule and promotes movement across the cell membrane. Finally, this should be a general method with which to compare the cell permeability of any class of peptidomimetics with the corresponding peptides.
Supplementary Material
The detailed procedures for the synthesis of compounds, GR binding assays and luciferase assays. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This project was funded with Federal funds as part of the NHLBI Proteomics Initiative of the National Heart, Lung & Blood Institute, National Institutes of Health, under contract No. NO1-HV-28185. We thank Dr. Xiangshu Xiao (UTSW) for providing the data for SDex-COOH.
References
- 1.(a) Adessi C, Soto C. Curr Med Chem. 2002;9:963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]; (b) Patch JA, Barron AE. Curr Opin Chem Biol. 2002;6:872–877. doi: 10.1016/s1367-5931(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 2.Miller SM, Simmon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Drug Dev Res. 1995;35:20–32. [Google Scholar]
- 3.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. J Am Chem Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 4.Reddy MM, Kodadek T. Proc Natl Acad Sci USA. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang K, Wu CW, Sanborn TJ, Patch JA, Kirshenbaum K, Zuckermann RN, Barron AE, Radhakrishnan I. J Am Chem Soc. 2006;128:1733–1738. doi: 10.1021/ja0574318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masip I, Pérez-Payá E, Messeguer A. Comb Chem High Throughput Screening. 2005;8:235–239. doi: 10.2174/1386207053764567. [DOI] [PubMed] [Google Scholar]
- 7.Hara T, Durell SR, Myers MC, Appella DH. J Am Chem Soc. 2006;128:1995–2004. doi: 10.1021/ja056344c. [DOI] [PubMed] [Google Scholar]
- 8.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu P, Liu B, Kodadek T. Nat Biotechnol. 2005;23:746–751. doi: 10.1038/nbt1099. [DOI] [PubMed] [Google Scholar]
- 10.Liu B, Yu P, Alluri PG, Kodadek T. Mol BioSyst. 2005;1:307–317. doi: 10.1039/b511514k. [DOI] [PubMed] [Google Scholar]
- 11.Olivos HJ, Alluri PG, Reddy MM, Salony D, Kodadek T. Org Lett. 2002;4:4057–4059. doi: 10.1021/ol0267578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The detailed procedures for the synthesis of compounds, GR binding assays and luciferase assays. This material is available free of charge via the Internet at http://pubs.acs.org.