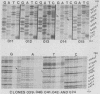
Abstract
Epidemiologic and genetic evidence suggests that influenza A viruses evolve more rapidly than other viruses in humans. Although the high mutation rate of the virus is often cited as the cause of the extensive variation, direct measurement of this parameter has not been obtained in vivo. In this study, the rate of mutation in tissue culture for the nonstructural (NS) gene of influenza A virus and for the VP1 gene in poliovirus type 1 was assayed by direct sequence analysis. Each gene was repeatedly sequenced in over 100 viral clones which were descended from a single virion in one plaque generation. A total of 108 NS genes of influenza virus were sequenced, and in the 91,708 nucleotides analyzed, seven point changes were observed. A total of 105 VP1 genes of poliovirus were sequenced, and in the 95,688 nucleotides analyzed, no mutations were observed. We then calculated mutation rates of 1.5 X 10(-5) and less than 2.1 X 10(-6) mutations per nucleotide per infectious cycle for influenza virus and poliovirus, respectively. We suggest that the higher mutation rate of influenza A virus may promote the rapid evolution of this virus in nature.
Full text
PDF

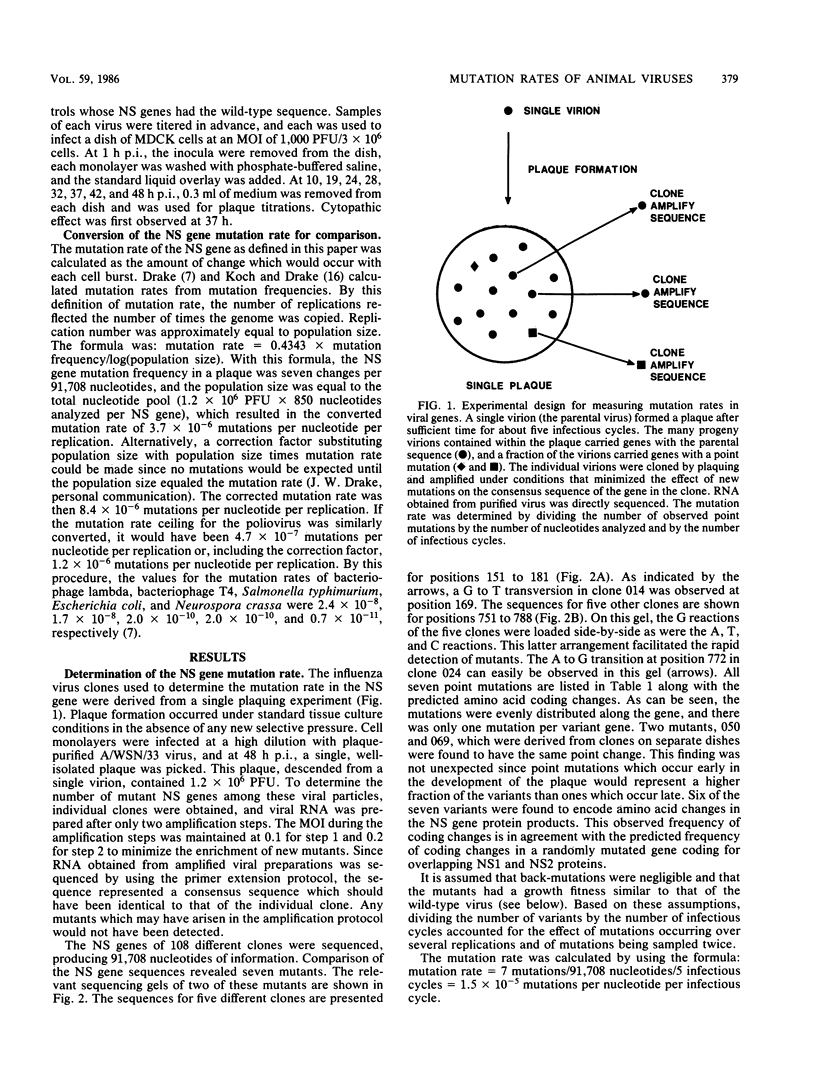
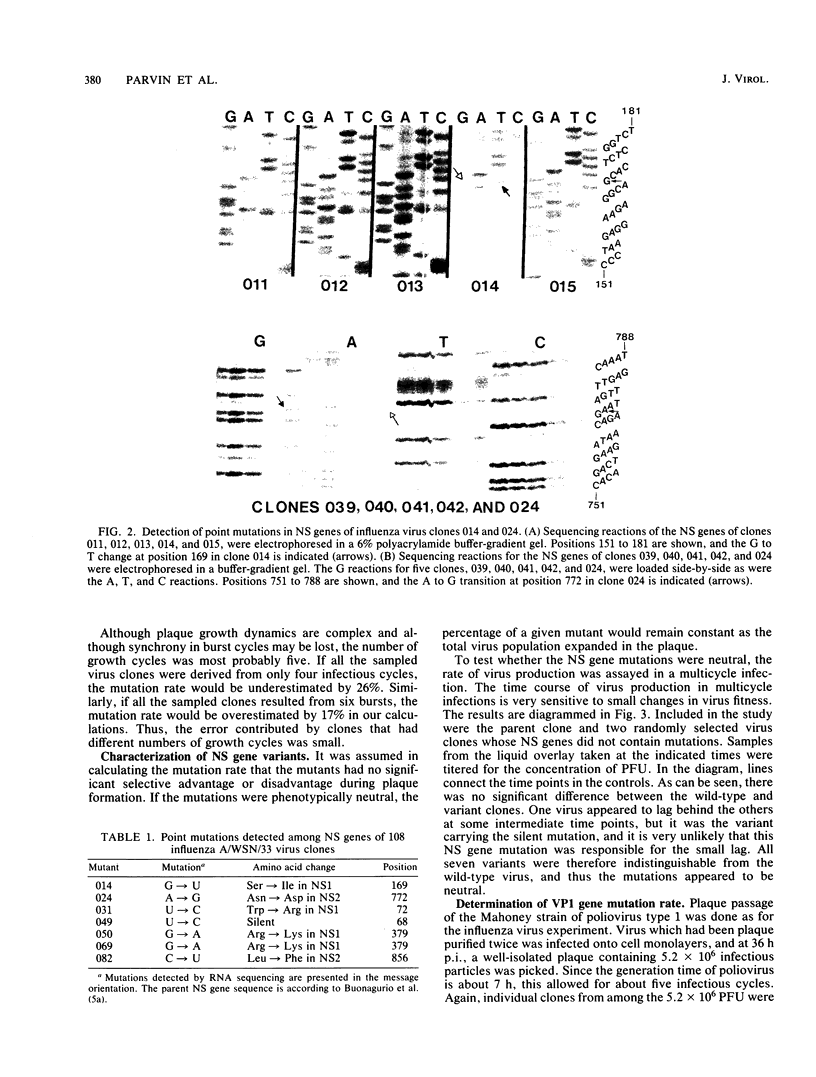
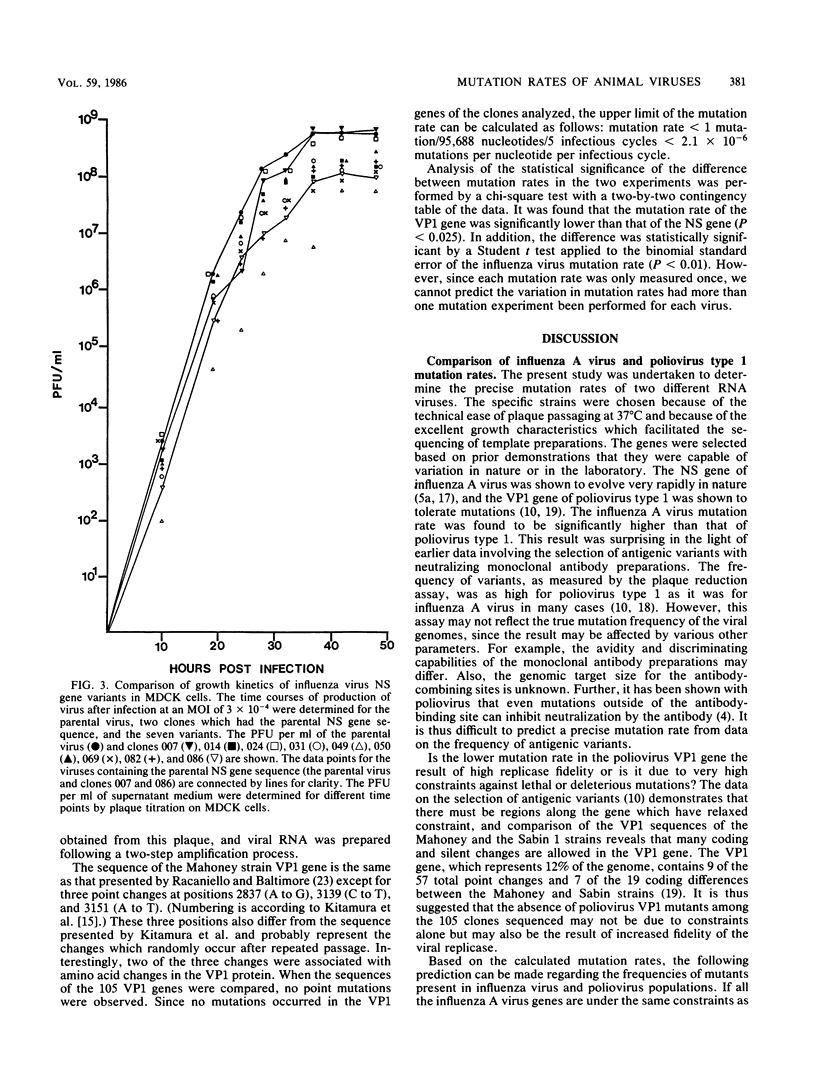


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Batschelet E., Domingo E., Weissmann C. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene. 1976;1(1):27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Sonenberg N., Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol Cell Biol. 1985 Nov;5(11):2913–2923. doi: 10.1128/mcb.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel B., Crainic R., Fichot O., Dufraisse G., Candrea A., Diamond D., Girard M., Horaud F. Mutations conferring resistance to neutralization with monoclonal antibodies in type 1 poliovirus can be located outside or inside the antibody-binding site. J Virol. 1986 Jan;57(1):81–90. doi: 10.1128/jvi.57.1.81-90.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand C., Palese P. Sequential passage of influenza virus in embryonated eggs or tissue culture: emergence of mutants. Virology. 1980 Dec;107(2):424–433. doi: 10.1016/0042-6822(80)90309-8. [DOI] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Parvin J. D., Krystal M., Palese P., Fitch W. M. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science. 1986 May 23;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- Domingo E., Sabo D., Taniguchi T., Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978 Apr;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Drake J. W. Comparative rates of spontaneous mutation. Nature. 1969 Mar 22;221(5186):1132–1132. doi: 10.1038/2211132a0. [DOI] [PubMed] [Google Scholar]
- Eigen M., Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977 Nov;64(11):541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971 Oct;58(10):465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Emini E. A., Kao S. Y., Lewis A. J., Crainic R., Wimmer E. Functional basis of poliovirus neutralization determined with monospecific neutralizing antibodies. J Virol. 1983 May;46(2):466–474. doi: 10.1128/jvi.46.2.466-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Coen D. M., Fisher B. L., Weisslitz M., Randall S., Almy R. E., Gelep P. T., Schaffer P. A. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984 Jan 15;132(1):26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Hayashida H., Toh H., Kikuno R., Miyata T. Evolution of influenza virus genes. Mol Biol Evol. 1985 Jul;2(4):289–303. doi: 10.1093/oxfordjournals.molbev.a040352. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Holland T. C., Marlin S. D., Levine M., Glorioso J. Antigenic variants of herpes simplex virus selected with glycoprotein-specific monoclonal antibodies. J Virol. 1983 Feb;45(2):672–682. doi: 10.1128/jvi.45.2.672-682.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Koch R. E., Drake J. W. Ligase-defective bacteriophage T4. I. Effects on mutation rates. J Virol. 1973 Jan;11(1):35–40. doi: 10.1128/jvi.11.1.35-40.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal M., Buonagurio D., Young J. F., Palese P. Sequential mutations in the NS genes of influenza virus field strains. J Virol. 1983 Feb;45(2):547–554. doi: 10.1128/jvi.45.2.547-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck M. D., Schulman J. L., Palese P. Antigenic variants of influenza viruses: marked differences in the frequencies of variants selected with different monoclonal antibodies. Virology. 1980 Apr 30;102(2):458–462. doi: 10.1016/0042-6822(80)90114-2. [DOI] [PubMed] [Google Scholar]
- Nomoto A., Omata T., Toyoda H., Kuge S., Horie H., Kataoka Y., Genba Y., Nakano Y., Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottay B. K., Kew O. M., Hatch M. H., Heyward J. T., Obijeski J. F. Molecular variation of type 1 vaccine-related and wild polioviruses during replication in humans. Virology. 1981 Jan 30;108(2):405–423. doi: 10.1016/0042-6822(81)90448-7. [DOI] [PubMed] [Google Scholar]
- Palese P., Schulman J. L. Mapping of the influenza virus genome: identification of the hemagglutinin and the neuraminidase genes. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2142–2146. doi: 10.1073/pnas.73.6.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portner A., Webster R. G., Bean W. J. Similar frequencies of antigenic variants in Sendai, vesicular stomatitis, and influenza A viruses. Virology. 1980 Jul 15;104(1):235–238. doi: 10.1016/0042-6822(80)90382-7. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. Polymorphism and evolution of influenza A virus genes. Mol Biol Evol. 1986 Jan;3(1):57–74. doi: 10.1093/oxfordjournals.molbev.a040381. [DOI] [PubMed] [Google Scholar]
- Salts Y., Ronen A. Neighbor effects in the mutation of ochre triplets in the T 4 rII gene. Mutat Res. 1971 Oct;13(2):109–113. doi: 10.1016/0027-5107(71)90002-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrino F., Dávila M., Ortín J., Domingo E. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology. 1983 Jul 30;128(2):310–318. doi: 10.1016/0042-6822(83)90258-1. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Direct method for quantitation of extreme polymerase error frequencies at selected single base sites in viral RNA. J Virol. 1986 Jan;57(1):219–228. doi: 10.1128/jvi.57.1.219-228.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Laver W. G., Air G. M., Schild G. C. Molecular mechanisms of variation in influenza viruses. Nature. 1982 Mar 11;296(5853):115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]