Abstract
L-serine inhibits the catalytic activity of E. coli D-3-Phosphoglycerate Dehydrogenase (PGDH) by binding to its regulatory domain. This domain is a member of the ACT domain family of regulatory domains that are modulated by small molecules. A comparison of the Phi and Psi torsional angle differences between the crystal structures of PGDH solved in the presence and absence of L-serine demonstrated a clustering of significant angle deviations in the regulatory domain. A similar clustering was not observed in either of the other two structural domains of PGDH. In addition, significant differences were also observed at the active site and in the Trp-139 loop. In order to determine if these residues were functionally significant and not just due to other factors such as crystal packing, mutagenic analysis of these residues was performed. Not unexpectedly, this analysis showed that residues that affected the kcat/Km were grouped around the active site and those that affected the serine sensitivity were grouped in the regulatory domain. However, more significantly, residues that affected the cooperativity of inhibition of activity were identified at both locations. These latter residues represent structural elements that participate in both the initial and ultimate events of the transfer of cooperative behavior from the regulatory domain to the active site. As such, their identification will assist in the elucidation of the pathway of cooperative interaction in this enzyme as well as in the elucidation of the regulatory mechanism of the ACT domain in general.
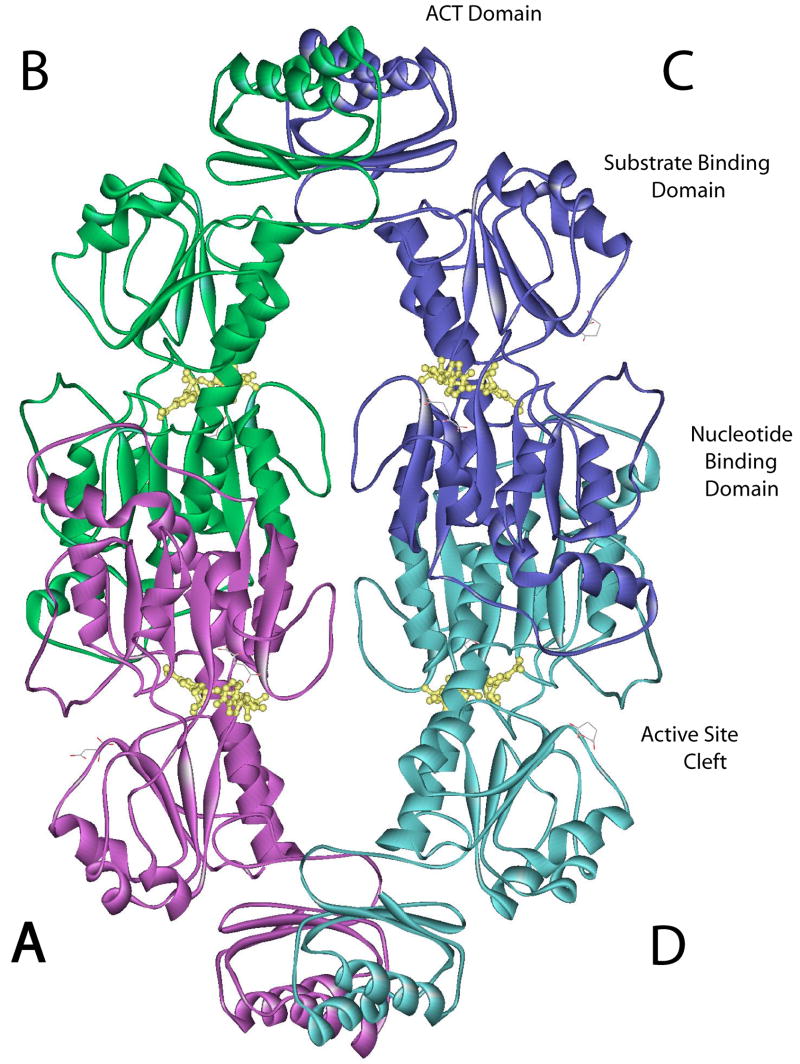
E. coli D-3-Phosphoglycerate Dehydrogenase (PGDH, EC 1.1.1.95) catalyzes the first committed step in L-serine biosynthesis. In turn, L-serine binds to the enzyme and inhibits its catalytic activity mainly by affecting the velocity of the enzyme. The crystal structure of E. coli D-3-Phosphoglycerate Dehydrogenase was originally solved in complex with L-serine (1) (PDB code: 1PSD) and is a tetramer of identical subunits that can be described as a dimer of dimers. Each subunit contains three structurally distinct domains referred to as the nucleotide binding domain, the substrate binding domain, and the regulatory or serine binding domain (Figure 1). The interface forming the fundamental dimer occurs between two nucleotide binding domains. Two such dimers form a tetramer through an interface between adjacent regulatory domains. The nucleotide binding domain is connected to the substrate binding domain by two strands of polypeptide that form a hinge near the active site (2, 3) (see Figure 8). There is a single polypeptide strand connecting the regulatory domain to the substrate binding domain that also appears to function as a hinge (2).
Figure 1. Ribbon diagram of Native of E. coli D-3-Phosphoglycerate Dehydrogenase.
The subunits are labeled A-D and are colored purple, green, blue, and aqua, respectively. The individual domains within a subunit are labeled, as is the active site cleft. NADH is shown in yellow ball and stick structure.
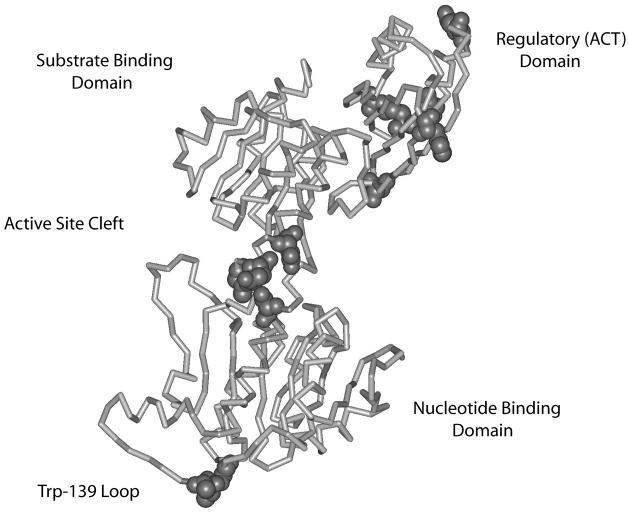
Figure 8. Depiction of the PGDH subunit indicating the location of mutations with a major effect on cooperativity.
A line diagram of the α-carbon chain of PGDH is depicted. The domains are labeled. CPK residues are those residues whose mutations show a change in the Hill coefficient for the cooperativity of L-serine inhibition of activity of equal to or greater than approximately ± 0.5. Note that residues in the Trp-139 loop are considered to be in the vicinity of the active site since they are near the active site of the adjacent subunit (not shown).
L-Serine binds between two adjacent regulatory domains and forms a hydrogen bonding network between the two domains. The regulatory domain of E. coli PGDH is a member of the ACT domain family of regulatory elements (4, 5). This domain, which consists of a ferredoxin-like βαββαβ fold, is being recognized as a major regulatory element in many proteins. It was originally associated with proteins functioning in amino acid metabolism (5) but it is now clear that it functions more generally as a small molecule binding element in the regulation of a variety of processes. The first structure of an ACT domain to be solved was, in fact, that of E. coli PGDH and is considered the archetypical ACT domain structure (1). More recently, additional structures have appeared that clearly show the ACT domain binding small regulatory molecules. These include the NikR transcription factor that is regulated by nickel binding (6), a thiamin binding protein from B. subtilus (7), the ATP phosphoribosyl transferase from M. tuberculsosis which functions in histidine biosysnthesis (8) and threonine deaminase (9) which is regulated by isoleucine and valine.
The mechanism of allosteric regulation of E. coli PGDH by means of effector binding to its regulatory domain has been the subject of extensive investigation (2, 3, 10, 11). However, it has not been until the recent elucidation of the crystal structure of native E. coli PGDH in the absence of L-serine (12) (PDB code: 1YBA) that direct comparison between the inhibited and active structures of this enzyme could be made. Overall, the differences between these structures are not large, but several areas were originally identified where significant differences were observed. These consisted of a rotation of approximately 14 ° about the polypeptide backbone at the hinge region near the active site and a rearrangement of amino acid side chains at the serine-binding site that revealed how L-serine enters the effector site. Since these structural rearrangements were manifested by major changes in the torsional angles of the polypeptide backbone, an analysis was performed to identify any additional torsional angle changes that may be present that weren’t initially identified (12). This manuscript presents a site-directed mutagenesis study of these regions as well as an assessment of the interaction between the two polypeptide chains that form the active site hinge.
METHODS
Site-directed mutagenesis was performed and mutated proteins were expressed and isolated as previously described (11,13). Protein purity was assessed by SDS PAGE gels and all mutants were present as predominate single bands of the appropriate size. All mutants described in this study are based on PGDH 4CA (11) and were verified by automated sequencing on an Applied Biosystems 3730 DNA sequencer. PGDH activity was measured by following the decrease in absorbance at 340 nm in the presence of NADH and α-ketoglutarate. Assays were performed in 20 mM potassium phosphate buffer, pH 7.5 containing 250 μM NADH and saturating levels of α-ketoglutarate. NADH and L-serine were purchased from Sigma Chemical Co. (St. Louis). All other reagents were analytical grade.
Protein quantitation was performed spectrophotometrically using an E1% at 280 nm of 6.7 (13). Data were fitted to the Michaelis-Menton equation for the determination of Km and kcat and to the Hill equation for the determination of the serine concentration at 50% inhibition (I 0.5) and the Hill coefficient (11). Curve fitting was performed with Kaleidograph from Synergy Software.
Absolute phi and psi angle differences were computed by subtracting the angles in the reference subunit from the other subunit, adjusting the values to represent a maximum of 180° and expressing them as positive numbers. These values were then plotted as a function of residue position.
Double mutant analysis was performed according to the “inverse thinking” method described by Mildvan (14). The parameters of the double mutant are used as the reference point with this method rather than those of the wild-type enzyme. The data are expressed in a logarithmic plot of the parameters relative to the double mutant that result from restoring each mutant singly, and both together. Additive effects indicate that each residue functions independently, whereas a departure from additivity indicates that the residues interact. Partial additivity indicates cooperativity between the two residues. Synergy, where both single mutants add up to more than the wild-type, indicates anti-cooperativity, and antagonism of the two residues indicates opposing structural effects of the two residues.
RESULTS
The original comparison of the crystal structures of E. coli PGDH with and without bound L-serine highlighted three areas of the structure where significant differences were apparent (12). These are the active site hinge region, the L-serine binding site, and a new inter-subunit contact involving Asn-190. Residues in these regions were subjected to site-directed mutagenesis to define their individual contributions to substrate binding, catalytic activity, serine sensitivity, and cooperativity of serine inhibition of activity. In addition, several double mutations were analyzed for linkage between the two polypeptide strands that comprise the active site hinge.
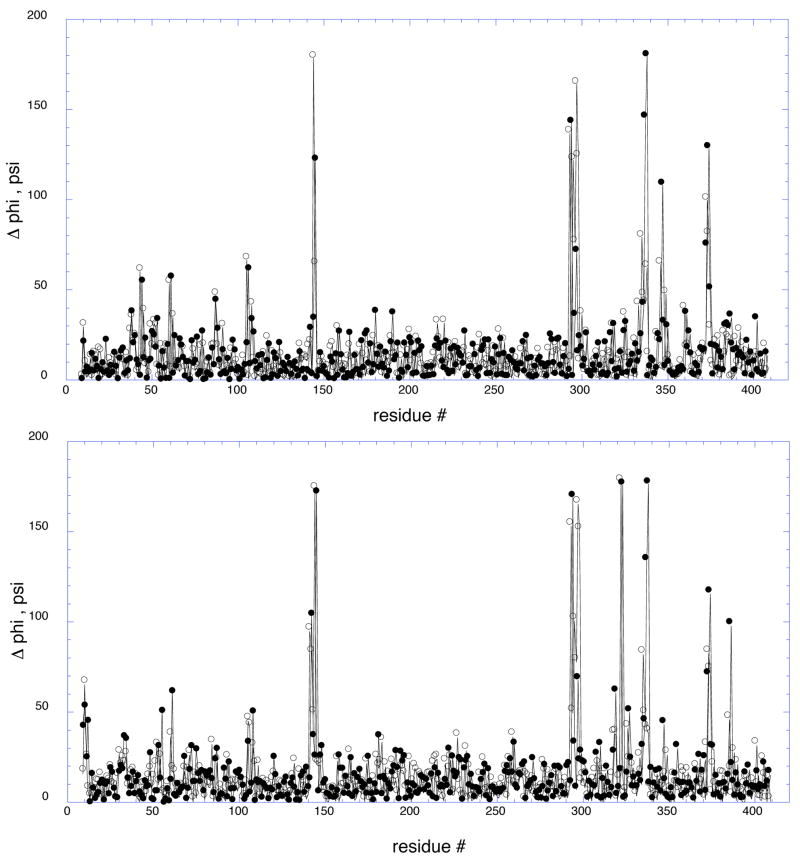
Potential additional areas of interest are identified by comparing the phi and psi angles of PGDH with and without bound L-serine. The structure with L-serine contains 2 subunits in the asymmetric unit, subunits “a” and “b”. The structure without serine contains all four subunits in the asymmetric unit, subunits “a, b, c, and d”. The 2 unique subunits of the inhibited enzyme, termed “ia” and “ib”, were compared to all four subunits of the uninhibited enzyme. The plots of the computed phi and psi angle differences between subunits, “ia” and “a” and between “ib” and “b are shown in Figure 2. All of the major angle differences found in the four subunits are represented in these plots and the results for each comparison are listed in Table 1. The three areas mentioned above that were originally identified are among those found in this analysis. It is also apparent from this figure that most of the other areas of significant torsional angle differences are clustered in the C-terminal portion of the enzyme. Of these, 4 are found in the regulatory or ACT domain and 2 are found in the loops between the regulatory domain and the substrate-binding domain. No differences of this magnitude occur in either the nucleotide binding domain or the subunit binding domain with the exception of the Trp-139 loop (residues 141–145) and the active site hinge (residues 293–298).
Figure 2. Plots of the Phi and Psi angle differences found in the crystal structures of E. coli D-3-Phosphoglycerate Dehydrogenase with and without bound L-serine.
The top panel compares inhibited subunit b (ib) with uninhibited subunit b and the bottom panel compares inhibited subunit a (ia) with uninhibited subunit a. The phi (●) and psi (○) angle differences are plotted versus residue number. The differences are expressed as positive numbers between 0 and 180°.
Table 1.
Major Areas of Divergence of Phi and Psi Angles between PGDH Structures Determined in the Absence and Presence of L-Serine
| Residue # | Sequence | Subunits a | Description b |
|---|---|---|---|
| 141–145 | KLAAG | ia - a, b, c, d | Trp-139 loop |
| ib - a, b, c, d | |||
| 293–298 | IGGSTQ | ia - a, b, c, d | Active site hinge |
| ib - a, b, c, d | |||
| 322–323 | LS | ia - a, b, c, d | 1st Loop between SBD and RD |
| 335–338 | HGGR | ia - a, b, c, d | 2nd Loop between SBD and RD |
| ib - a, b, c, d | |||
| 346–347 | NR | ia - b | Loop between β1 and α1 of RD containing serine binding site residues |
| ib - a, b, c | |||
| 360–363 | EQGV | ia - c | Loop between α1 and β2 of RD |
| ib - c | |||
| 373–374 | SA | ia - a, b, c | Loop between β2 and β3 of RD |
| ib - a, b, c | |||
| 385–387 | ADE | ia - a, b, c, d | Loop between β3 and α2 of RD |
| ib - a |
Denotes the subunit comparisons where differences were observed expressed as a subunit with bound serine (ia, ib) minus a subunit without bound serine (a, b, c, d)
NBD, nucleotide binding domain; SBD, substrate binding domain; RD, regulatory domain; αi or βi, α helices and β strands are numbered in succession in the direction of the N to C termini.
Four of the six new regions identified by this analysis are found in the regulatory or ACT domain in loop regions connecting β-strands and helices. Although it would not be unexpected to see these kinds of differences predominantly in loop regions, it is curious that they are clustered in the regulatory domain. However, since the ACT domains form multiple crystal contacts these differences may represent local changes introduced by crystal packing forces. Therefore, the functions of these regions were also probed using site-directed mutagenesis in an effort to distinguish between changes due to crystal packing or other factors and residues that exhibit a functional role.
Active Site Hinge and Domain Connecting Strands
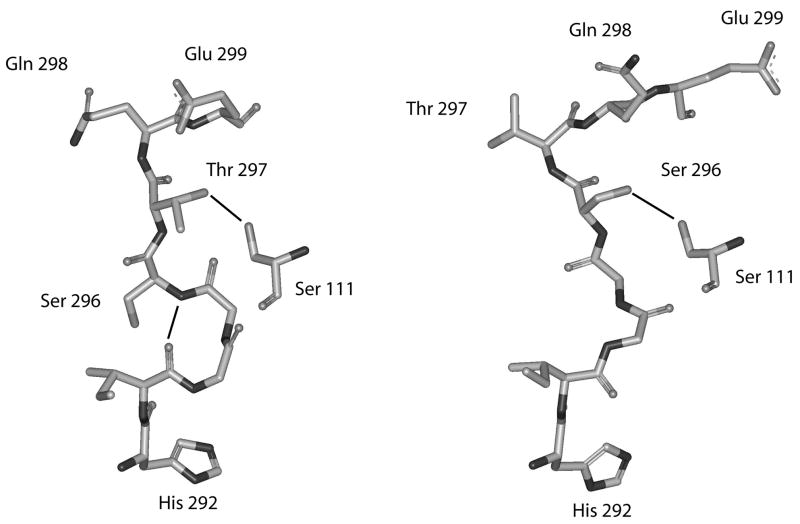
A pronounced change observed with the binding of L-serine is an inversion of the polypeptide chain at the Gly-294, Gly-295 hinge as illustrated in Figure 3. This causes the side chains of Thr-297 and Ser-296 to invert, causing a switch in potential hydrogen bonding to Ser-111. This inversion also causes the inter-chain hydrogen bond between the amide nitrogen of Gln-301 to switch from the carbonyl oxygen of Thr-297 to the carbonyl oxygen of Ser-296 and also breaks an inter-chain hydrogen bond between the carbonyl of Ile-293 and the amide nitrogen of Ser-296. In order to assess the potential contributions of residues in this region and along the two chains connecting this site to the regulatory domain (Figure 4), residues whose side chains could form hydrogen bonds other than internal to the α-helix were mutated to alanine residues. In addition, glycine residues were converted to valine residues. The results are listed in Table 2. It can be seen from the data that mutations to these chains variably effect catalytic efficiency, sensitivity to serine, and the cooperativity of serine inhibition. As much as a seventy-fold decrease in catalytic efficiency (kcat/Km) is seen when Thr-297 or Lys-311 are mutated. This decrease seems to be due mainly to an effect on substrate binding since the greatest change in Vmax seen for any residue in this series is only 3 fold. The catalytic efficiencies of most other mutations also decrease, with the main effect also being on the Km. The only exception is Glu-302, whose catalytic efficiency is slightly increased because of a slightly better Km than the native enzyme.
Figure 3. Depiction of the area where structural inversion occurs near the active site of E. coli D-3-Phosphoglycerate Dehydrogenase.
Residues shown are from His-292 to Glu-299 from one strand and residue Ser-111from the adjacent strand. Hydrogen bonds are shown as solid lines. The structure on the left is from the enzyme in the absence of L-serine and the structure on the right is the enzyme in the presence of L-serine.
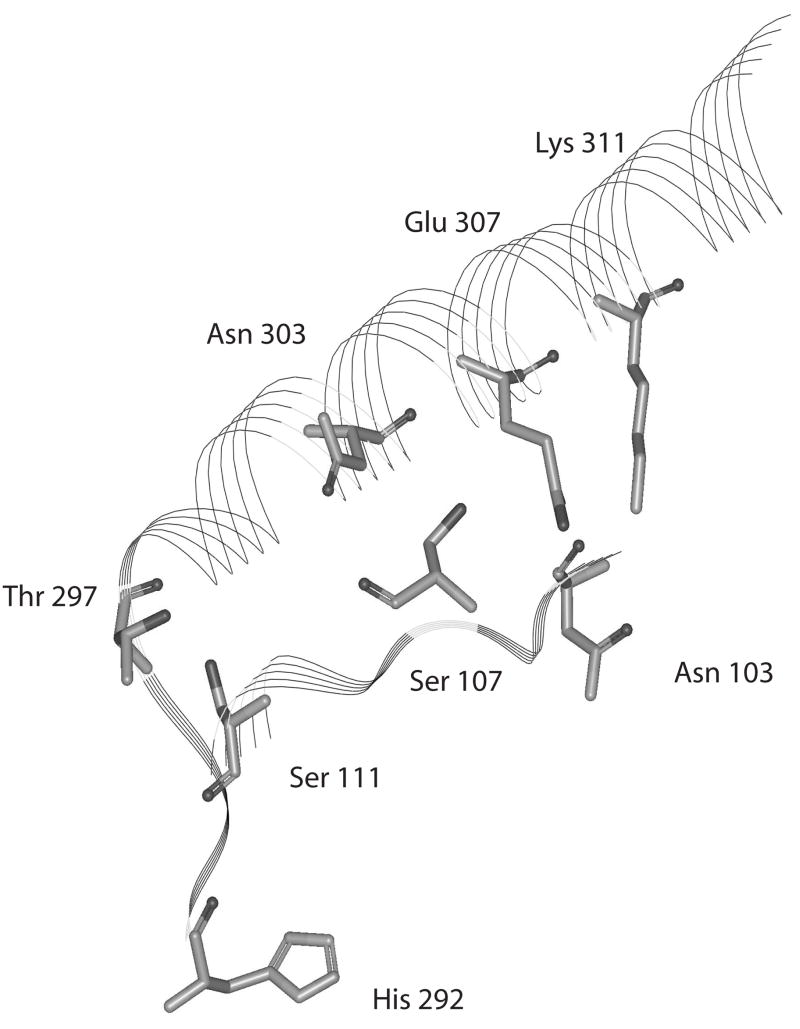
Figure 4. Depiction of the orientation of selected residues from the connecting strands between the substrate binding domain and the ACT domain of E. coli D-3-Phosphoglycerate Dehydrogenase.
The area depicted is from His-292 to Asp-317 from one strand and from Asn-103 to Ser-107 from the adjacent strand. The other residues do not participate in hydrogen bonding between strands and are left out for clarity. The structure on the left is from the enzyme in the absence of L-serine and the structure on the right is the enzyme in the presence of L-serine.
Table 2.
Kinetic Parameters of PGDH with mutated Residues in the Polypeptides Connecting the Substrate Binding Domain and the Regulatory Domain
| Mutant | Km (mM) |
Vm (μmol min−1 mg−1) |
kcat (sec−1) |
kcat / Km (sec−1 M−1) |
I 0.5(μM) | nH |
|---|---|---|---|---|---|---|
| Native (4CA) | 0.49 ± 0.03 | 4.61 ± 0.06 | 13.5 | 2.8 × 104 | 2.34 ± 0.4 | 1.88 ± 0.07 |
| S107A | 0.97 ± 0.07 | 3.07 ± 0.06 | 9.0 | 0.9 × 104 | 2.58 ± 0.08 | 1.54 ± 0.09 |
| S111A | 4.33 ± 0.29 | 2.56 ± 0.07 | 7.5 | 0.17 × 104 | 2.23 ± 0.10 | 1.26 ± 0.08 |
| S296A | 1.37 ± 0.14 | 2.51 ± 0.09 | 7.4 | 0.54 × 104 | 14.28 ± 0.19 | 2.34 ± 0.09 |
| T297A | 16.9 ± 1.1 | 2.38 ± 0.10 | 7.0 | 0.04 × 104 | 8.83 ± 0.20 | 1.36 ± 0.05 |
| Q298A | 1.41 ± 0.18 | 3.93 ± 0.14 | 11.5 | 0.82 × 104 | 3.45 ± 0.13 | 1.62 ± 0.11 |
| E299A | 1.99 ± 0.2 | 1.66 ± 0.06 | 4.9 | 0.25 × 104 | 7.43 ± 0.34 | 1.23 ± 0.07 |
| Q301A | 4.78 ± 0.41 | 3.61 ± 0.16 | 10.6 | 0.22 × 104 | 4.31 ± 0.22 | 1.49 ± 0.13 |
| E302A | 0.30 ± 0.05 | 3.81 ± 0.14 | 11.2 | 3.7 × 104 | 3.79 ± 0.18 | 1.49 ± 0.11 |
| N303A | 1.41 ± 0.09 | 4.22 ± 0.09 | 12.4 | 0.89 × 104 | 1.50 ± 0.08 | 1.14 ± 0.09 |
| E307A | 1.77 ± 0.13 | 4.21 ± 0.11 | 12.3 | 0.69 × 104 | 5.65 ± 0.4 | 1.83 ± 0.06 |
| K311A | 12.0 ± 1.2 | 1.77 ± 0.11 | 5.2 | 0.04 × 104 | 5.74 ± 0.18 | 1.56 ± 0.09 |
| S316A | 0.63 ± 0.06 | 3.93 ± 0.08 | 11.5 | 1.8 × 104 | 2.10 ± 0.07 | 1.62 ± 0.07 |
| D317A | 1.60 ± 0.14 | 1.57 ± 0.04 | 4.6 | 0.29 × 104 | 2.19 ± 0.08 | 1.52 ± 0.09 |
| N303A/K311A | 13.1 ± 0.8 | 3.28 ± 0.06 | 9.6 | 0.07 × 104 | 4.83 ± 0.16 | 1.42 ± 0.07 |
| S111A/K311A | 320 ± 16 | 1.42 ± 0.05 | 4.2 | 0.0026 × 104 | 17.8 ± 0.4 | 1.63 ± 0.07 |
| S107A/S111A | 6.6 ± 0.8 | 3.88 ± 0.23 | 11.4 | 0.17 × 104 | 2.62 ± 0.04 | 1.70 ± 0.06 |
Sensitivity to serine varies over a relatively narrow range (from 0.65 to 2.5 fold) with the exception of Ser-296 and Thr-297, which show a 4 and 6 fold decrease in sensitivity, respectively. In most cases, the cooperativity of serine inhibition is decreased, with the greatest differences seen for Ser-111, Thr-297, and Asn-303. That for Glu-307 is essentially unaffected. Interestingly the Ser-296 mutation increases cooperativity relative to the native enzyme while the others decrease cooperativity.
Double Mutants
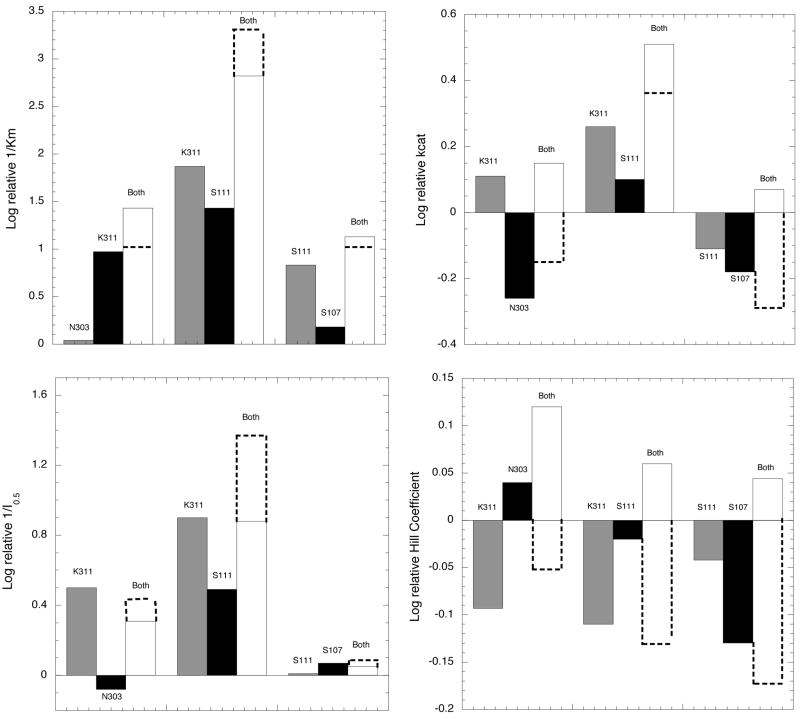
A series of double mutants were produced to assess the interaction of residues within the two polypeptide strands connecting the active site with the regulatory domain. The parameters are listed in Table 2 and the analyses using the double mutant as the reference point are shown in Figure 5. Each bar represents the change in that particular parameter when the designated residue is restored to the double mutant, which is set at zero. For instance, in the case of N303A- K311A, the bar designated N303 represents the parameter upon restoration of that residue, which would be the K311A single mutant. Likewise, K311 represents the N303A single mutant and the bar designated as “Both” represents the wild-type enzyme relative to the double mutant.
Figure 5. Bar plots of the double mutant data according to the method of Mildvan (15).
The values for the double mutants are set to zero and the parameters of the single mutants are expressed as the effect of restoring each one individually or both together. The magnitude of the bars is expressed as the log of the relative parameter. The dotted lines represent the magnitude of restoring both single mutants together if their respective effects were simply additive, i.e. no interaction between residues. Deviation from this additive value indicates either cooperativity, anti-cooperativity, or antagonism in the interaction of the two residues.
For the Km values, the residues on the same strand (N303/K311 and S111/S107) interact cooperatively while the residues on opposite strands (K311/S111) interact anti-cooperatively. On the other hand, for the kcat parameter the residues on opposite strands interact cooperatively while those on the same strand are antagonistic. For the serine interaction parameters, all of the double mutants interact anti-cooperatively for I 0.5 and antagonistically in reference to the Hill coefficient. It should be noted that this analysis is not strictly appropriate for the Hill coefficient since it does not represent a binding parameter per se. As such, thermodynamic values cannot be derived from the data. However, it is included here for purposes of illustration and serves to show the directionality of the effect relative to the double mutant. The difference in the additive value and that of the wild type (both) indicates the magnitude of the cooperative, anti-cooperative, or antagonistic effect of the interaction. It should be noted that in no case is the cooperative effect of these mutants greater than approximately 100.5 or about 3 fold.
The Tryptophan Loop
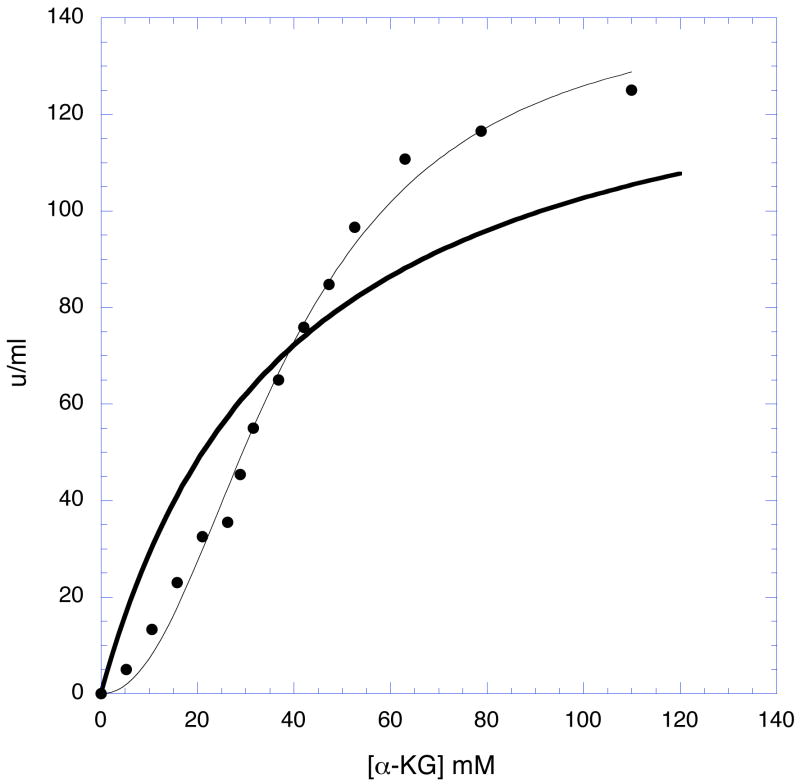
Residues 141–145 are in a loop that borders the active site of the adjacent subunit. This loop also contains Trp-139 that inserts itself into a pocket at the base of the catalytic residues of the adjacent subunit. Trp-139 is thought to function in the transmission of cooperativity from one subunit to another (3, 15). The results of the mutations are presented in Table 3. Three of the residues in this region possess small side chains. Mutation of these residues to more bulky side chains may restrict rotation of the polypeptide at this point. When Ala-143, Ala-144, and Gly-145 are converted to valine residues, significant decreases in the cooperativity of serine inhibition are observed, suggesting that rotational flexibility in this area is necessary for cooperativity. The side chain amino group of Lys-141 also lies within the substrate binding pocket of the adjacent subunit and has been shown by crystallographic analysis to participate in an intermediate stage of substrate binding (12). Mutation of Lys-141 to alanine not only causes significant changes in activity but also lowers the cooperativity of serine inhibition. The most striking effect of this mutation is that it produces a homotropic cooperative effect in the dependence of activity on substrate concentration (Figure 6). All of the other mutations studied retain the typical hyperbolic behavior of the native enzyme.
Table 3.
Kinetic Parameters of PGDH with mutated Residues in the Trp-139 loop and in the Regulatory Domain
| Mutant | Km (mM) |
Vm (mmol min−1 mg−1) |
kcat (sec−1) |
kcat / Km (sec−1 M−1) |
I 0.5 (μM) |
nH |
|---|---|---|---|---|---|---|
| Native(4CA) | 0.49 ± 0.03 | 4.61 ± 0.06 | 13.5 | 2.8 × 104 | 2.34 ± 0.4 | 1.88 ± 0.07 |
| K141A | 29.1 ± 1.9 | 1.10 ± 0.06 a | 3.2 | 0.011 × 104 | 1.58 ± 0.05 | 1.48 ± 0.07 |
| A143V | 1.19 ± 0.12 | 2.92 ± 0.08 | 8.6 | 0.72 × 104 | 2.09 ± 0.08 | 1.39 ± 0.09 |
| A144V | 0.26 ± 0.04 | 4.70 ± 0.10 | 13.8 | 5.3 × 104 | 1.60 ± 0.07 | 1.16 ± 0.07 |
| G145V | 7.20 ± 0.60 | 2.97 ± 0.12 | 8.7 | 0.12 × 104 | 1.78 ± 0.08 | 1.22 ± 0.07 |
| N190A | No detectable protein | |||||
| S323A | 0.68 ± 0.05 | 3.76 ± 0.06 | 11.0 | 1.6 × 104 | 2.92 ± 0.05 | 1.73 ± 0.06 |
| H335A | 0.79 ± 0.11 | 2.67 ± 0.09 | 7.8 | 0.98 × 104 | 0.96 ± 0.06 | 1.13 ± 0.10 |
| G336V | 0.19 ± 0.03 | 3.39 ± 0.09 | 9.9 | 5.2 × 104 | 47.9 ± 0.9 | 1.82 ± 0.07 |
| G337V | 0.64 ± 0.09 | 1.66 ± 0.05 | 4.9 | 0.77 × 104 | 4.42 ± 0.3 | 1.79 ± 0.21 |
| R338A | 0.33 ± 0.07 | 1.97 ± 0.08 | 5.8 | 1.80 × 104 | 34.0 ± 1.0 | 1.97 ± 0.14 |
| E360A | 0.66 ± 0.06 | 3.68 ± 0.08 | 10.8 | 1.6 × 104 | 2.6 ± 0.1 | 1.08 ± 0.07 |
| Q361A | 0.47 ± 0.03 | 3.07 ± 0.03 | 9.0 | 1.9 × 104 | 2.92 ± 0.06 | 1.94 ± 0.08 |
| G362V | 0.68 ± 0.12 | 2.17 ± 0.09 | 6.4 | 0.94 × 104 | 29.1 ± 1.4 | 1.82 ± 0.20 |
| S373A | 0.72 ± 0.04 | 4.04 ± 0.06 | 11.9 | 1.7 × 104 | 1.47 ± 0.9 | 1.17 ± 0.09 |
| A374V | 0.35 ± 0.03 | 4.00 ± 0.07 | 11.7 | 3.3 × 104 | 3.55 ± 0.07 | 1.99 ± 0.08 |
| Q375A | 0.49 ± 0.04 | 6.36 ± 0.13 | 18.7 | 3.8 × 104 | 2.13 ± 0.10 | 1.65 ± 0.11 |
| D386A | 0.35 ± 0.04 | 4.48 ± 0.10 | 13.1 | 3.7 × 104 | 4.82 ± 1.0 | 1.92 ± 0.09 |
| E387A | 0.28 ± 0.04 | 4.36 ± 0.12 | 12.8 | 4.6 × 104 | 3.37 ± 0.07 | 2.12 ± 0.11 |
Displays sigmoidal kinetics as a function of substrate concentration with a Hill coefficient of 2.3 ± 0.2.
Figure 6. A plot showing the Homotrophic Cooperative behavior of the K-141A mutant enzyme.
Enzyme activity in u/ml is plotted versus the concentration of substrate. The symbols (●) represent the data and the thin solid line is a fit of the data to a modified Michaelis-Menton equation to account for the non-hyperbolic nature of the plot (v = (Vm × Sn) / (Kmn + Sn)). The dark solid line shows a plot representing a hyperbolic (non-cooperative) curve with the same Vm (116 u/ml) and Km (29 mM) determined from the data. The fit produces a value for nH = 2.28 ± 0.24.
Asn-190
The structure of PGDH in the absence of L-serine shows a new inter-subunit contact involving Asn-190 that is not apparent in the structure determined in the presence of L-serine. Mutation of Asn-190 to alanine results in the inability to isolate detectable levels of protein.
ACT Domain - Serine Binding Site
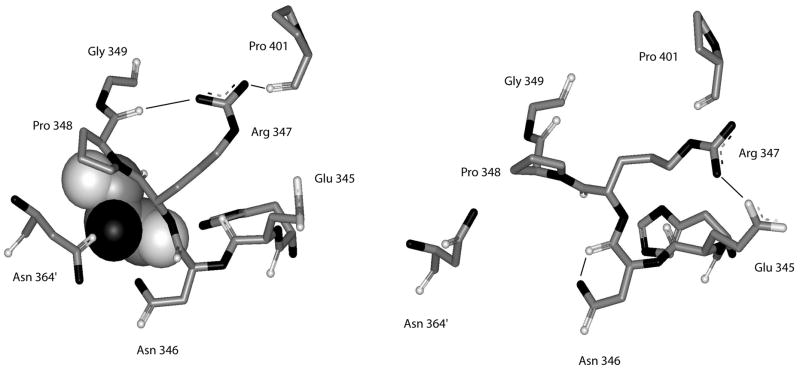
The structure of the native enzyme in the absence of serine suggests that serine enters its binding site between adjacent regulatory domains through a small opening between the side chains of Asn-346, Asn-364′, and Pro-348 (12). When serine binds, these side chains move to cover the serine and close the binding site cavity. Figure 7 shows the conformation of the serine-binding site in the presence and absence of L-serine. The rotation of the polypeptide at Asn-346 and Arg-347 opens and closes the binding site. In the process, the hydrogen-bonding pattern of these two residues also changes. Upon serine binding, a hydrogen bond between the side chain nitrogen of Asn-346 and its main chain carbonyl carbon is broken and the side chain of Asn-346 interacts with the bound serine. In addition, a hydrogen bond between the side chains of Arg-374 and Glu-345 is broken and new hydrogen bonds are formed between the side chain if Arg-347 and the carbonyl carbons of Pro-401 and Pro-348. However, this shift in hydrogen bonding of Arg347 is seen in only 2 of the 4 subunits, 1 subunit in each of the two dimeric asymmetric units. Although the phi/psi shift in Asn-346 and Arg-347 occurs in all subunits, the angles differ by as much as 25° between the two subunits and the hydrogen bonding network between Arg-347, Pro-348, and Pro-401 occurs only in subunits b and d, diagonally across from each other in the tetramer. Thus, the two serine binding sites at each regulatory domain interface have a different hydrogen bonding pattern.
Figure 7. Depiction of the structure of the residues forming the serine binding site of E. coli D-3-Phosphoglycerate Dehydrogenase.
Residues shown are from His-344 to Gly-349 from one subunit and residue Asn-364′ from the adjacent subunit. Hydrogen bonds are shown as solid lines. The structure on the left is from the enzyme in the presence of L-serine and the structure on the right is the enzyme in the absence of L-serine. L-serine is depicted as a CPK structure.
None of the mutations at this site have more than a two-fold effect on the Km or the kcat (Table 4). However, profound effects on cooperativity and sensitivity to serine are observed. As expected, the three residues that form hydrogen bonds directly with the L-serine effector, His-344, Asn 346, and Asn-364′, have the greatest effect on serine sensitivity. A relatively large effect is also seen when a bulky side chain is placed at residue 349, suggesting that rotation of the polypeptide may be functional in this region. Two of these residues, Arg-347 and Gly-349 have a significant effect on the cooperativity of serine inhibition.
Table 4.
Kinetic Parameters of PGDH with mutated Residues at the Serine Binding Site
| Mutant | Km (mM) |
Vm (mmol min−1 mg−1) |
kca (sec−1) |
kcat / Km (sec−1 M−1) |
I 0.5 (μM) |
nH |
|---|---|---|---|---|---|---|
| Native (4CA) | 0.49 ± 0.03 | 4.61 ± 0.06 | 13.5 | 2.8 × 104 | 2.34 ± 0.4 | 1.88 ± 0.07 |
| H344A | 0.23 ± 0.03 | 3.35 ± 0.10 | 9.8 | 4.3 × 104 | 147.2 ± 4.4 | 1.70 ± 0.11 |
| E345A | 0.76 ± 0.06 | 4.07 ± 0.08 | 11.9 | 1.6 × 104 | 7.02 ± 0.11 | 1.80 ± 0.06 |
| N346A | 0.39 ± 0.05 | 5.08 ± 0.15 | 14.9 | 3.8 × 104 | 3138 ± 51 | 1.99 ± 0.07 |
| R347A | 0.45 ± 0.08 | 4.11 ± 0.08 | 12.1 | 2.7 × 104 | 5.5 ± 0.3 | 1.20 ± 0.10 |
| P348A | 0.58 ± 0.07 | 3.16 ± 0.08 | 9.3 | 1.6 × 104 | 10.11 ± 0.15 | 1.99 ± 0.07 |
| G349V | 0.69 ± 0.06 | 2.06 ± 0.04 | 6.0 | 0.9 × 104 | 238 ± 23 | 0.97 ± 0.11 |
| N364A | 0.29 ± 0.03 | 4.02 ± 0.09 | 11.8 | 4.1 × 104 | 15618 ± 313 | 2.12 ± 0.08 |
Regulatory (ACT) Domain -Loop Residues
At the end of the substrate binding domain, two loops provide a transition into the βαββαβ fold of the ACT domain. Two areas of significant backbone angle differences are seen in these transition loops at residues 322–323 and residues 335 to 338. Residues 335 to 338 encompass a putative hinge area previously shown to have an effect on the serine sensitivity of inhibition of activity (2). None of the mutations in these areas had more than a three fold effect on either Km or kcat (Table 3). However, much more significant effects were observed for serine sensitivity and cooperativity. A significant decrease in serine sensitivity is seen for G336V and for R338A. On the other hand, the cooperativity decreased significantly for H335A and the serine sensitivity increased.
Within the regulatory domain itself, none of the mutations caused more than a two-fold difference in Km or kcat while three showed significant differences in serine sensitivity and cooperativity of inhibition. Serine sensitivity decreased an order of magnitude when a bulky side chain was introduced at Gly-362, and the cooperativity of serine inhibition was reduced to nearly 1.0 when Glu-360 and Ser-373 were converted to alanyl residues. Glu-360, Gly-362 and Ser-373 are at the ends of the α-helix and β-sheet that flank the regulatory domain interface where serine binds.
DISCUSSION
A comparison of the phi and psi angles of E. coli PGDH with and without bound L-serine showed a limited number of areas where these angles underwent a relatively large change reflecting significant rotations of the polypeptide backbone. Two of these regions, residues 293–298 (2) and residues 335–338 (2, 16) were areas already identified as being critical to the mechanism of serine inhibition of the enzyme. Unexpectedly, most of the other major angle changes occurred in, or very near the regulatory domain. There could be several reasons for this observation. It could be that the precision of the determination of the dihedral angles could be low because they occur in potentially flexible regions. However, if this were the case, one would expect to see dihedral angle changes of this magnitude throughout the molecule. They could be due to crystal contact effects that influence the regulatory domain more than the other domains of the enzyme. They could also represent real changes that occur upon effector binding. In an effort to distinguish between these possibilities and to further elucidate the mechanism of regulation of E. coli PGDH and of the regulatory (ACT) domain, the specific areas identified by crystallographic analysis to undergo rotational change were probed by residue-specific mutagenesis. As the data shows, upon mutation several of the residues identified by their torsional angle differences display significant effects on cooperativity.
Those mutations that demonstrate a significant alteration of behavior can be grouped into three general categories. Those that affect mainly the selectivity of the active site (kcat/Km), those that effect mainly the serine sensitivity or serine binding (I 0.5) and those that effect mainly the cooperativity of serine inhibition of catalytic activity (nH). A few mutations belong to more than one of these groups. Those that affect serine sensitivity tend to be grouped in the regulatory domain around the serine binding site. Those that affect catalytic activity tend to be grouped around the active site. Neither of these observations is unexpected. However, the significant observation is those residues that affect cooperativity of inhibition are found both in the regulatory domain as well as around the active site. The locations of these residues are shown in figure 8. Conceptually, these residues comprise part of the cooperativity network representing critical elements at the initiation of the cooperative event (regulatory or ACT domain) and those at the culmination of the cooperative event (active site).
One of the most dramatic differences seen upon serine binding is the inversion of the polypeptide chain centering on Ser-296 and Thr-297. The interaction between Thr-297 and Ser-111 is critical for optimal substrate interaction as well as cooperativity of inhibition. Studies with hybrid tetramers (11) have shown that E. coli PGDH displays half-the-sites activity and that the first serine to bind completely inhibits the active site of the subunit to which it binds as well as the second functional active site by approximately 33%. Thus, the Thr-297/Ser-111 interaction is also necessary for the conformational change that occurs at adjacent active sites upon the binding of the first effector molecule.
Asn-303 and Glu-299 appear to play a similar role in the mechanism of cooperativity. Interestingly, although its side chain appears to make a large rotational swing when serine binds, the crystal structure shows no apparent interaction of Glu-299 with another residue. On the other hand, a new hydrogen bond between Asn-303 and Ser-107 is apparent and may serve to stabilize the cooperative effect.
In contrast, Ser-296 which changes places with Thr-297 as a hydrogen binding partner with Ser-111 subsequent to serine binding, displays the opposite effect on the cooperativity of inhibition as judged by the increase in cooperativity in its absence. The mutation also shows a 6-fold decrease in serine sensitivity. Thus, Ser-296 serves to stabilize the serine bound structure but somewhat at the expense of the degree of cooperativity realized.
The double mutants demonstrate that there is interaction among the residues of the two adjacent polypeptide segments that join the substrate binding domain to the ACT domain, with the major effect being on the stabilization of ligand binding. The largest interaction effect is seen for the Km, followed by the I 0.5 for serine and the least is seen for kcat. For Km, the effect between adjacent polypeptide strands is anti-cooperative while that within the same strand is cooperative. Thus, the interaction between strands is actually slightly detrimental to the Km. For kcat, on the other hand, the effect between polypeptides is cooperative while the effect on the same polypeptide is antagonistic in this case. Thus, interaction between the strands favors kcat. For the I0.5 of serine, the effect is anti-cooperative for both situations with the interaction between Ser-111 and Ser-107 essentially negligible. The effect on the Hill coefficient is antagonistic in all cases since restoring the double mutant antagonistically reverses the negative effect of a single mutant restoration. According to Mildvan (14), anti-cooperative effects may be viewed as strain between the two residues in their function regarding a particular parameter and antagonism may result from opposing structural effects. Antagonism is very much like cooperativity since the effect of restoring both mutants is greater than the sum of the individual restorations. In the case of cooperative residues, each single mutant abolishes an intrinsic non-cooperative effect as well as the same cooperative contribution.
The effect of mutations at the serine-binding site is mainly on serine binding and cooperativity. It is not surprising that mutations to His-344, Asn-346, and Asn-364 cause a large decrease in serine sensitivity since these are the residues that hydrogen bond directly to L-serine. G349V affects both serine affinity as well as cooperativity. It may be that the presence of a valine side chain at this position prevents the binding site from adopting an optimal conformation. However, the binding site of R347A has near normal affinity for serine and also a significant decrease in cooperativity. Thus, it can be concluded that the side chain of Arg-347 plays a critical role in the initial transfer of the cooperativity of inhibition. The observation that the hydrogen bonds between Arg-347, Pro-401, and Pro-348 occur in only one of the effector sites at each regulatory domain interface, may also contribute directly to the negative cooperativity observed for the binding of the second serine at the same regulatory domain interface if one structure is more stable than the other with respect to serine binding.
Mutation of the other residues in the regulatory domain primarily affect serine sensitivity and cooperativity of inhibition. Interestingly, the mutations that primarily affect cooperativity are located in loops that connect elements of secondary structure that border the regulatory domain interface. Unfortunately, the existing crystal structures don’t provide any obvious clues as to what the specific interaction of these side chains are during the transformation from active to inhibited enzyme.
Mutations of the tryptophan loop residues show a significant effect on both the cooperativity of inhibition and the kcat/Km but not on serine sensitivity. This is consistent with the interpretation that the tryptophan loop, which interacts with the active site of the adjacent subunit, is functional in both optimal substrate interaction and in transferring the cooperative effect of inhibition across subunits. A very interesting observation here is that removal of the Lys-141 side chain introduces a homotropic cooperative effect. Thus, removal of this side chain creates a new enzyme state that undergoes a conformational change upon binding of the substrate, a conformational shift that apparently does not take place in the native enzyme.
The site specific mutations that have been undertaken in this investigation have identified new residues in the regulatory domain and near the active site that exert significant effect on cooperativity of inhibition by L-serine. These residues represent some of the structural entities that function in the initial and final stages of the transmission of the allosteric effect from the effector site to the active site. The crystal structures of E. coli PGDH have demonstrated that serine binding ultimately produces a 14° rotational transformation at the active site hinge. This rotation is initiated by serine binding and is propagated initially through conformational change in the regulatory domain. The rotational event is likely responsible for the reduction of catalytic velocity in this V-type enzyme by rearrangement of the active site residues. The cooperativity of inhibition comes about because the velocity effect propagated from a single effector-binding event affects both functional active sites but the effect is not equal at both active sites.
Elucidation of how the ACT (regulatory) domain functions in the regulation of enzyme activity involves not only an understanding of how it inhibits the active site within its own subunit, but also how inhibition is propagated across subunits. This investigation has identified some key residues in this process that will advance our efforts in addressing these questions as well as the larger more general question of whether or not ACT domains in other enzymes function in a similar way.
Abbreviations
- PGDH
D-3-Phosphoglycerate Dehydrogenase
- I 0.5
concentration that produces 50% inhibition
Footnotes
Supported by grant # GM 56676 (G. A. G.) from the National Institutes of Health.
References
- 1.Schuller DJ, Grant GA, Banaszak LJ. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nature Struct Biol. 1995;2:69–75. doi: 10.1038/nsb0195-69. [DOI] [PubMed] [Google Scholar]
- 2.Grant GA, Hu Z, Xu XL. Amino acid residue mutations uncouple cooperative effects in Escherichia coli D-3-phosphoglycerate dehydrogenase. J Biol Chem. 2001;276:17844–17850. doi: 10.1074/jbc.M009957200. [DOI] [PubMed] [Google Scholar]
- 3.Bell JK, Grant GA, Banaszak LJ. Multiconformational states in phosphoglycerate dehydrogenase. Biochemistry. 2004;43:3450–3458. doi: 10.1021/bi035462e. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 5.Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol. 2001;11:694. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- 6.Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, Drennan CL. Crystal structure of the nickel-responsive transcription factor NikR. Nature Structural Biology. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 7.Devedjiev Y, Surendranath Y, Derewenda U, Gabrys A, Cooper DR, Zhang R-g, Lezondra L, Joachimiak A, Derewenda ZS. The structure and ligand binding properties of the B. subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J Mol Biol. 2004;343:395–406. doi: 10.1016/j.jmb.2004.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho Y, Sharma V, Sacchettini JC. Crystal structure of ATP phosphoribosyltransferase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:8333–8339. doi: 10.1074/jbc.M212124200. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher DT, Gilliland GL, Xiao G, Zondlo J, Fisher KE, Chinchilla D, Eisenstein E. Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure. 1998;6:465–475. doi: 10.1016/s0969-2126(98)00048-3. [DOI] [PubMed] [Google Scholar]
- 10.Grant GA, Hu Z, Xu XL. Hybrid tetramers reveal elements of cooperativity in Escherichia coli D-3-phosphoglycerate dehydrogenase. J Biol Chem. 2003;278:18170–18176. doi: 10.1074/jbc.M213050200. [DOI] [PubMed] [Google Scholar]
- 11.Grant GA, Xu XL, Hu Z. Quantitative relationships of site to site interaction in Escherichia coli D-3-phosphoglycerate dehydrogenase revealed by asymmetric hybrid tetramers. J Biol Chem. 2004;279:13452–13460. doi: 10.1074/jbc.M313593200. [DOI] [PubMed] [Google Scholar]
- 12.Thompson JR, Bell JK, Bratt J, Grant GA, Banaszak LJ. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry. 2005;44:5763–5773. doi: 10.1021/bi047944b. [DOI] [PubMed] [Google Scholar]
- 13.Al-Rabiee R, Lee EJ, Grant GA. The mechanism of velocity modulated allosteric regulation in D-3-phosphoglycerate dehydrogenase. Cross-linking adjacent regulatory domains with engineered disulfides mimics effector binding. J Biol Chem. 1996;271:13013–13017. doi: 10.1074/jbc.271.22.13013. [DOI] [PubMed] [Google Scholar]
- 14.Mildvan AS. Inverse Thinking about Double Mutant Enzymes. Biochemistry. 2004;43:14517–14520. doi: 10.1021/bi048052e. [DOI] [PubMed] [Google Scholar]
- 15.Grant GA, Xu XL, Hu Z. Removal of the tryptophan 139 side chain in Escherichia coli D-3-phosphoglycerate dehydrogenase produces a dimeric enzyme without cooperative effects. Archives Biochem Biophys. 2000;375:171–174. doi: 10.1006/abbi.1999.1652. [DOI] [PubMed] [Google Scholar]
- 16.Grant GA, Xu XL, Hu Z. Role of an interdomain Gly-Gly sequence at the regulatory-substrate domain interface in the regulation of Escherichia coli. D-3-phosphoglycerate dehydrogenase. Biochemistry. 2000;39:7316–7319. doi: 10.1021/bi000218y. [DOI] [PubMed] [Google Scholar]