Abstract
Polypeptides targeted to the yeast endoplasmic reticulum (ER) posttranslationally are thought to be kept in the cytoplasm in an unfolded state by Hsp70 chaperones before translocation. We show here that Escherichia coli β-lactamase associated with Hsp70, but adopted a native-like conformation before translocation in living Saccharomyces cerevisiae cells. β-Lactamase is a globular trypsin-resistant molecule in authentic form. For these studies, it was linked to the C terminus of a yeast polypeptide Hsp150Δ, which conferred posttranslational translocation and provided sites for O-glycosylation. We devised conditions to retard translocation of Hsp150Δ-β-lactamase. This enabled us to show by protease protection assays that an unglycosylated precursor was associated with the cytoplasmic surface of isolated microsomes, whereas a glycosylated form resided inside the vesicles. Both proteins were trypsin resistant and had similar β-lactamase activity and Km values for nitrocefin. The enzymatically active cytoplasmic intermediate could be chased into the ER, followed by secretion of the activity to the medium. Productive folding in the cytoplasm occurred in the absence of disulfide formation, whereas in the ER lumen, proper folding required oxidation of the sulfhydryls. This suggests that the polypeptide was refolded in the ER and consequently, at least partially unfolded for translocation.
INTRODUCTION
Translocation of newly synthesized precursor proteins into the yeast endoplasmic reticulum (ER) occurs cotranslationally or posttranslationally, depending on the hydrophobicity of the signal peptides (Brodsky and Schekman, 1994; Ng et al., 1996; Rapoport et al., 1996). In the cotranslational pathway, the signal recognition particle binds to the signal peptide emerging from the ribosome, translation halts, and the nascent chain–ribosome complex is targeted to the trimeric Sec61 translocon complex (Sec61p, Sbh1p, and Sss1p) embedded in the ER membrane (Panzner et al., 1995). The polypeptide traverses the aqueous translocon channel simultaneously with elongation, apparently in an extended form, whereafter it adopts its native structure in the ER lumen. For instance, in cotranslational translocation of the Escherichia coli Lep protein into canine microsomes, 65 amino acids bridge the ribosomal P site and the luminal surface of the ER membrane (Whitley et al., 1996). In contrast, posttranslational translocation is signal recognition particle independent. Translation of the polypeptide is completed on free ribosomes, whereafter the preprotein traverses the ER membrane via the translocon complex associated with the Sec62–63 complex (Sec62p, Sec63p, Sec71p, and Sec72p; Deshaies and Schekman, 1989; Rothblatt et al., 1989; Deshaies et al., 1991; Brodsky and Schekman, 1993; Feldheim and Schekman, 1994; Panzner et al., 1995). Because the amino acid sequence primarily dictates the three-dimensional structure of proteins, polypeptides could fold in the cytoplasm unless they were prevented from folding. Depletion of two of the four predominant Hsp70 homologues of the yeast cytosol, Ssa1 and Ssa2, prevented translocation of pre-pro-α factor, suggesting that the Hsp70s prevent precursor proteins from folding and keep them in a translocation-competent form (Chirico et al., 1988; Deshaies et al., 1988).
We wanted to study whether secretory proteins in living yeast cells could fold to stable conformations in the cytoplasm before posttranslational translocation into the ER. To this end, E. coli TEM1 β-lactamase was chosen as a marker, because it is trypsin resistant in authentic form and has a globular crystal structure (Jelsch et al., 1992). In E. coli, β-lactamase is translocated posttranslationally by an N-terminal signal peptide across the cytoplasmic membrane into the periplasm, where it acquires one disulfide bond (Koshland and Botstein, 1982). We fused β-lactamase to the C terminus of an N-terminal fragment (Hsp150Δ) of the natural secretory yeast protein Hsp150 (see Figure 3A). The Hsp150 signal peptide was anticipated, according to its hydrophobicity, to confer posttranslational translocation. The rest of the Hsp150Δ polypeptide provided O-glycosylation sites that enabled clear distinction of the unglycosylated and glycosylated molecules. Moreover, the Hsp150Δ polypeptide does not adopt any regular secondary structure (Jämsäet al., 1995a), allowing the β-lactamase portion to fold to an enzymatically active conformation in the yeast ER (Simonen et al., 1994). Here we show that the β-lactamase portion of Hsp150Δ-β-lactamase folded in the cytoplasm of living yeast cells to a stable, enzymatically active conformation before translocation into the ER.
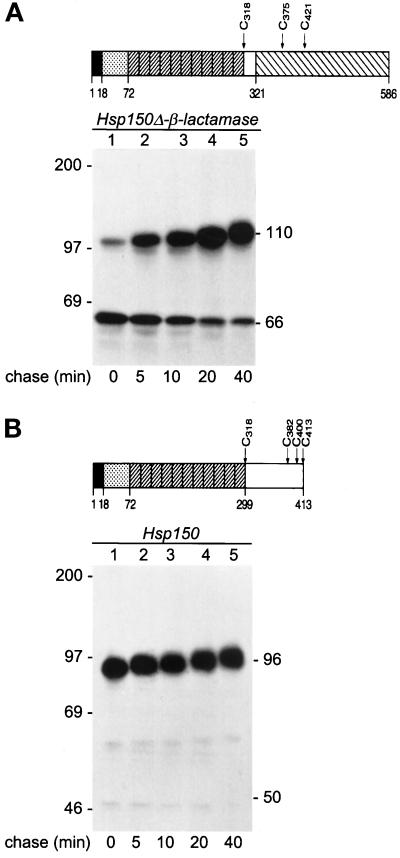
Figure 3.
Kinetics of translocation. Schematic presentations of Hsp150Δ-β-lactamase (A) and Hsp150 (B). Amino acids 1–18, Hsp150 signal peptide. Amino acids 19–72, subunit I of Hsp70. Diagonally striped boxes, 11 tandem repeats of a homologous peptide of subunit II of Hsp70. Amino acids 300–413, unique C-terminal domain of subunit II. Amino acids 322–586, mature β-lactamase. The cysteine residues (C) are indicated. The Hsp150 fragment of amino acids 19–321 contains 96 potential O-glycosylation sites, many of which obtain glycans consisting of 2–5 mannose residues (Jämsäet al., 1995a). H393 cells (A, sec18–1 cells with HSP150Δ-β-lactamase) and H4 cells (B, sec18–1 without the recombinant gene) were preincubated under high cell density conditions at 37°C for 10 min and 35S labeled for 5 min (lane 1), followed by chase in the presence of CHX for the indicated times (lanes 2–5). The lysed cell samples were immunoprecipitated with anti-β-lactamase (A) or anti-Hsp150 (B) antiserum and analyzed by SDS-PAGE and fluorography. The decreasing electrophoretic migration was apparently due to increasing O-glycosylation. Hsp150Δ-β-lactamase (A) and Hsp150 (B) molecules are indicated on the right (kDa), and molecular weight markers on the left.
MATERIALS AND METHODS
Yeast Strains and Media
The following yeast strains were grown at 24°C in shake flasks overnight to early logarithmic phase in YPD medium for β-lactamase activity measurements and for [3H]mannose labeling experiments, and in synthetic complete medium lacking methionine and cysteine for 35S-labeling experiments: sec18–1 (H393, Matα sec18–1 ura3–52 trp1–289 leu2–3,112 URA3::HSP150Δ-β-lactamase; Simonen et al., 1994); H4 (isogenic with H393 but lacks HSP150Δ-β-lactamase); wild type (WT) (H335, Mata ade2–101 ura3–52 leu2–3, 112 suc2–9 gal2 URA3::HSP150Δ-β-lactamase; Simonen et al., 1994); sec63–1 (H482, Matα sec63–1 ura3–52 leu2–3, 112 URA3::HSP150Δ-β-lactamase); sec65–1 (H664, Matα sec65–1 his3 ura3–52 ade2 trp1–1 leu2–3, 112 URA3::HSP150Δ-β-lactamase); sec62–101 (H694, Matα sec62–101 ura3Δ99 ade2–101ochre trpΔ99 leu2Δ1 LEU2::HSP150Δ-β-lactamase); and sec63–201 (H695, Matα sec63–201 ura3Δ99 ade2–101ochre trpΔ99 leu2Δ1 LEU2::HSP150Δ-β-lactamase). Strains H482, H664, H778, and H779 were created by integrating the HSP150Δ-β-lactamase gene in plasmid pKTH4544 (Simonen et al., 1994) into the URA3 locus of the parental strains. Strains H694 and H695 were created by integrating the HSP150Δ-β-lactamase gene in plasmid pKTH4660 into the LEU2 locus of the parental strains H681 and H682. pKTH4660 was constructed by isolating the Hsp150Δ-β-lactamase fragment from pKTH4544 with BamHI, and transferring it to pFL26 (Bonneaud et al., 1991). pKTH4660 was linearized with AlfII before integration into the parental strains H681 and H682.
Metabolic Labeling and Immunoprecipitation
Metabolic labeling of cells was performed with 20 μCi of [35S]methionine/cysteine (1000 Ci/mmol, Amersham, United Kingdom) per ml of synthetic complete medium lacking methionine and cysteine, or after a 15-min preincubation in low-glucose YPD (0.1%), with 100 μCi of 2-[3H]mannose (11.5 Ci/mmol, Amersham) per ml of low-glucose YPD medium. In pulse-label experiments, chase was performed by adding cycloheximide (CHX) to the final concentration of 100 μg/ml. The incubations were terminated by adding NaN3 (Sigma Chemical, St. Louis, MO) to the final concentration of 10 mM, and the culture medium and cell lysate samples were immunoprecipitated as described (Simonen et al., 1994). Briefly, cells were lysed mechanically with glass beads in NET buffer (0.05 M Tris-HCl, pH 8.0, containing 0.4 M NaCl, 5 mM EDTA, 1% Nonidet P-40, and 100 U/ml of aprotinin) in the presence of 2 mM phenylmethylsulfonyl fluoride (Sigma) and 2% SDS. The lysates were boiled and precleared for 1 h at 4°C with protein A-Sepharose (Pharmacia, Piscataway, NJ). Immunoprecipitation was performed with anti-β-lactamase antiserum (1:100) or anti-Hsp150 antiserum (1:100) and protein A-Sepharose for 2 h at 4°C. After washing with diluted NET buffer (1:1), wash buffer (0.1 M Tris-HCl, pH 7.5, containing 0.2 M NaCl, 2 M urea, and 0.5% Tween 20), and with 0.1% SDS, the precipitates were analyzed by SDS-PAGE.
Isolation of Microsomes
The isolation was performed essentially as described by Sanderson and Meyer (1994). Briefly, cells harvested by centrifugation were suspended at 0.2 mg/ml of 10 mM Tris-HCl (pH 7.5) containing 10 mM CaCl2, 1.2 M sorbitol, 10 mM dithiothreitol (DTT), and 10 mM NaN3. Zymolyase Z 100-T (Seikagaku Kogyo, Rockville, MD) was added to a final concentration of 15 U/ml, and the suspension was incubated for 2 h at 37°C. The spheroplasts were underlayed with 10 ml of 0.8 M sucrose containing 1.5 g of Ficoll/100 ml of 20 mM HEPES (pH 7.6). The spheroplasts were pelleted by centrifugation at 5000 rpm for 15 min at 4°C (Beckman JS 13.1) and were resuspended at 0.5 g/ml in lysis buffer (20 mM HEPES, pH 7.6, containing 1 mM DTT, 2 mM EDTA, and 50 mM potassium acetate, pH 7.5), and homogenized in a Dounce device using 15–20 strokes. The homogenate was transferred to a Corex tube that held an equal volume of a solution containing 0.5 M sucrose, 50 mM potassium acetate (pH 7.5), 2 mM EDTA, and 1 mM DTT; 20 mM HEPES (pH 7.6) was added. The homogenate was underlayed with the same buffer but containing 1 M sucrose, and centrifuged at 8000 rpm for 15 min at 4°C (Beckman JS 13.1). The supernatant was removed and centrifuged in a Beckman’s ultraclear tube at 14,600 rpm for 15 min at 4°C (SW 51.1). The pellet was resuspended in membrane buffer containing 0.25 M sucrose, 50 mM potassium acetate (pH 7.5), 1 mM DTT, and 20 mM HEPES (pH 7.6), frozen in liquid nitrogen, and stored at −70°C until it was used.
Proteolytic Digestions
Isolated unlabeled microsomes were digested with trypsin (Sigma; 25 μg/ml) for 30 min at 10°C, followed by addition of phenylmethylsulfonyl fluoride to the final concentration of 1 mM and heating in a boiling water bath for 5 min. Proteinase K (Merck, St. Louis, MO; 16 μg/ml) digestion occurred for 90 min at 0°C. When Triton X-100 was present, it was used at a concentration of 0.5%. 35S-labeled cells were lysed mechanically as above, but with 2% Triton X-100 in the absence of SDS and heating. After immunoprecipitation of 35S-Hsp150Δ-β-lactamase, the precipitates were washed twice with NET buffer and twice with 1% NH4HCO3, and resuspended in 1% NH4HCO3 for trypsin and proteinase K digestion, which were performed as above except that 25 μg/ml proteinase K was used.
Coimmunoprecipitation and Western Blot Analysis
Cells were lysed with glass beads in the presence of protease inhibitors in the same manner as for immunoprecipitation, but in the presence of Triton X-100 and absence of SDS and boiling. Apyrase (Sigma) was added to the final concentration of 30 U/ml. The lysates were precleared with 5% protein A-Sepharose in NET buffer containing 10 mM CaCl2 and lacking EDTA, and incubated for 2 h at 4°C with anti-β-lactamase antiserum (1:100), or with preimmune serum (1:100) at 4°C, and 5% protein A-Sepharose in NET buffer containing 10 mM CaCl2 and lacking EDTA. The precipitates were washed twice with the same NET buffer and twice with the same buffer diluted 1:1 and subjected to SDS-PAGE. Proteins were blotted from the gel onto nitrocellulose membrane (Hybond-C Extra, Amersham) and immunostained with anti-β-lactamase antiserum (1:6000) and alkaline phosphatase-conjugated anti-rabbit antibody (1:7500, Promega, Madison, WI), or with monoclonal anti-Hsp70 antibody (1:1000, Stressgen, Victoria, British Columbia, Canada) and alkaline phosphatase-conjugated anti-mouse antibody (1:3000, Promega).
β-Lactamase Activity and Km Values
For determination of β-lactamase activity, duplicate cell samples (5 × 107/ml of YPD medium) were incubated for the indicated times. After addition of NaN3, the cells were separated from the culture medium and lysed under mild detergent conditions, followed by determination of the β-lactamase activity of the lysates and the media samples using nitrocefin as a substrate as described (Simonen et al., 1994). For determination of the Km values (Lineweaver-Burke plot), yeast cells were incubated for 1.5 h at 37°C. The strains used were H482 (sec63–1) for cytoplasmic activity, H393 (sec18–1) for ER-located activity, and H335 (WT) for secreted activity.
Other Methods and Materials
Hydrophobicity plots were constructed according to Kyte and Doolittle (1982). SDS-polyacrylamide gel was used in reducing 8% gels, and DTT (Sigma) was used in the final concentration of 20 mM, if not otherwise stated.
RESULTS
Posttranslational Translocation of Hsp150Δ-β-lactamase
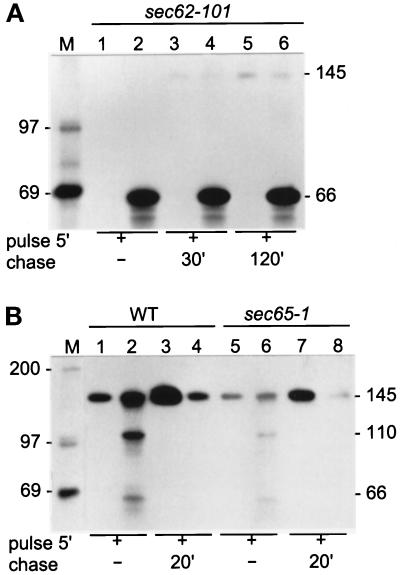
First we established the mode of translocation of Hsp150Δ-β-lactamase, taking advantage of the sec62–101 mutant in which posttranslational translocation is constitutively defective (see Table 1 for key for mutant yeast strains). Strain sec62–101 was labeled with [35S]methionine/cysteine for 5 min (Figure 1A, lanes 1 and 2). A parallel cell sample was chased for 30 min (lanes 3 and 4) or 120 min (lanes 5 and 6) in the presence of CHX. The culture media samples (lanes 1, 3, and 5) and lysed cell samples (lanes 2, 4, and 6) were subjected to immunoprecipitation with anti-β-lactamase antiserum followed by SDS-PAGE analysis. No protein could be detected in the culture medium before (lane 1) or after (lanes 3 and 5) chase, whereas a protein of 66 kDa was found in all cell lysates (lanes 2, 4, and 6). The same results were obtained for the sec63–201 mutant also defective in posttranslational translocation (Table 1; our unpublished results). We have shown before that the ER form of Hsp150Δ-β-lactamase migrates in SDS-PAGE in the same manner as a 110-kDa protein, and is primary O-glycosylated with single mannose residues at multiple serine and threonine residues on the HspΔ150 fragment. Thus, the 66-kDa form must have been the unglycosylated precursor. During passage through the Golgi, the O-glycans are extended up to pentamannosides and a 53-amino acid N-terminal propeptide (subunit I) is released at a Kex2p recognition site, yielding the mature protein of 145 kDa (Simonen et al., 1994; Jämsäet al., 1995a; see top of Figure 3A). A wild-type (WT) strain was labeled and chased for reference. After a pulse of 5 min, some mature Hsp150Δ-β-lactamase (145 kDa) was detected in the culture medium (Figure 1B, lane 1). The cell lysate contained mature protein, some ER form (110 kDa), and very little of the 66-kDa form (lane 2). After chase, all of the 66-kDa and 110-kDa forms and most of the 145-kDa form had disappeared from the cells (lane 4), and the mature protein was found in the medium (lane 3), demonstrating efficient secretion. Similar experiments were performed using the signal recognition particle mutant sec65–1 blocking specifically cotranslational translocation at 37°C (Table 1). Under restrictive conditions, Hsp150Δ-β-lactamase was secreted in this strain as it was in the WT strain (lanes 5–8), though the expression level was somewhat lower. The hydrophobicity index of the Hsp150 signal peptide was less than 2, as in the case of procarboxypeptidase Y, which is translocated posttranslationally (Ng et al., 1996). We conclude that Hsp150Δ-β-lactamase used the posttranslational pathway to reach the ER lumen.
Table 1.
Defects of the mutant yeast strains
| Mutant | Defect | Reference |
|---|---|---|
| sec62-101 | Posttranslational ER translocation constitutive | Ng et al. (1996) |
| sec63-201 | Posttranslational ER translocation constitutive | Ng et al. (1996) |
| sec63-1 | Posttranslational ER translocation conditional | Rothblatt et al. (1989) |
| sec65-1 | Cotranslational ER translocation conditional | Stirling et al. (1992) |
| sec18-1 | Membrane traffic from ER to Golgi conditional | Kaiser and Schekman (1990) |
Figure 1.
Secretion of Hsp150Δ-β-lactamase in translocation-defective mutants. Sec62–101 (A, lanes 1–6), WT (B, lanes 1–4), and sec65–1 (B, lanes 5–8) cells (5 × 108/ml) were preincubated for 15 min and labeled with [35S]methionine/cysteine for 5 min. Parallel similarly pulse-labeled cells received CHX and were chased for the indicated times. In A all incubations were performed at 30°C and in B at 37°C. The growth media (lanes with uneven numbers) were separated from the cells (lanes with even numbers), which were lysed. All samples were subjected to immunoprecipitation with anti-β-lactamase antiserum followed by SDS-PAGE analysis. The Hsp150Δ-β-lactamase proteins (kDa) are indicated on the right, and the molecular weight markers (lane M; kDa) on the left.
Retardation of Translocation In Vivo
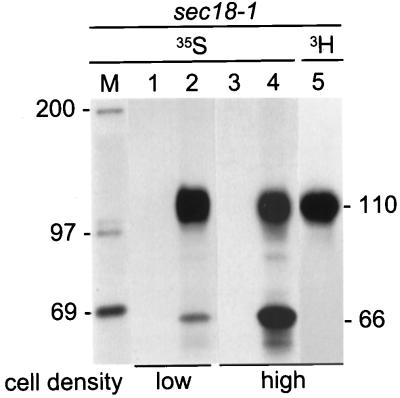
We wanted to find conditions in which translocation of Hsp150Δ-β-lactamase would not be inhibited but would be retarded, to confirm simultaneously the topologies of the 66-kDa and 110-kDa forms respective to the ER membrane in cells that were WT for translocation. For these experiments, Hsp150Δ-β-lactamase was expressed in an sec18–1 mutant, in which ER-derived secretory vesicles are unable to fuse with the Golgi at 37°C, preventing escape of secretory proteins from the pre-Golgi compartment (Table 1). Sec18–1 cells were 35S-labeled at 37°C at two cell densities, 5 × 107 cells/ml as above, and 5 × 108 cells/ml. The cell lysates and culture media were immunoprecipitated, followed by SDS-PAGE analysis. The low cell density lysate contained primarily the 110-kDa species, indicating efficient translocation (Figure 2, lane 2). In the high-density lysate, both the 66- and 110-kDa forms could be detected (Figure 2, lane 4), and only the 110-kDa form could be labeled with [3H]mannose (lane 5). No protein was found in the culture media samples (Figure 2, lanes 1 and 3).
Figure 2.
Retardation of translocation of Hsp150Δ-β-lactamase. (A) Sec18–1 cells were preincubated for 15 min at 37°C and labeled for 1 h at the same temperature with [35S]methionine/cysteine under low cell density conditions (5 × 107/ml; lanes 1 and 2) or high cell density conditions (5 × 108/ml; lanes 3 and 4). One sample was labeled with [3H]mannose under high cell density conditions (lane 5). The media (lanes 1 and 3) were separated from the cells, which were lysed (lanes 2, 4, and 5). The samples were immunoprecipitated with anti-β-lactamase antiserum and analyzed by SDS-PAGE. The Hsp150Δ-β-lactamase proteins (kDa) are indicated on the right and the molecular weight markers (lane M; kDa) on the left. Lanes 3 and 4 were exposed four times longer than were lanes 1 and 2.
Translocation Kinetics
To study whether cytoplasmic Hsp150Δ-β-lactamase could be chased into the ER, sec18–1 cells were 35S-labeled for 5 min at 37°C (Figure 3A, lane 1). After addition of CHX, most of the protein was converted to the ER form of 110 kDa in about 20 min (Figure 3, lanes 2–4), with some of the 66-kDa form still persisting after a 40-min chase (lane 5). Authentic Hsp150 (schemes in Figure 3B) was translocated much faster, because after the 5-min pulse (Figure 3B, lanes 1–5), and even after a 1-min pulse (our unpublished results), it occurred exclusively in the ER-specific primary O-glycosylated form of 93–97 kDa. The cytoplasmic Hsp150 precursor accumulated in sec63–201 migrated in SDS-PAGE in the same manner as a 50-kDa protein (our unpublished results).
Topology and Trypsin Resistance of Hsp150Δ-β-lactamase Precursors
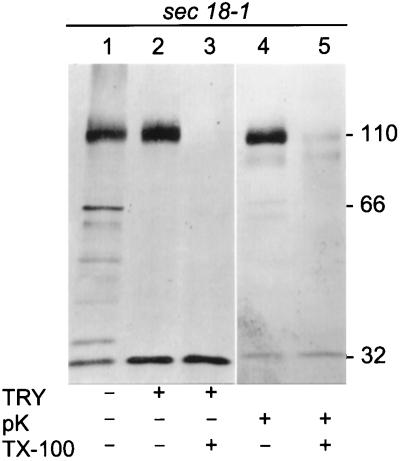
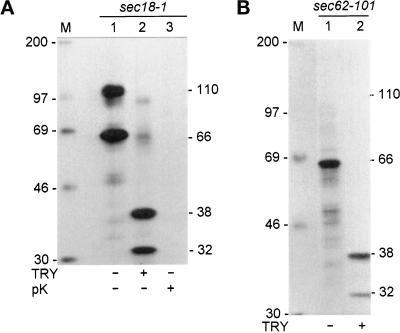
Sec18–1 cells were incubated at 37°C under high cell density conditions to retard translocation of Hsp150Δ-β-lactamase, followed by mechanical cell lysis and isolation of microsomes. SDS-PAGE and Western blot analysis using anti-β-lactamase antiserum revealed both the 66-kDa and the 110-kDa forms in the microsomal preparation, plus putative degradation products with higher electrophoretic mobility, among them a 32-kDa protein (Figure 4, lane 1). Authentic mature β-lactamase migrates in SDS-PAGE like a 29-kDa protein and is resistant to trypsin and sensitive to proteinase K (Minsky et al., 1986). The Hsp150Δ fragment is likely to be trypsin sensitive, because it contains multiple recognition sites and adopts no regular secondary structure (Jämsäet al., 1995a). When the microsomal preparation was subjected to trypsin digestion before SDS-PAGE, the 110-kDa form remained intact, whereas the 66-kDa form disappeared concomitantly with an increase of the 32-kDa band material (Figure 4, lane 2). When Triton X-100 was added to solubilize the membranes, trypsin destroyed both the 66-kDa and the 110-kDa forms (Figure 4, lane 3). When the intact microsomal preparation was subjected to proteinase K digestion, the 110-kDa form persisted, but the 66-kDa form disappeared (Figure 4, lane 4). In the presence of Triton X-100, both forms disappeared (Figure 4, lane 5). This shows that the 66-kDa form was exposed on the cytoplasmic aspect of microsomal membranes, whereas the 110-kDa form resided inside the microsomes. The data also suggest that the β-lactamase portion was resistant to trypsin. However, the occurrence of the immunoreactive 32-kDa band even in the untreated microsomes (Figure 4, lane 1) weakened this conclusion. Thus, we repeated the trypsin digestion experiments using metabolically labeled whole cell lysates.
Figure 4.
Trypsin sensitivity of microsome-associated Hsp150Δ-β-lactamase. Sec18–1 cells were incubated for 2 h at 37°C under high cell density conditions, followed by isolation of microsomes. The microsomal preparation was divided in equal portions, which received trypsin (TRY), proteinase K (pK), and Triton X-100 (TX-100), as indicated. The digests were subjected to SDS-PAGE in 7.5–15% gels and Western blotting using anti-β-lactamase antiserum. The β-lactamase-related proteins (kDa) are indicated.
Sec18–1 cells were labeled with [35S]methionine/cysteine under high cell density conditions at 37°C to retard translocation, lysed under nondenaturing conditions in mild detergent to disrupt the membranes, and subjected to immunoprecipitation. As before, both the 110-kDa and the 66-kDa form could be detected in SDS-PAGE analysis after the labeling (Figure 5A, lane 1). When the immunoprecipitate was digested with trypsin, SDS-PAGE analysis revealed a 38-kDa and a 32-kDa band, with concomitant disappearance of the 66-kDa and 110-kDa forms (lane 2). The 35S label was mostly in the β-lactamase portion since it contains nine methionines and two cysteines and the Hsp150Δ fragment has only one cysteine. Proteinase K digestion destroyed the fusion protein completely (Figure 5A, lane 3). To confirm that the β-lactamase portion of the 66-kDa form was trypsin resistant, sec62–101 cells were 35S labeled to accumulate the fusion protein in the cytoplasm (Figure 5B, lane 1). Trypsin digestion of the 66-kDa form yielded 32-kDa and 38-kDa products (Figure 5B, lane 2), as with the mixture of the 66-kDa and 110-kDa forms. The difference between the 32-kDa and 38-kDa products was not studied. Perhaps the most C-terminal part of the Hsp150Δ portion was protected in part of the molecules by cytoplasmic components, which were present in the whole lysate, but absent from isolated microsomes. We conclude that the β-lactamase portions of both cytoplasmic and ER-confined Hsp150Δ-β-lactamase were resistant to trypsin.
Figure 5.
Trypsin sensitivity of 35S-Hsp150Δ-β-lactamase. (A) Sec18–1 cells (5 × 108/ml) were preincubated for 15 min at 37°C and 35S labeled for 30 min. (B) Sec62–101 cells (5 × 108/ml) were preincubated for 15 min at 30°C and labeled for 5 min. The cells were lysed under nondenaturing conditions and immunoprecipitated with anti-β-lactamase antiserum. The precipitates were divided in equal portions and digested with trypsin or proteinase K, as indicated, and analyzed by SDS-PAGE. The β-lactamase-related proteins (kDa) are indicated on the right and the molecular weight markers (kDa) on the left.
Productive Folding of Hsp150Δ-β-lactamase in the Cytoplasm and in the ER
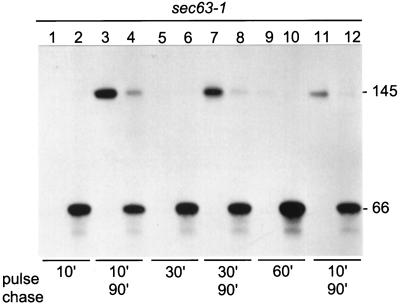
To learn whether the cytoplasmic, trypsin-resistant β-lactamase portion was enzymatically active, Hsp150Δ-β-lactamase was expressed in a sec63–1 mutant, where translocation is defective at 37°C (Toyn et al., 1988; Table 1). First we characterized how the sec63–1 mutation affected our reporter protein. To this end, sec63–1 cells were 35S labeled at 37°C for 10 min. Immunoprecipitation and SDS-PAGE analysis revealed the 66-kDa form in the cell lysate (Figure 6, lane 2) and no protein in the medium (lane 1). When similarly pulse-labeled cells were chased in the presence of CHX at 24°C for 90 min, about half of Hsp150Δ-β-lactamase could be detected in the medium in the mature form of 145 kDa (Figure 6, lane 3), with the other half remaining in the cells mostly as the 66-kDa form (lane 4). When the labeling period was extended to 30 min (Figure 6, lanes 5 and 6), even less of the reporter was secreted during chase (lanes 7 and 8), and after 60 min of labeling (lanes 9 and 10) very little of it resumed secretion (lanes 11 and 12). Thus, the sec63–1 translocation block was efficient for our reporter protein, but only partially reversible. The longer the cells spent under nonpermissive conditions, the less efficient was the reversal of the block.
Figure 6.
Translocation of Hsp150Δ-β-lactamase in sec63–1 cells. Sec63–1 cells (5 × 107/ml) were preincubated at 37°C for 15 min and 35S labeled for 10, 30, or 60 min as indicated (lanes 1, 2, 5, 6, 9, and 10). Parallel samples were chased with CHX for 90 min (lanes 3, 4, 7, 8, 11, and 12). The media were separated from the cells, which were lysed. The media and lysate samples were immunoprecipitated and subjected to SDS-PAGE analysis. The Hsp150Δ-β-lactamase proteins are indicated on the right (kDa).
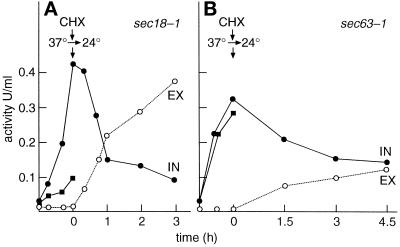
To study folding in the cytoplasm, we incubated first sec18–1 cells at 37°C for reference. The intracellular activity increased to >0.4 U/ml in 1 h (Figure 7A, closed circles), whereas no activity could be detected in the medium (open circles). The activity was secreted to the medium once the cells were shifted to 24°C in the presence of CHX. When the sec63–1 mutant was incubated similarly at 37°C to block the reporter protein in the cytoplasm, 0.33 U/ml β-lactamase activity accumulated inside the cells (Figure 7B, closed circles) and nothing was found in the culture medium (open circles). Thus, the reporter acquired biological activity even in the cytoplasm, although the disulfide most probably was not formed. To confirm the lack of disulfide formation in the cytoplasm, the incubations at 37°C were repeated in the presence of the reducing agent DTT, which diffuses across membranes and prevents disulfide formation (Jämsäet al., 1994). In the ER, DTT prevented productive folding of the reporter protein in sec18–1 cells (Figure 7A, closed squares). In contrast, DTT had no effect on the folding in the cytoplasm (Figure 7B, closed squares). The expression level or stability of the reporter protein in the two mutants was not affected by DTT (our unpublished results). These data show that β-lactamase acquired not only a trypsin-resistant conformation but also a biologically active one in the yeast cytoplasm.
Figure 7.
Translocation of β-lactamase activity. Sec18–1 (A) and Sec63–1 (B) cells were incubated at 37°C, followed by addition of CHX and shift of the cells to 24°C. Intracellular (IN, filled circles) and secreted (EX, open circles) β-lactamase activity was determined. Parallel samples received DTT for the 37°C incubation. The intracellular activity (filled squares) was determined. No secreted activity was found in the presence of DTT.
Next, we verified that properly folded biologically active β-lactamase molecules, and not incompletely folded inactive copies, were translocated from the cytoplasm into the ER. To this end sec63–1 cells, first incubated at 37°C to retain β-lactamase activity in the cytoplasm, were chased at 24°C with CHX. The intracellular activity declined slowly (Figure 7B, closed circles), concomitantly with an increase of the activity in the medium (open circles). Though the reversal of the sec63–1 block was incomplete, these results show that molecules that were catalytically active in the cytoplasm were translocated into the ER and secreted to the medium in active form. Dependence of proper folding in the ER on disulfide formation suggests a refolding step after translocation.
Finally, we determined the kinetic parameter Km of different Hsp150Δ-β-lactamase forms using nitrocefin as substrate. The Km values of the cytoplasmic and ER-confined Hsp150Δ-β-lactamase precursors, and the mature protein harvested from the culture medium, were similar to that of authentic β-lactamase from E. coli (Table 2).
Table 2.
Km values for nitrocefin of β-lactamase variants
| Protein | Km (μM) |
|---|---|
| Cytoplasmic Hsp150Δ-β-lactamase | 43 |
| ER-located Hsp150Δ-β-lactamase | 55 |
| Secreted Hsp150Δ-β-lactamase | 47 |
| E. coli β-lactamase | |
| This study | 49 |
| Lamineth and Plückthun (1989) | 49 |
See MATERIALS AND METHODS for production of β-lactamase variants.
Association of Hsp70 with Cytoplasmic Hsp150Δ-β-Lactamase
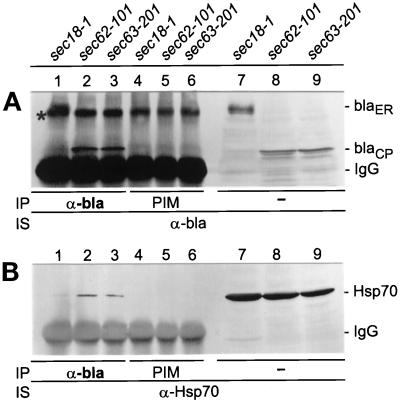
To study whether Hsp150Δ-β-lactamase was in association with Hsp70s, sec18–1 cells were incubated for 1 h at 37°C under low cell density conditions, followed by a 30-min chase with CHX to promote translocation into the ER, and sec62–101 and sec63–201 cells were incubated for 1 h at 30°C to accumulate the reporter in the cytoplasm. The cells were lysed under mild detergent conditions and the lysates were divided in thirds. One batch was subjected to immunoprecipitation with anti-β-lactamase antiserum (α-bla; Figure 8, lanes 1–3) and another with preimmune serum (PIM; lanes 4–6). The immunoprecipitates were subjected to SDS-PAGE and Western blot analysis using anti-β-lactamase antiserum (Figure 8A) or anti-Hsp70 antibody (Figure 8B). The 110-kDa ER form could be detected in the sec18–1 lysate (Figure 8A, lane 1) and the cytoplasmic 66-kDa form in the sec62–101 and sec63–201 lysates (lanes 2 and 3, respectively). Preimmune serum did not precipitate these proteins (Figure 8A, lanes 4–6). The protein migrating slightly faster than the 110-kDa form, marked with an asterisk (lanes 1–6), is unrelated to Hsp150Δ-β-lactamase, as it was detected in similar coimmunoprecipitation experiments even in cells lacking the HSP150Δ-β-lactamase gene (our unpublished results). It associates with protein A-Sepharose beads in the absence of denaturation, as shown before (Jämsäet al., 1995b). Immunostaining with anti-Hsp70 antibody revealed very little Hsp70 (70 kDa) in the sec18–1 lysate (Figure 8B, lane 1). This shows that after solubilization of the membranes, the ER-specific glycosylated 110-kDa form did not significantly associate with cytoplasmic Hsp70. In contrast, Hsp70 was coimmunoprecipitated with the cytoplasmic 66-kDa fusion protein (Figure 8B, lanes 2 and 3). Preimmune serum did not precipitate Hsp70 (Figure 8B, lanes 4–6). The third batch of lysates was subjected directly to SDS-PAGE and Western blot analysis omitting immunoprecipitation. The total amounts of Hsp150Δ-β-lactamase (Figure 8A, lanes 7–9) and Hsp70 (Figure 8B, lanes 7–9) show that immunoprecipitation of the reporter protein was efficient, whereas only a small fraction of total Hsp70 was coimmunoprecipitated with Hsp150Δ-β-lactamase. Thus, cytoplasmic Hsp150Δ-β-lactamase was found in association with Hsp70 chaperones.
Figure 8.
Coimmunoprecipitation of Hsp70 with cytoplasmic Hsp150Δ-β-lactamase. The indicated yeast strains were incubated under low cell density conditions for 1 h at 37°C (sec18–1) or 30°C (sec62–101 and sec63–201). The sec18–1 cells received CHX and were incubated for an additional 30 min 37°C. NaN3 was added and the cells were lysed under mild detergent conditions. After addition of apyrase, the lysates were immunoprecipitated (IP) with anti-β-lactamase antiserum (α-bla; lanes 1–3, A and B), or with preimmune serum (PIM; lanes 4–6, A and B). The precipitates, and parallel unprecipitated lysate samples (lanes 7–9, A and B), were subjected to SDS-PAGE, followed by Western blotting and immunostaining (IS) with either anti-β-lactamase antiserum (A) or anti-Hsp70 antiserum (B). On the right, ER-located (blaER; 110 kDa) and cytoplasmic (blaCP; 66 kDa) Hsp150Δ-β-lactamase forms, and IgG bands, are indicated. An unspecific band is indicated with an asterisk (A).
DISCUSSION
We show here that the β-lactamase portion of newly synthesized Hsp150Δ-β-lactamase acquired a trypsin-resistant and enzymatically active structure in the yeast cytosol. Thereafter, the fusion protein was translocated into the ER, where the Hsp150Δ fragment was primary O-glycosylated and the β-lactamase portion acquired a disulfide and adopted a similarly trypsin-resistant and active form as in the cytoplasm, followed by secretion of the fusion protein to the culture medium (Figure 9). The cytoplasmic and ER forms had similar Km values for nitrocefin as did authentic E. coli β-lactamase, demonstrating that the structural features critical for catalytic activity were assumed on both sides of the ER membrane. Authentic β-lactamase has a tight globular structure (Jelsch et al., 1992), whereas the Hsp150Δ fragment adopts no regular secondary structure but occurs as a random coil (Jämsäet al., 1995a). Earlier it has been noted that inhibition of translocation resulted in accumulation of a protease-resistant form of pre-pro-α factor at the cytoplasmic face of the yeast ER membrane (Nguyen et al., 1991). The authors speculated this to be due to aggregation or tight association with microsomal membrane, but did not consider folding. We could detect the cytoplasmic Hsp150Δ-β-lactamase precursor in mutants defective in posttranslational translocation, as well as in normal cells under high cell density conditions, where translocation was slowed down. Translocation of authentic Hsp150, whose C-terminal domain (amino acids 300–413, Figure 3B) consists largely of β-sheet (Jämsäet al., 1995a), was not retarded under these conditions. Penetration through the translocon was thus not retarded under high cell density conditions, and slow translocation of the fusion protein must have been due to cytoplasmic events concerning the β-lactamase portion.
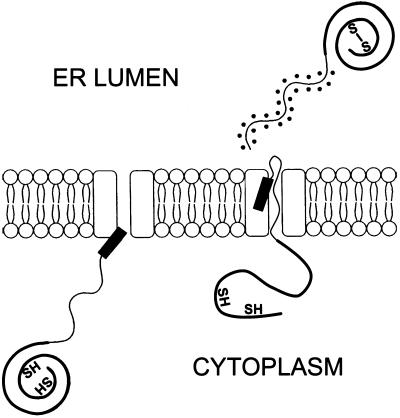
Figure 9.
Model for cytoplasmic and ER folding of Hsp150Δ-β-lactamase. The β-lactamase portion of newly synthesized Hsp150Δ-β-lactamase assumes a native-like conformation in the yeast cytosol on release from the ribosomes, because it is trypsin resistant and enzymatically active. No disulfide is formed between the two cysteine residues. The protein chain is translocated into the ER through the translocon complex (rectangular structures), with concomitant release of the signal peptide (black bar). The Hsp150Δ fragment is O-glycosylated (dots), the sulfhydryls of the β-lactamase portion (not drawn at proper positions) are oxidized to form a disulfide bond, and the protein assumes a conformation with similar catalytic properties as the cytosolic form. To what extent the β-lactamase portion unfolds for translocation is not known. The Hsp150Δ fragment occurs as a random coil (Jämsäet al., 1995a).
We found by coimmunoprecipitation experiments that Hsp70 was in association with cytoplasmic Hsp150Δ-β-lactamase. The interaction appeared to be specific, because after solubilization of the membranes, very little (if any) Hsp70 associated with the ER form of Hsp150Δ-β-lactamase. The ER form in turn was associated with BiP/Kar2p (Jämsäet al., 1995b), an ER-located Hsp70 homologue that is generally required for translocation (Vogel et al., 1990; Sanders et al., 1992). Although BiP/Kar2p and the predominant cytosolic Hsp70 member Ssa1p are >60% identical and Hsp70 members bind to hydrophobic peptides promiscuously (Gething and Sambrook, 1992), Ssa1p and BiP/Kar2p have specific functions, as they could not substitute for one another in ER translocation (Brodsky et al., 1993). Glick (1995) has proposed that BiP/Kar2p could drive simultaneously unfolding and translocation of precursor proteins. As our reporter protein was not kept in an extended conformation but assumed a native-like structure before translocation, the role of Hsp70 in its fate remains open. Bush and Meyer (1996) showed that immunodepletion of Ssa1p and SSa2p had no effect on translocation of pre-pro-α factor in vitro, nor on folding of nascent luciferase, whereas refolding of chemically denatured luciferase was defective. This led them to propose that nascent chains could assume translocation-incompetent conformations in the yeast cytosol, whereafter Ssa1/2p would unfold or refold them for translocation. Our data demonstrate directly, for the first time, that a polypeptide chain indeed can assume a stable conformation in the yeast cytosol before ER translocation in vivo.
Cotranslocational oxidation of sulfhydryls of the β-lactamase portion in the ER is obligatory for acquisition of a biologically active and secretion-competent structure. In the presence of the reducing agent DTT, the newly synthesized molecules are inactive and retained permanently in the ER (Simonen et al., 1994). In the yeast cytosol, productive folding of Hsp150Δ-β-lactamase occurred in the absence of disulfide formation. The disulfide bond of authentic β-lactamase is buried in the interior of the tight globular molecule (Jelsch et al., 1992). As the sulfhydryls of a properly folded β-lactamase portion cannot be reached by glutathione or disulfide isomerizing enzymes, they must have been exposed when emerging in the ER lumen, whereafter they were oxidized and folding was completed. This scenario would require at least partial unfolding before translocation. Also, authentic β-lactamase can adopt an enzymatically active disulfide-free form in the E. coli cytosol (Plückthun and Knowles, 1987). Moreover, it undergoes a refolding step after translocation across the E. coli membrane, occuring first as a membrane-bound, trypsin-sensitive intermediate, which is then converted to a soluble, trypsin-resistant, and bioactive form (Minsky et al., 1989). The requirement of the disulfide for β-lactamase activity in the ER but not in the cytosol of S. cerevisiae highlights the difference of these milieus as folding compartments. In addition to preventing or allowing disulfide formation, the reducing and oxidating conditions of the cytosol and ER lumen, respectively, apparently affect other amino acids besides cysteines, and consequently folding of the polypeptide chain.
Also, mitochondrial precursor proteins are thought to be kept in an unfolded or loosely folded state by the cytosolic Hsp70 chaperones before import, but more recently, artificial reporter proteins and authentic mitochondrial cytochrome b2 have been found to fold stably in the cytosol (Deshaies et al., 1988; Glick et al., 1993; Langer and Neupert, 1994; Wachter et al., 1994). The tightly folded heme-binding cytochrome b2 domain unfolds during translocation (Voos et al., 1993; Stuart et al., 1994). Unexpectedly, recent data have shown that the diameter of the translocon pore is about 20 Å (Hanein et al., 1996), or even 40–60 Å (Hamman et al., 1997), large enough to accommodate folded protein domains. Moreover, the same translocon is used to direct misfolded glycosylated proteins back to the cytoplasm for proteosomal degradation (Hiller et al., 1996; Pilon et al., 1997; Plemper et al., 1997). It has been hypothesized that precursor proteins could be translocated as “molten globules” that possess native-like secondary structure but lack rigid tertiary structure (Bychkova et al., 1988). Properly folded β-lactamase measures 32 × 37 × 53 Å (Jelsch et al., 1992). Whether the β-lactamase portion unfolds for translocation, and if it does, by what mechanism, remain to be studied.
ACKNOWLEDGMENTS
This work was supported by the Academy of Finland (grant 38017). M.M. is a senior researcher of the Academy of Finland and a Biocentrum Helsinki fellow. We thank Dr. Leevi Kääriäinen and Ms. Nina Saris, M.Sc., from our institute for critical reading of the article, and Ms. Anna Liisa Nyfors for excellent technical assistance. Dr. Randy Schekman (University of California, Berkeley, CA) and Dr. Peter Walter (University of California, San Francisco, CA) generously provided yeast strains.
REFERENCES
- Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastian L, Lacoute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:608–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. Heat shock cognate proteins and polypeptide translocation across the endoplasmic reticulum membrane. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 85–109. [Google Scholar]
- Bush GL, Meyer DI. The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their role in protein translocation. J Cell Biol. 1996;135:1229–1237. doi: 10.1083/jcb.135.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bychkova VE, Pain RH, Ptitsyn OB. The “molten globule” state is involved in the translocation of proteins across membranes. FEBS Lett. 1988;238:231–234. doi: 10.1016/0014-5793(88)80485-x. [DOI] [PubMed] [Google Scholar]
- Chirico WJ, Waters MG, Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation onto the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. Sec62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol. 1994;126:935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–44. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Glick BS, Wachter C, Reid GA, Schatz G. Import of cytochrome b2 to the mitochondrial intermembrane space: the tightly folded heme-binding domain makes import dependent upon matrix ATP. Prot Sci. 1993;2:1901–1917. doi: 10.1002/pro.5560021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman BD, Cheng J-C, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KES, Jungnickel B, Plath K, Kalies K-U, Miller KR, Rapoport TA, Akey CW. Olicomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Jämsä E, Holkeri H, Vihinen H, Wikström M, Simonen M, Walse B, Kalkkinen N, Paakkola J, Makarow M. Structural features of a polypeptide carrier promoting secretion of a β-lactamase fusion protein in yeast. Yeast. 1995a;11:1381–1391. doi: 10.1002/yea.320111406. [DOI] [PubMed] [Google Scholar]
- Jämsä E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Jämsä E, Vakula N, Affman A, Kilpeläinen I, Makarow M. In vivo reactivation of heat-denatured protein in the endoplasmic reticulum of yeast. EMBO J. 1995b;14:6028–6033. doi: 10.1002/j.1460-2075.1995.tb00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsch C, Lenfant F, Masson JM, Samama JP. β-Lactamase TEM1 of E. coli crystal structure determination at 2.5 Å resolution. FEBS Lett. 1992;299:135–142. doi: 10.1016/0014-5793(92)80232-6. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Schekman R. Distinct set of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Koshland D, Botstein D. Evidence for posttranslational translocation of β-lactamase across the bacterial inner membrane. Cell. 1982;30:893–902. doi: 10.1016/0092-8674(82)90294-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laminet AA, Plückthun A. The precursor of β-lactamase: purification, properties and folding kinetics. EMBO J. 1989;8:1469–1477. doi: 10.1002/j.1460-2075.1989.tb03530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T, Neupert W. Chaperoning mitochondrial biogenesis. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 53–83. [Google Scholar]
- Minsky A, Summers RG, Knowles JR. Secretion of β-lactamase into the periplasm of Escherichia coli: evidence for distinct release step associated with a conformational change. Proc Natl Acad Sci USA. 1986;83:4180–4184. doi: 10.1073/pnas.83.12.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DTW, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Law DTS, Williams DB. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:1565–1569. doi: 10.1073/pnas.88.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Römisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Böhmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Plückthun A, Knowles J. The consequences of stepwise deletions from the signal-processing site of β-lactamase. J Biol Chem. 1987;262:3951–3957. [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2642–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S, Wheatfield KM, Vogel JL, Rose M, Schekman R. Sec63p and BiP directly facilitate polypeptide translocation into ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- Sanderson C, Meyer D. Functional identification of membranes derived from the rough endoplasmic reticulum of yeast. Cell Biology: A Laboratory Handbook. J. Celis, New York: Academic Press; 1994. pp. 531–533. [Google Scholar]
- Simonen M, Jämsä E, Makarow M. The role of the carrier protein and disulfide formation in the folding of β-lactamase fusion proteins in the endoplasmic reticulum of yeast. J Biol Chem. 1994;269:13889–13892. [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RA, Cyr DM, Craig EA, Neupert W. Mitochondrial molecular chaperones: their role in protein translocation. TIBS. 1994;19:87–92. doi: 10.1016/0968-0004(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Toyn J, Hibbs AR, Sanz P, Crowe J, Meyer DI. In vivo and in vitro analysis of ptl1, a yeast ts mutant with a membrane-associated defect in protein translocation. EMBO J. 1988;7:4347–4353. doi: 10.1002/j.1460-2075.1988.tb03333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Misra MD, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Gambill D, Guiard B, Pfanner N, Craig EA. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J Cell Biol. 1993;123:119–126. doi: 10.1083/jcb.123.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter C, Schatz G, Glick BS. Protein import into mitochondria: the requirement for external ATP is precursor-specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol Biol Cell. 1994;5:465–474. doi: 10.1091/mbc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley P, Nilsson I-M, von Heijne G. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J Biol Chem. 1996;271:6241–6244. doi: 10.1074/jbc.271.11.6241. [DOI] [PubMed] [Google Scholar]