Abstract
The QM/MM MD and free energy simulations show that the dynamics involving a His residue at the P1 site of the substrate may play an important role in substrate-assisted catalysis and specificity for a serine-carboxyl peptidase.
Substrate-assisted catalysis (SAC) is the process in which one or more functional groups from the substrate, in addition to those from the enzyme, contribute to the rate acceleration for the enzyme-catalyzed reaction. There is growing evidence that SAC may exist in many naturally occurring or engineered catalysts.1–2 The existence of product-assisted catalysis has also been suggested recently.3 Understanding the role of SAC in enzyme specificity is of considerable interest, as it may provide new insights into how substrates compete with each other for enzymes. Many of the previous studies of SAC1–2 have focused on the catalytic effects resulting from the well-positioned substrate groups that can participate in SAC directly, without undergoing significant conformational changes. A fundamental question is whether some substrate groups which are not located at such ideal positions in enzyme-substrate complexes would be able to undergo conformational changes and participate in SAC as well. In this Communication, we report the results of computer simulations and show that the dynamics involving distant substrate groups that are triggered by bond breaking and making events of enzyme-catalyzed reactions may indeed play an important role in SAC. The importance of this type of SAC involving conformational changes of substrates might have been overlooked in some enzymes due in part to the dynamic nature of the effects that may not be well reflected in the X-ray structures.
The system used to demonstrate the importance of dynamics in substrate-assisted catalysis is kumamolisin-As, a member of a recently characterized sedolisin family of proteolytic enzymes.4–7 The QM(SCC-DFTB)/MM molecular dynamics (MD) and free energy simulations have been performed in this study.8 Sedolisins have a fold resembling that of subtilisin and a maximal activity at low pH (~3.9).4–5 The defining features of kumamolisin-As and other members of this family are a unique catalytic triad, Ser278-Glu78-Asp82 (kumamolisin-As numbering), as well as the presence of an aspartic acid residue, Asp164, in the active site. Asp164 replaces Asn155 of subtilisin, a residue that creates the oxyanion hole.6 Unlike Asn155 that provides electrostatic stabilization of the tetrahedral intermediate during the catalysis,6 Asp164 seems to act as a general acid catalyst to protonate the tetrahedral intermediate and assist in the nucleophilic attack by Ser278.7 The substrates used in the present work are RPXaa*FR (here * designates the scissile peptide bond and Xaa denotes the residue at the P1 site). The formation of the tetrahedral intermediate (TI) is examined, as the efficiency of this step of the reaction is crucial for high substrate specificity of proteolytic enzymes.6 The models were built using the X-ray structure of kumamolisin-As complexed with an inhibitor N-acetyl-isoleucylprolyl-phenylalaninal (AcIPF)5b and the substrate coordinates were obtained based on those from the corresponding residues in the Ser278Ala mutant of pro-kumamolisin.5f We demonstrate that there is a conformational transition of the His residue at the P1 site for RPH*FR and that a salt bridge is formed between His and Asp164 as the system changes from the substrate to the TI complex. It is suggested that this conformational change and the formation of the salt bridge make it possible for the His residue to participate in SAC during the formation of the tetrahedral intermediate. Our suggestion for the existence of dynamic SAC is supported by the comparison of the free energy profiles of the TI formation for RPH*FR and RPF*FR and is consistent with experimental data.5
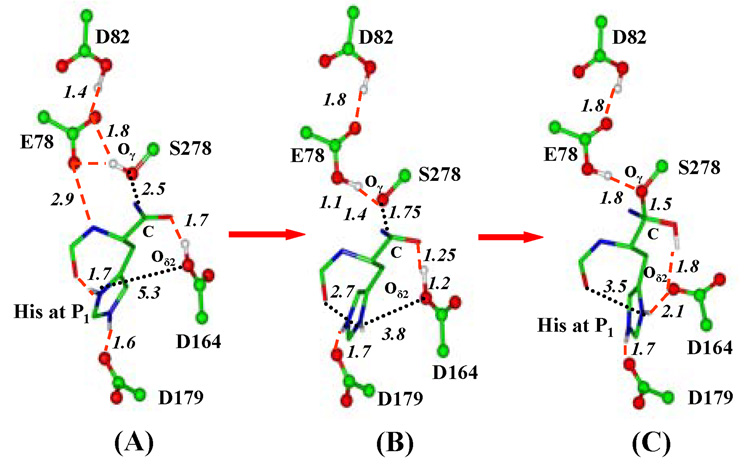
The average structures of the active site of the kumamolisin-As complex during the nucleophilic attack by Ser278 on the RPH*FR substrate are given in Figure 1. As is evident from Figure 1A, Ser278 is well aligned for the nucleophilic attack on C in the substrate complex. Glu78 and Asp164 are well positioned to act as the general base and acid catalysts, respectively, to assist in the nucleophilic attack. His at the P1 site forms two hydrogen bonds (H-bonds) to the C=O group of Pro at P2 and the carboxylate of Asp179, respectively. Moreover, there is an H-bond between the backbone N-H group of the P1 residue and Glu78. Figure 1B shows that the interaction of the His sidechain with the C=O group is significantly weakened as the nucleophilic attack proceeds to the transition state, making the His sidechain available for other interaction. The weakening of the interaction involving His seems to be triggered by the breaking of the H-bond between the backbone N-H group at P1 and Glu78, as Glu78 accepts the proton from Ser278 and becomes uncharged during the nucleophilic attack. Thus, there appears to be the cooperative effects of hydrogen bonding17 in the stabilization of the H-bond between the His sidechain and the P2 C=O group in the substrate complex, although other factors might be involved as well. Figure 1C shows that the histidine sidechain has rotated significantly (mainly around the Cβ—Cγ bond) to interact with the unprotonated Asp164 residue during the formation of the TI complex. Since Asp164 is the general acid catalyst,7 the formation of the salt bridge between His and Asp164 may make the TI complex more stable (see below).
Figure 1.
Average structures of kumamolisin-As complex during the nucleophilic attack by Ser278 on substrate RPH*FR obtained from the QM/MM MD and free energy simulations. The enzyme is plotted in ball and stick and the ligand in stick. Hydrogen bonds (H-bonds) are indicated by red dashed lines. Ser278-Glu78-Asp82 is the catalytic triad. Only the sidechain of the P1 (His) residue for the substrate is shown for clarity. (A) The substrate complex. His sidechain at P1 donates an H-bond to the C=O group of Pro at P2, in addition to the carboxylate of Asp179. Glu78 accepts an H-bond from the backbone N-H group of the P1 residue (the H-bond distance shown here is between the non-hydrogen atoms). (B) An average structure near the transition state of the nucleophilic attack. The interaction of the His sidechain with the C=O group is significantly weakened and the H-bond between Glu78 and the N-H group is broken. (C) The TI complex. His at P1 has undergone a conformational change to interact with the unprotonated Asp164 residue and the H-bond to the C=O group observed in the substrate complex is broken.
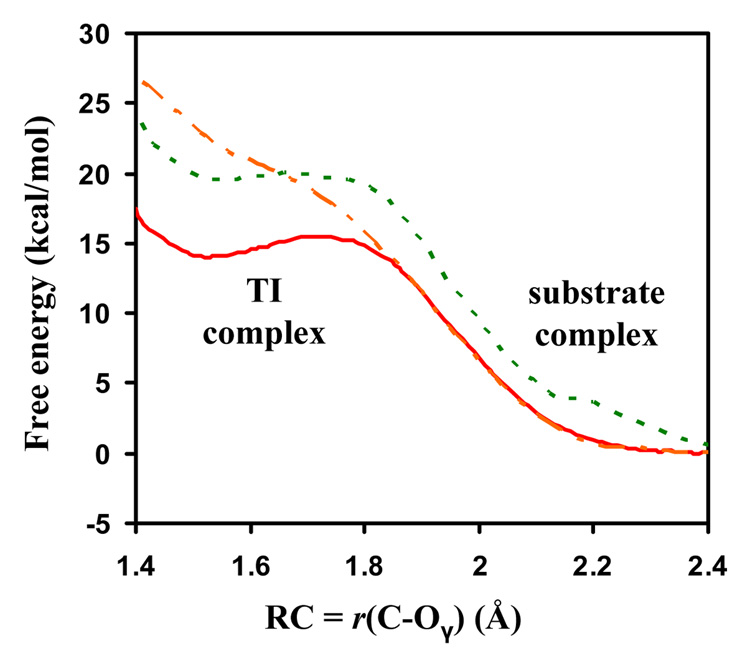
The changes of free energy (potential of mean force) as functions of the reaction coordinate (ξ) for the nucleophilic attack by Ser278 on the RPH*FR and RPF*FR substrates are given in Figure 2. The free energy change for the RPH*FR complex in which the proton on Asp164 is fixed by the Shake algorithm9 (to prevent Asp164 to act as the general acid) is also given. Figure 2 shows that the free energy barrier of the TI formation is ~ 15 kcal/mol for RPH*FR, and this barrier increases to ~ 20 kcal/mol when the substrate is changed to RPF*FR. The major difference for the two substrates is that His at the P1 site in RPH*FR is able to form a salt bridge with Asp164 through a conformational transition (see above), and this salt bridge may help to stabilize the charge formation on Asp164 and make it a better general acid. In contrast, Phe at P1 for RPF*FR is unable to interact with Asp164 and the stabilization of the TI through this mechanism is therefore impossible. It is of interest to note that the TI for RPH*FR becomes considerably less stable when the proton transfer from Asp164 to the ligand is prevented by using the Shake algorithm, supporting the suggestion that Asp164 acts as a general acid catalyst.7
Figure 2.
The free energy change from the substrate to TI complex as a function of ξ = r(C–Oγ), the reaction coordinate for the nucleophilic attack. Red solid line: the RPH*FR substrate without use of the Shake algorithm; Orange dot-dashed line: the RPH*FR substrate with use of the Shake algorithm to fix the proton on Asp164; Blue dashed line: the RPF*FR substrate.
The results of the simulations reported here are consistent with the available experimental data.5 It has been shown that kumamolisin-As can cleave the –Pro–Xaa*Yaa– sequence and that this enzyme generally has higher specificity for the substrates with a positively charged residue at the P1 site than for those they do not. For instance, it was found that the kcat (KM) value for a substrate containing the –Pro–Arg*Gly sequence is ~ 65-fold (~ 6.5-fold) higher than for a substrate containing the –Pro–Gly*Gly sequence, leading to 10-fold increase of kcat/KM.5a Moreover, specificity profile analysis5b using a peptide library showed that kumamolisin-As generally displays higher specificity for the substrates with His, Lys or Arg at P1 than for the other substrates. Quantitative comparisons of the experimental data and theoretical results require determinations of the catalytic mechanism and rate limiting step which are poorly understood.
We suggest that the dynamics may play an important role in SAC and specificity of kumamolisin-As. The conformational change observed is triggered by the deprotonation/protonation events during the general acid-base catalysis. It is of interest to examine whether this would also occur for other enzymes.
Supplementary Material
A description of the computational methods, additional and more complete structures obtained from the simulations and complete ref. 12. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
We thank Professor Martin Karplus for a gift of the CHARMM program and an anonymous reviewer of the earlier paper for pointing out a possible role of dynamics in aspartic protease catalysis. This work was supported in part by the Center of Excellence for Structural Biology, the University of Tennessee and the Petroleum Research Fund (HG) and in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (AW).
References
- 1.For reviews of earlier work, see Dall’Acqua W, Carter P. Protein Sci. 2000;9:1–9. doi: 10.1110/ps.9.1.1. Kosloff M, Selinger Z. Trends Biochem. Sci. 2001;26:161–166. doi: 10.1016/s0968-0004(00)01748-5.
- 2.(a) Weinger JS, Parnell KM, Doner S, Green R, Strobel SA. Nat. Struct. Mol. Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]; (b) Jogl G, Tong L. Cell. 2003;112:113–122. doi: 10.1016/s0092-8674(02)01228-x. [DOI] [PubMed] [Google Scholar]; (c) Ryan M, Liu T, Dahlquist FW, Griffith OH. Biochem. 2001;40:9743–9750. doi: 10.1021/bi010958m. [DOI] [PubMed] [Google Scholar]; (d) Breldenbach MA, Brunger AT. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 3.Fromme JC, Bruner SD, Yang W, Karplus M, Verdine GL. Nat. Struct. Mol. Biol. 2003;10:204–210. doi: 10.1038/nsb902. [DOI] [PubMed] [Google Scholar]
- 4.(a) Wlodawer A, Li M, Gustchina A, Oyama H, Dunn BM, Oda K. Acta Biochim. Pol. 2003;50:81–102. [PubMed] [Google Scholar]; (b) Vandeputte-Rutten L, Gros P. Curr. Opin. Struct. Biol. 2002;12:704–708. doi: 10.1016/s0959-440x(02)00393-7. [DOI] [PubMed] [Google Scholar]; (c) Powers JC, Asgian JL, Ekici OD, James KE. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]; (d) Wlodawer A. Structure. 2004;12:1117–1119. doi: 10.1016/j.str.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 5.(a) Tsuruoka N, Nakayama T, Ashida M, Hemmi H, Nakao M, Minakata H, Oyama H, Oda K, Nishino T. Appl. Environ. Microbiol. 2003;69:162–169. doi: 10.1128/AEM.69.1.162-169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wlodawer A, Li M, Gustchina A, Tsuruoka N, Ashida M, Minakata H, Oyama H, Oda K, Nishino T, Nakayama T. J. Biol. Chem. 2004;279:21500–21510. doi: 10.1074/jbc.M401141200. [DOI] [PubMed] [Google Scholar]; (c) Wlodawer A, Li M, Dauter Z, Gustchina A, Uchida K, Oyama H, Dunn BM, Oda K. Nature Struct. Biol. 2001;8:442–446. doi: 10.1038/87610. [DOI] [PubMed] [Google Scholar]; (d) Wlodawer A, Li M, Gustchina A, Dauter Z, Uchida K, Oyama H, Goldfarb NE, Dunn BM, Oda K. Biochemistry. 2001;40:15602–15611. doi: 10.1021/bi011817n. [DOI] [PubMed] [Google Scholar]; (e) Comellas-Bigler M, Fuentes-Prior P, Maskos K, Huber R, Oyama H, Uchida K, Dunn BM, Oda K, Bode W. Structure. 2002;10:865–876. doi: 10.1016/s0969-2126(02)00772-4. [DOI] [PubMed] [Google Scholar]; (f) Comellas-Bigler M, Maskos K, Huber R, Oyama H, Oda K, Bode W. Structure. 2004;12:1313–1323. doi: 10.1016/j.str.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 6.For reviews, see: Hedstrom L. Chem. Rev. 2002;102:4501–4523. doi: 10.1021/cr000033x. Vandeputte-Rutten L, Gros P. Curr. Opin. Struct. Biol. 2002;12:704–708. doi: 10.1016/s0959-440x(02)00393-7. Powers JC, Asgian JL, Ekici OD, James KE. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. Perona JJ, Craik CS. J. Biol. Chem. 1997;272:29987–29990. doi: 10.1074/jbc.272.48.29987.
- 7.Guo HB, Wlodawer A, Guo H. J. Am. Chem. Soc. 2005;127:15662–15663. doi: 10.1021/ja0520565. [DOI] [PubMed] [Google Scholar]
- 8.A semi-empirical density-functional approach (SCC-DFTB)10 recently implemented in the CHARMM program11 was used for QM/MM MD and free energy simulations. The initial coordinates were obtained from the crystal structure (PDB ID:1SIO) of kumamolisin-As which has AcIPF covelantly bound to Ser278.5b The substrate coordinates were obtained based on those from the corresponding residues in pro-kumamolisin.5f A part of the substrate and the side chains of Glu32, Asp82, Glu78, Ser278 and Asp164 were treated by QM and the rest of the system by MM. The all-hydrogen potential function (PARAM22)12 was used for MM atoms. A modified TIP3P water model13–14 was employed for the solvent. The stochastic boundary MD method9 was used for the simulations. A 1-fs time step was used for integration of the equations of motion. The umbrella sampling method15 along with Weighted Histogram Analysis Method16 was applied to determine the changes of the free energy.
- 9.Brooks CL, III, Brunger A, Karplus M. Biopolymers. 1985;24:843. doi: 10.1002/bip.360240509. [DOI] [PubMed] [Google Scholar]
- 10.Cui Q, Elstmer M, Kaxiras E, Frauenheim T, Karplus M. J. Phys. Chem. B. 2001;105:569–585. [Google Scholar]
- 11.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 12.Karplus M, et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen WL. J. Am. Chem. Soc. 1981;103:335–340. [Google Scholar]
- 14.Neria E, Fisher S, Karplus M. J. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
- 15.Torrie GM, Valleau JP. Chem. Phys. Lett. 1974;28:578–581. [Google Scholar]
- 16.Kumar M, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 17.Guo H, Salahub DR. Angew. Chem. Int. Ed. 1998;37:2985–2990. doi: 10.1002/(SICI)1521-3773(19981116)37:21<2985::AID-ANIE2985>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A description of the computational methods, additional and more complete structures obtained from the simulations and complete ref. 12. This material is available free of charge via the Internet at http://pubs.acs.org.