Abstract
We have used unbiased phosphoproteomic approaches, based on quantitative mass spectrometry using stable isotope labeling with amino acids in cell culture (SILAC), to identify tyrosine phosphorylated proteins in isogenic human bronchial epithelial cells (HBECs) and human lung adenocarcinoma cell lines, expressing either of the two mutant alleles of EGFR (L858R and Del E746-A750), or a mutant KRAS allele, which are common in human lung adenocarcinomas. Tyrosine phosphorylation of signaling molecules was greater in HBECs expressing the mutant EGFRs than in cells expressing WT EGFR or mutant KRAS. Receptor tyrosine kinases (such as EGFR, ERBB2, MET, and IGF1R), and Mig-6, an inhibitor of EGFR signaling, were more phosphorylated in HBECs expressing mutant EGFR than in cells expressing WT EGFR or mutant RAS. Phosphorylation of some proteins differed in the two EGFR mutant-expressing cells; for example, some cell junction proteins (β-catenin, plakoglobin, and E-cadherin) were more phosphorylated in HBECs expressing L858R EGFR than in cells expressing Del EGFR. There were also differences in degree of phosphorylation at individual tyrosine sites within a protein; for example, a previously uncharacterized phosphorylation site in the nucleotide-binding loop of the kinase domains of EGFR (Y727), ERBB2 (Y735), or ERBB4 (Y733), is phosphorylated significantly more in HBECs expressing the deletion mutant than in cells expressing the wild type or L858R EGFR. Signaling molecules not previously implicated in ERBB signaling, such as polymerase transcript release factor (PTRF), were also phosphorylated in cells expressing mutant EGFR. Bayesian network analysis of these and other datasets revealed that PTRF might be a potentially important component of the ERBB signaling network.
Keywords: proteomics, tyrosine phosphorylation
Aberrant signaling generated by mutant protooncogenes drives tumorigenesis. The two protooncogenes currently known to be most commonly mutated in human lung adenocarcinoma are KRAS and epidermal growth factor receptor (EGFR). Approximately 90% of the lung cancer-specific EGFR mutations are equally distributed between a Leucine to Arginine substitution at position 858 (L858R) and deletion mutants in exon 19 that affect the conserved sequence LREA (e.g.delE746-A750) (1–4). Although the mutations in EGFR correlate with sensitivity of the tumors to tyrosine kinase inhibitors (TKIs), KRAS mutations are associated with primary resistance to TKIs (5). Expression of the lung cancer-specific EGFR mutants leads to carcinogenesis in the lung epithelium of transgenic mice or to transformation in fibroblasts or Ba/F3 cells (6–9). However, aberrant signaling pathways downstream of oncogenic EGFR and KRAS have been only partially characterized, and differences between signaling events downstream of the two most common EGFR mutants are not known. However, patients with exon 19 deletions may respond better to TKI therapy than those with the L858R mutation (10, 11), underscoring the importance of elucidating the differences in signaling downstream of the mutant receptors.
Lung cancer-specific EGFR mutations result in constitutive activation of EGFR and downstream signaling components, such as AKT and STAT5 (8, 9, 12). Global surveys of phosphotyrosine containing proteins in human lung adenocarcinoma cell lines have identified hundreds of sites with phosphorylated tyrosine (13, 14). Phosphorylation at a substantial number of these sites is inhibited by treatment of gefitinib in the TKI sensitive cell line H3255, which harbors the L858R mutation (14). However, the adenocarcinoma cell lines were derived from tumors from different patients and, hence, have heterogeneous genetic backgrounds, compromising comparisons among the lines. It is difficult to determine which of the many observed differences in patterns of phosphorylation is the consequence of a particular oncogenic mutation.
To circumvent the problems of heterogeneous genetic background, we have focused our studies of phosphotyrosine-mediated signaling to isogenic human bronchial epithelial cells (HBECs), immortalized by hTERT and CDK4 (15, 16), which stably express wild type EGFR (WT EGFR), KRAS G12V, L858R EGFR, or Del E746-A750 EGFR. In this study, we undertook global phosphoproteomic approaches, involving metabolic labeling of cells, immunoprecipitation of proteins or peptides with anti-phosphotyrosine antibodies, followed by liquid chromatography and tandem mass spectrometry (LC-MS/MS), to identify and quantify tyrosine phosphorylated proteins and proteins strongly associated with them in cells expressing WT EGFR, either of the two EGFR mutants and mutant KRAS.
Results
As a preliminary survey of phosphotyrosine-based signaling activity stimulated by oncogenic alleles of KRAS and EGFR, we used immunoprecipitation and Western blot analysis with anti-phosphotyrosine antibodies to compare proteins in isogenic HBECs expressing WT EGFR, KRAS G12V, L858R EGFR, or DelE746-A750 EGFR, and in several nonisogenic human adenocarcinoma cell lines with KRAS and EGFR mutations [supporting information (SI) Fig. S1]. The HBECs expressing EGFR mutants and the adenocarcinoma cells with an EGFR mutation constitutively displayed more tyrosine phosphorylated proteins than did the other cell lines, and there was minimal further stimulation of tyrosine phosphorylation upon EGF administration in these cells. However, AKT and ERK were activated, suggesting that overexpression of the mutant receptors did not alter canonical EGFR signaling in these cells (Fig. S1C).
Differences in Abundance of Proteins in Phosphotyrosine IPs from Cells Expressing Mutant EGFR and Mutant KRAS.
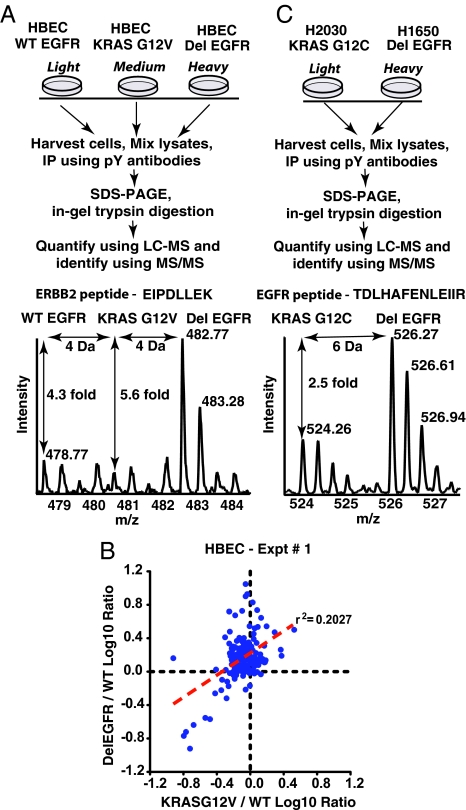
We first identified and measured the abundance of proteins obtained in phospho-tyrosine immunoprecipitates (IPs) from lysates of HBECs expressing WT EGFR, KRAS G12V, or Del E746-A750 EGFR using mass spectrometry. The cells were metabolically labeled with “light,” “medium,” or “heavy” isotope forms of arginine and lysine [stable isotope labeling with amino acids in cell culture (SILAC)] (see Materials and Methods) to perform relative quantitation of tyrosine phosphorylation (Fig. 1A). A representative MS spectrum shows that an ERBB2 peptide is approximately five times more abundant in IPs from HBECs expressing Del EGFR than in IPs from HBECs expressing WT EGFR or mutant KRAS (Fig. 1A). Based on pairwise comparisons with extracts of HBECs producing WT EGFR, extracts from cells expressing mutant EGFR and mutant KRAS were associated with very different profiles of proteins in phosphotyrosine IPs (Fig. 1B). The abundance of identified proteins from the IPs was very similar using extracts from HBECs expressing WT EGFR and extracts from cells expressing KRAS G12V. However, 75 of 175 identified proteins were more abundant in the phosphotyrosine IPs from HBECs expressing Del EGFR, than from cells expressing WT EGFR (Table S1 and Fig. S2). The complete list of proteins and tryptic peptides identified in the phosphotyrosine immunoprecipitates of intact proteins is shown in Table S2 and Table S3.
Fig. 1.
Increased tyrosine phosphorylation of proteins in HBECs or adenocarcinoma cells expressing mutant EGFR compared with cells expressing mutant KRAS. Schematic of experimental design and representative MS spectra of a peptide of ERBB2 (A) and a peptide of EGFR (C) identified in the phosphotyrosine immunoprecipitates of proteins from lysates of HBECs and adenocarcinoma cell lines, respectively. (B) The degree of tyrosine phosphorylation of proteins in mutant RAS expressing cells does not correlate with that in mutant EGFR-expressing cells as demonstrated in the log-log plot of the two ratios obtained in this experiment (Del EGFR/WT EGFR and Mut RAS/WT EGFR). Log 10 transformation of SILAC ratios for individual proteins and linear regression analysis yields a coefficient of determination (r2) of 0.20, and a Pearson product-moment correlation (r) of 0.45.
In similar experiments using two lung adenocarcinoma cell lines, H1650 and H2030, that harbor DelE746-A750 EGFR and KRAS G12C mutations, respectively (Fig. 1C), 47 proteins were more abundant in the phosphotyrosine IPs from H1650 than from H2030 cells. Surprisingly, 53 proteins were less abundant in phosphotyrosine IPs from H1650 cells than in IPs from H2030 cells (Table S1 and Fig. S2). Some of the proteins that were less abundant in the IPs may be expressed at lower levels in H1650 than in H2030 cells, as shown for the p66 isoform of SHC and the RAS inhibitor 1 (RIN1) (Fig. S3). These observations reflect a significant limitation of comparisons of phosphoproteomic data generated with nonisogenic cell lines. For this reason, most of our experiments were conducted with isogenic HBECs expressing oncogenic alleles of EGFR and KRAS.
Differences in Tyrosine Phosphorylation of Proteins in HBECs Expressing Del EGFR or L858R EGFR.
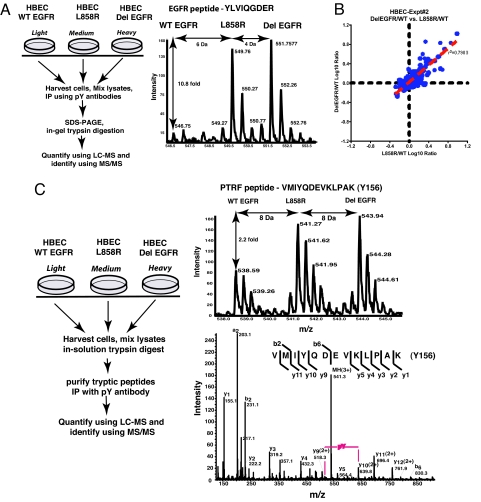
Differences in the pathophysiological effects of the two common EGFR mutants in lung adenocarcinoma might be attributed in part to differential use of targets for phosphorylation. We therefore sought to determine the differences in the amounts or sites of protein-tyrosine phosphorylation in HBECs expressing L858R or Del EGFR. The MS spectrum of a representative EGFR-peptide (Fig. 2A) shows that this peptide was at least 10-fold more abundant in the phosphotyrosine IPs from lysates of HBECs, expressing either of the EGFR mutants than in material from HBECs expressing WT EGFR. We obtained quantitative data on 245 proteins from phosphotyrosine IP of proteins from the lysates of HBECs expressing WT EGFR, L858R, or Del EGFR (Table S1, Table S2, and Table S4). Of these, only nine were at least 1.5 times more abundant in the phosphotyrosine IPs from HBECs expressing Del EGFR than in IPs from cells expressing L858R EGFR. Eight of these were at least 1.5 times more abundant in IPs from HBECs expressing L858R EGFR than in IPs from cells expressing Del EGFR (Table S1 and Fig. S2). Interestingly, junctional proteins such as β-catenin, plakoglobin, and E-cadherin were more abundant in the phosphotyrosine IPs from L858R-expressing HBECs than in those from Del EGFR-expressing HBECs. The similar abundance of proteins in phosphotyrosine IPs from lysates of HBECs expressing L858R and from those expressing Del EGFR is reflected in the high correlation coefficients of ratios of protein abundance in phosphotyrosine IPs from lysates of these cells compared with those with cells expressing WT EGFR (Fig. 2B).
Fig. 2.
Extent of tyrosine phosphorylation is similar between HBECs expressing L858R EGFR or Del EGFR. (A) Experimental design and a representative MS spectrum of EGFR shows the increased abundance of this peptide in phosphotyrosine immunoprecipitates of proteins from the EGFR mutant-expressing cell lines compared with HBECs expressing WT EGFR. (B) The degree of tyrosine phosphorylation of proteins is similar in the HBECs expressing the two EGFR mutants as shown by the log-log plot of the two ratios obtained in this experiment (Del EGFR/WT EGFR and L858R/WT EGFR). Log 10 transformation of SILAC ratios for individual proteins and linear regression analysis yields a coefficient of determination (r2) of 0.79, and a Pearson product-moment correlation coefficient (r) of 0.89. (C) Schematic of the experimental design (left panel) to compare the extent of site-specific phosphorylation. The MS spectrum of the peptide VMIYQDEVKLPAK of PTRF in the right upper panel shows that the abundance of this peptide is higher in each of the two mutant EGFR-expressing cell lines compared with HBECs expressing WT EGFR. The MS/MS spectrum of the same peptide in right lower panel shows that Y156 of PTRF is phosphorylated. Because tyrosine phosphorylated peptides have specifically been enriched in this experiment, it can be inferred that the phosphorylation is more in the EGFR mutant-expressing lines.
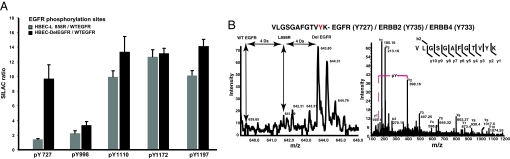
Proteins containing SH2 and PTB domains recognize phosphotyrosines in specific sequence contexts. As a result, the extent of phosphorylation of different tyrosines, in the same protein, may have implications for protein interactions and effects on cell signaling. To quantify the extent of phosphorylation in individual tyrosine-containing peptides in cells expressing WT EGFR, L858R EGFR, or Del EGFR, we immunoprecipitated tyrosine phosphorylated peptides, after digestion of mixed lysates with trypsin in solution, followed by LC MS/MS (Fig. 2C). A representative peptide of polymerase transcript release factor (PTRF) containing tyrosine Y156 is more abundant in phosphotyrosine IPs from HBECs expressing either of the two EGFR mutants, than in the IPs from HBECs expressing WT EGFR. The MS/MS spectrum of fragments from the same peptide shows that Y156 is the phosphorylated residue (Fig. 2C). We identified and quantified phosphorylation at 47 tyrosine residues from 38 different proteins (Table S6). Four known tyrosine autophosphorylation sites in the C-terminal domain of EGFR were more heavily phosphorylated in HBECs expressing L858R or Del EGFR than in HBECs expressing WT EGFR. However, a fifth site, Y727, in the nucleotide-binding loop of the kinase domain, was highly phosphorylated in the Del EGFR-expressing cells, minimal in L858R EGFR-expressing HBECs, but showed no phosphorylation in WT EGFR-expressing cells (Fig. 3). An identical tryptic peptide is predicted to be present in ERBB2 and ERBB4, but EGFR is overexpressed in the HBEC lines; thus, it is likely that the phosphorylated tryptic peptide is exclusively from EGFR (Fig. S4).
Fig. 3.
Differential extent of phosphorylation at individual tyrosine sites in EGFR between the two mutant EGFR-expressing HBECs and cells expressing WT EGFR. (A) Ratio of abundance of peptides containing each of the indicated phosphorylated tyrosine residues in L858R or Del EGFR compared with WT EGFR. Note that the tryptic peptide containing Y727 of EGFR is also present in ERBB2 (Y735) and ERBB4 (Y733). Mean and STDV are shown from three independent peptide-level phosphotyrosine IPs and three separate mass spectrometry analyses. (B) MS and MS/MS spectra of the tryptic peptide containing Y727 of EGFR show that Y727 is phosphorylated in Del-EGFR but not in WT EGFR and minimally in L858R EGFR. The locations of possible peaks of this peptide from WTEGFR and L858R are shown in the MS spectrum.
We also identified proteins involved in receptor recycling, such as Caveolin 1, Rab7, and Rab8, which were more phosphorylated in cells expressing mutant EGFRs than cells expressing WT EGFR or mutant KRAS (Table S2). In addition, tyrosine phosphorylation of MIG6, a protein involved in inhibition of ERBB signaling, was greater in cells expressing mutant EGFRs than in those expressing WT EGFR or mutant KRAS. Phosphorylation of Mig-6 at Y394 was greater in HBECs expressing Del EGFR than in HBECs expressing L858R EGFR (Table S2 and Table S6). However, Mig-6 interacted both with wild type and mutant EGFRs, as measured by co-IP (Fig. S5).
Validation of Mass Spectrometry-Based “Phosphorylation Ratios” by IP-Western Blot Experiments.
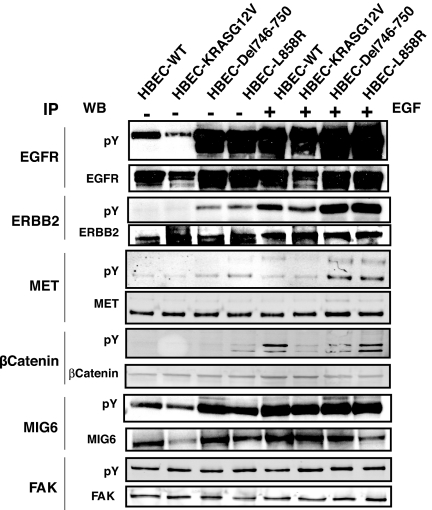
We have validated the tyrosine phosphorylation of a number of proteins by immunoprecipitation from the lysates of the isogenic HBECs (Fig. 4) and a panel of lung adenocarcinoma cell lines (Fig. S6) with protein-specific antibodies, followed by Western blot analysis with an anti-phosphotyrosine antibody. As expected, EGFR, ERBB2, MET, β-catenin, and Mig-6 were all more heavily phosphorylated on tyrosine in HBECs expressing mutant EGFR than in cells expressing mutant RAS or WT EGFR (Fig. 4). Another signaling molecule that was highly tyrosine-phosphorylated, FAK, was present at similar levels and tyrosine-phosphorylated to similar degrees in HBECs expressing mutant RAS or WT EGFR, and in adenocarcinoma cell lines, confirming the ratios of relative abundance derived from the mass spectrometry results.
Fig. 4.
Validation of mass spectrometry-based quantitation by immunoprecipitation and Western blots of representative proteins. Immunoprecipitation was done with antibodies to the indicated proteins from lysates of HBECs, and Western blots done with anti-phosphotyrosine and protein-specific antibodies.
Functional Classification of Identified Proteins by DAVID.
The complete list of proteins that we identified in the experiments with phosphotyrosine IPs of proteins is summarized in Table S2. A partial list of those proteins known and not known to be involved in EGFR signaling is summarized in Table 1. To group the proteins identified in phosphotyrosine IPs into functional classes, we performed functional annotation using DAVID, the Database for Annotation, Visualization and Integrated Discovery (17), and then filtered, named and ranked the resulting clusters (see SI Methods for details). Overall, 26 clusters contained one or more significant overlaps with Gene Ontology (GO) biological process categories, and 14 of these clusters were considered significant after correction based on the method of Benjamini and Hochberg (18) (Fig. S7). Notably, 17 proteins identified in at least one of the large-scale experiments were in receptor tyrosine kinase (RTK) signaling pathways. Seven of these, EGFR, ERBB2, SRC, SHC, BCAR1, PXN, and EPHA2, were more abundant in phosphotyrosine IPs from cells expressing mutant EGFR compared with those from cells expressing WT EGFR or mutant KRAS.
Table 1.
Representative list of proteins identified, and the ratio of their relative abundance in phosphotyrosine immunoprecipitates in the three “large-scale” phosphoproteomic studies
| Protein name | HBEC Experiment #1 |
HBEC Experiment #2 |
H1650 (DelEGFR) /H2030 (KRASG12C) | ||
|---|---|---|---|---|---|
| DelEGFR /WTEGFR | KRASG12V /WTEGFR | DelEGFR /WTEGFR | L858R /WTEGR | ||
| Proteins implicated in EGFR signaling pathway | |||||
| EGFR | 5.2 | 0.6 | 10.4 | 9.6 | 3.8 |
| ERBB2 | 3.5 | 1.0 | 3.5 | 3.9 | |
| MET | 3.2 | 3.4 | 2.3 | 1.7 | — |
| SRC | 1.8 | 0.6 | — | — | — |
| SHC1 | 2.2 | 0.9 | 1.8 | 1.9 | 0.7 |
| BCAR1 | 1.4 | 0.6 | 1.2 | 1.3 | 1.5 |
| PXN | 1.5 | 0.8 | 1.4 | 1.2 | 1.6 |
| PIK3R1 | 1.4 | 1.5 | — | — | 1.1 |
| GRB2 | 1.0 | 1.1 | 1.2 | 0.6 | 0.6 |
| CTNND1 | 1.9 | 1.0 | 3.1 | 2.4 | 8.2 |
| CTNNB1 | 2.3 | 0.7 | 1.9 | 3.3 | 7.2 |
| CAV1 | 6.8 | 1.1 | 5 | 5.2 | 4.5 |
| CAV2 | — | — | 6.8 | 7.2 | 3.5 |
| ERRFI1 | 8.0 | 0.9 | — | — | — |
| Proteins not previously implicated in EGFR signaling pathway | |||||
| IGF1R | 1.5 | 1.0 | 0.9 | 1.3 | — |
| CTNNA1 | 1.5 | 0.8 | — | — | 5.7 |
| CDH1 | 1.8 | 1.1 | 1.9 | 4.8 | 15.6 |
| JUP | 2.5 | 0.8 | 2.0 | 4.4 | 5.1 |
| RAB7A | 11.2 | 0.9 | 7.2 | 2.2 | 1.9 |
| PRKCDBP | 4.7 | 0.7 | 3.5 | 4.1 | |
| SLC25A5 | 2.5 | 0.7 | 3.2 | 1.7 | 0.3 |
| BICD2 | 8.5 | 0.9 | 6.7 | 4.4 | — |
| PTRF | 5.3 | 0.8 | 4.9 | 5.4 | — |
| EMD | 4.4 | 1.4 | 1.6 | 2.5 | — |
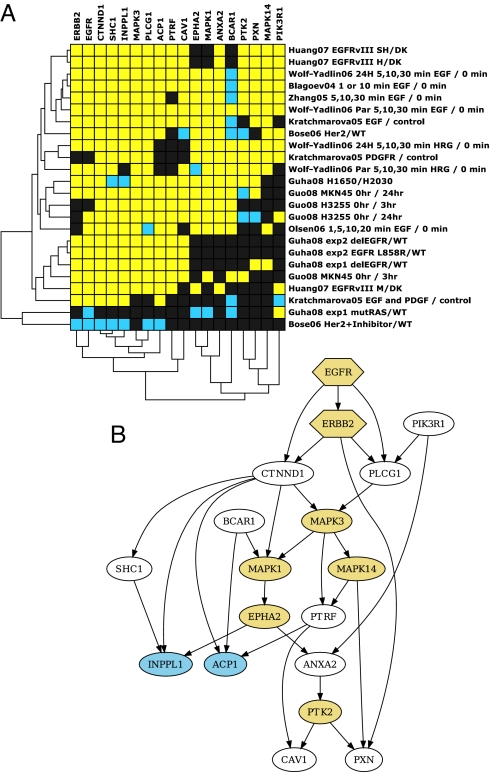
Bayesian Network Modeling of ERBB Signaling-Related Mass Spectrometry Datasets.
We compared our datasets with eight other published proteomic studies on ERBB signaling pathways to gain a better understanding of tyrosine phosphorylation signaling networks (14, 19–25). Each study contained one or more experiments performed under different conditions to study an EGFR-related signaling network. The phosphorylation ratio obtained from each of these experiments was considered as an observation. This resulted in 24 total observations of EGFR-related signaling networks for our analysis. We developed a “heat map” that reflects the phosphorylation status of 50 proteins that were identified in at least half of these 24 experiments (Fig. S8A). Bayesian network (BN) structure learning was used as a machine learning method to propose a network of interactions that are responsible for the tyrosine phosphorylation of 18 proteins identified in at least 70% or more of the studies (Fig. S8B) after estimation of missing data by a “nearest neighbors” algorithm (Fig. 5A). The highest scoring network generated from this data showed known features of EGFR and ERBB2 signaling, such as EGFR phosphorylation having a positive influence on phosphorylation of ERBB2 and PLCG1. The network also included PTRF as an integral member of the ERBB signaling (Fig. 5B), and we confirmed that EGFR interacts with PTRF by IP Western experiments (Fig. S5).
Fig. 5.
Bayesian network modeling of phosphorylation data from the current study and eight other published ERBB signaling related tyrosine phosphorylation data. (A) The heatmap of the 18 proteins with the approximations for the missing data generated by the “nearest neighbor method” using a discrete distance measure. (B) The top scoring Bayesian network generated from the above phosphorylation data. The nodes are obtained from the phosphorylation ratios from the datasets. The edges represent direct and indirect causal influence. The arrows indicate the direction of causality. Nodes connected by black edges have the same phosphorylation level more often than not, indicating a positive influence. EGFR and ERBB2, were forced source nodes in the network; they were only allowed outgoing edges except between each other. Nodes were restricted to have no more than three parents. Yellow nodes are kinases and blue nodes phosphatases.
Discussion
We used mass spectrometry to identify proteins in phosphotyrosine IPs from whole cell extracts, an approach that has been used extensively to probe the dynamics of tyrosine phosphorylation and tyrosine phosphorylation-dependent interaction after growth factor stimulation (19, 26). In addition, we enriched tyrosine phosphorylated peptides by immunoprecipitation to quantify differences in degree of tyrosine phosphorylation, at specific sites, within a protein.
Our survey of tyrosine phosphorylated proteins showed that either of the common forms of oncogenic EGFR causes increased tyrosine phosphorylation of numerous signaling molecules, either directly or indirectly, compared with WT EGFR or mutant KRAS. In addition, some proteins enriched in antiphosphotyrosine immunoprecipitates may not themselves be phosphorylated, but may be more likely to associate with proteins that are more efficiently tyrosine phosphorylated in cells expressing mutant EGFR than in cells expressing WT EGFR or mutant KRAS.
In some systems, activated RAS proteins appear to augment RTK signaling, for example, by increasing the production of EGFR ligands, such as TGFα (27). We did not find evidence for this in HBECs expressing oncogenic KRAS. However, 52 proteins were more abundant in phosphotyrosine IPs from the adenocarcinoma cell line H2030 (harboring mutant KRAS), than in the cell line H1650 (with mutant EGFR). We contend that the likely explanation for these unexpected findings is the difference in concentrations of such proteins in the two adenocarcinoma cell lines, as exemplified by the lower levels of RIN1 or the p66 isoform of SHC in the H1650 cell line. These findings underscore the importance of studying differences in the status of signaling proteins by using isogenic cells (such as the HBEC lines) that express different oncogenes or by comparing a single cell line under different conditions (e.g., with and without growth factors or in the presence and absence of inhibitors).
We identified 13 proteins that were hyperphosphorylated in HBECs expressing mutant EGFR and in the adenocarcinoma cell line H1650. Among these were RTKs, such as EGFR, ERBB2, and MET, suggesting a role for lateral signaling or cross talk between various RTKs, with subsequent signaling through multiple receptors. In support of this possibility, multiple RTKs have been shown to be active in glioblastoma cell lines, and combinations of RTK inhibitors can reduce cell survival and downstream signaling in these cells (28). MET has been shown to interact with EGFR (14), and combination therapy with EGFR and MET inhibitors is better than EGFR inhibitors alone in glioblastoma cells expressing the oncogenic EGFR-VIII mutant (21).
Cell junction proteins that have been implicated in ERBB signaling, such as β-catenin, E-cadherin, δ-catenin, and junctional plakoglobin, were also more abundant in phosphotyrosine IPs from cells expressing mutant EGFR. Others have reported that β-catenin-ERBB complexes are present in breast tumors in MMTV-Wnt1 and MMTV-neu transgenic mice, and in infiltrating ductal breast carcinomas in women (29). Recently, AKT-mediated phosphorylation of β-catenin downstream of EGFR signaling has been linked to increased transcriptional activity of β-catenin and tumor cell invasion (30), suggesting that increased tyrosine phosphorylation of β-catenin and E-cadherin may be important in transducing signals from the oncogenic EGFRs.
We also identified proteins that were hyperphosphorylated in mutant EGFR-expressing cell lines, but not previously implicated in EGFR signaling (see Table 1). One such protein, PTRF, co-localizes with caveolin 1 in caveolae in adipocytes and has been implicated in the formation of caveolae (31, 32, 39). Bayesian network modeling of proteins, based on their degree of tyrosine phosphorylation in various ERBB signaling and mass spectrometry-based studies, including ours, reveals that PTRF may be an integral member of ERBB signaling network (Fig. 5B).
The overall level of tyrosine phosphorylation in EGFR or ERBB2 was similar in HBECs expressing L858R EGFR and Del EGFR. However, there were interesting differences at individual sites of phosphorylation within EGFR. The most significant of these sites was Y727 of EGFR, the site corresponding to Y735 of ERBB2 or Y733 of ERBB4. This site was phosphorylated in the Del-EGFR-expressing cells but not in L858R or WT EGFR-expressing HBEC cells. Although this site was found to be phosphorylated in adenocarcinoma cell lines that harbor the L858R mutation (H3255) and a line that carries the Del EGFR allele (HCC827) (14), the degree of phosphorylation at this site was not compared between the cell lines expressing the two EGFR mutants. Ward et al. have shown that a synthetic peptide of EGFR, phosphorylated at Y727, binds to purified SHC SH2 domain in vitro (33). In addition, phosphorylated peptides containing Y727 of EGFR, Y735 of ERBB2, or Y733 of ERBB4 have been shown to bind SHC in lysates of HeLa cells (34). We speculate that phosphorylation at the above site in EGFR, ERBB2, or ERBB4 occurs in a specific signaling context, and phosphorylation may recruit certain signaling molecules having SH2 domains. Phosphorylation at this site may also alter the structure of the catalytic site in the ERBBs to affect kinase activity.
A few proteins involved in ERBB receptor recycling, such as Caveolins, Rab7, Rab8, and the protein Mig-6, a known inhibitor of ERBB signaling had increased phosphorylation in cells expressing either of the mutant EGFRs. Mig-6 has been shown to act as a tumor suppressor, and germ-line disruption of Mig-6 results in adenoma or adenocarcinomas in the lung (35). The crystal structure of a fragment of Mig-6 and the EGFR kinase domain has shown that Mig-6 inhibits the formation of an activating dimer interface in EGFR. The C-terminal fragment of Mig-6 containing the Y394 site increases the potency of inhibition of the L858R EGFR kinase activity in solution (36). From these studies, we speculate that Y394 of Mig-6 is a direct substrate of EGFR. The significance of increased phosphorylation at this site by the mutant EGFRs remains to be determined.
Materials and Methods
Cell Culture.
HBECs were cultured as described before (16). SILAC methods were used as described in ref. 37. See SI Methods for details.
Mass Spectrometry.
Details of sample preparation and mass spectrometry can be found in SI Methods. Peptide samples were analyzed using nanoscale reversed phase liquid chromatography on a QSTAR Pulsar (Applied Biosystems). LC-MS/MS data were acquired using AnalystQS 1.1 (MDS Sciex) and were searched using Mascot v2.2.0 (Matrixscience). Relative quantitation of the abundance of peptides in phosphotyrosine IPs was performed using MSQuant (http://msquant.sourceforge.net) along with manual verification of all peptides quantified.
Bayesian Network Analysis.
We used the general method of data discretization and network modeling as outlined in ref. 20. Data from our study and the eight published ERBB signaling-related phosphoproteomic studies were used for network modeling (14, 19–25). Bayesian networks (BN) were generated using the BN structure learning software Banjo 2.0.0 with a simulated annealing edge searcher that considered one billion potential networks (38). (see SI Methods for details).
Supplementary Material
Acknowledgments.
We thank Marjan Gucek (Johns Hopkins University School of Medicine Proteomics core facility) for mass spectrometry operation; Henrik Molina for advice regarding mass spectrometry analysis; Bruce E. Johnson (Dana-Farber Cancer Institute, Boston, MA) for the H3255 cell line; William Pao, Katerina Politi, and Phil Cole for careful reading of the manuscript. This work was supported by National Institutes of Health grants PO1 CA129243–01 (H.E.V. and A.E.L.), Labrecque foundation grant and American Cancer Society postdoctoral fellowship (U.G.), NIH Road map Grant US4RR020839 (A.P., J.B., and H.E.V.), National Science Foundation Career Award 0546446 (J.B.), National Cancer Institute P50CA70907, and Department of Defense VITAL grants (J.D.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806158105/DCSupplemental.
References
- 1.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Politi K, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji H, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Greulich H, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. Plos Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang J, et al. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib -sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 10.Jackman DM, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 11.Riely GJ, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 12.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 13.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Guo A, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser. 1995;B57:289–300. [Google Scholar]
- 19.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 20.Bose R, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci USA. 2006;103:9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang PH, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 23.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 24.Wolf-Yadlin A, et al. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Blagoev B, et al. A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 27.Gangarosa LM, et al. A raf-independent epidermal growth factor receptor autocrine loop is necessary for Ras transformation of rat intestinal epithelial cells. J Biol Chem. 1997;272:18926–18931. doi: 10.1074/jbc.272.30.18926. [DOI] [PubMed] [Google Scholar]
- 28.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder JA, et al. ErbB-beta-catenin complexes are associated with human infiltrating ductal breast and murine mammary tumor virus (MMTV)-Wnt-1 and MMTV-c-Neu transgenic carcinomas. J Biol Chem. 2002;277:22692–22698. doi: 10.1074/jbc.M201975200. [DOI] [PubMed] [Google Scholar]
- 30.Fang D, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem. 2007;282:11221–11229. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aboulaich N, Vainonen JP, Stralfors P, Vener AV. Vectorial proteomics reveal targeting, phosphorylation and specific fragmentation of polymerase I and transcript release factor (PTRF) at the surface of caveolae in human adipocytes. Biochem J. 2004;383:237–248. doi: 10.1042/BJ20040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Pilch PF. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J Biol Chem. 2008;283:4314–4322. doi: 10.1074/jbc.M707890200. [DOI] [PubMed] [Google Scholar]
- 33.Ward CW, et al. Systematic mapping of potential binding sites for Shc and Grb2 SH2 domains on insulin receptor substrate-1 and the receptors for insulin, epidermal growth factor, platelet-derived growth factor, and fibroblast growth factor. J Biol Chem. 1996;271:5603–5609. doi: 10.1074/jbc.271.10.5603. [DOI] [PubMed] [Google Scholar]
- 34.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005 doi: 10.1038/msb4100012. msb4100012-E1–E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YW, et al. Evidence that MIG-6 is a tumor-suppressor gene. Oncogene. 2007;26:269–276. doi: 10.1038/sj.onc.1209790. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 38.Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- 39.Hill MM, et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.