Abstract
Myeloproliferative disorders (MPDs) are stem cell-derived clonal diseases arising as a consequence of acquired aberrations in c-ABL, JAK2, and PDGFR, that generate oncogenic fusion tyrosine kinases (FTKs) including BCR/ABL, TEL/ABL, TEL/JAK2, and TEL/PDGFβR. Here we show that FTKs stimulate the formation of reactive oxygen species (ROS) and DNA double-strand breaks (DSBs) both in hematopoietic cell lines and in CD34+ leukemic stem/progenitor cells from patients with chronic myeloid leukemia (CML). Single-strand annealing (SSA) represents a relatively rare, but very unfaithful DSB repair mechanism causing chromosomal aberrations. Using a specific reporter cassette integrated into genomic DNA, we found that BCR/ABL and other FTKs stimulated SSA activity. Imatinib-mediated inhibition of BCR/ABL abrogated this effect implicating a kinase-dependent mechanism. Y253F, E255K, T315I, and H396P mutants of BCR/ABL that confer imatinib resistance also stimulated SSA. Increased expression of either non-mutated or mutated BCR/ABL kinase, as is typical of blast phase cells and very primitive chronic phase CML cells, was associated with higher SSA activity. BCR/ABL-mediated stimulation of SSA was accompanied by enhanced nuclear co-localization of RAD52 and ERCC1, which play a key role in the repair. Taken together, these findings suggest a role of FTKs in causing disease progression in MPDs by inducing chromosomal instability through the production of DSBs and stimulation of SSA repair.
Keywords: SSA, FTK, leukemia, genomic instability
INTRODUCTION
Myeloproliferative disorders (MPDs) are stem cell-derived clonal proliferative diseases whose shared and diverse phenotypic characteristics can be attributed to dysregulated signal transduction events caused by acquired somatic aberrations in genes encoding tyrosine kinases such as c-ABL, JAK2, and PDGFR (1). For example, chromosomal translocations are responsible for the appearance of BCR/ABL and related oncogenic fusion tyrosine kinases (FTKs) including TEL/ABL, TEL/JAK2, and TEL/PDGFβR, all of which have been associated with MPDs. BCR/ABL is derived from a relocation of the TK-containing portion of the c-ABL gene from chromosome 9 to a portion of the BCR gene locus on chromosome 22 [t(9;22)] and is now considered a defining feature of chronic myelogenous leukemia (CML). TEL/ABL results from a t(9;12) translocation reported in atypical CML (aCML) cases and consists of the amino terminal fragment of the TEL domain fused in-frame with exon 2 of ABL. TEL/JAK2 was also found in aCML and is a product of a t(9;12) translocation which includes the TEL oligomerization domain and the JAK2 catalytic domain. TEL/PDGFβR results from a t(5;12) translocation which juxtaposes the amino terminal region of TEL to the transmembrane and TK domains of the platelet derived growth factor β receptor (PDGFβR) and is found in chronic myelomonocytic leukemia (CMML).
FTKs contribute to oncogenesis in two complementary ways: they activate signaling pathways that render cells less dependent on their environment and they modulate responses to DNA damage, thereby promoting both resistance to genotoxic therapies and chromosomal instability (2). MPDs expressing FTKs, if not treated, tend to accumulate additional genetic aberrations eventually leading to the appearance of more malignant subclones and evolution of the disease into a more aggressive phase.
This is well documented for the transition of CML from a relatively benign chronic phase (CP) to the rapidly fatal blast crisis (BC), a transition that is usually also accompanied by an increased level of expression of the BCR/ABL kinase in the predominant circulating population, the emergence of imatinib-resistant BCR/ABL cells and accumulation of chromosomal aberrations (3–5). The frequency of additional chromosomal abnormalities is around 7% in CML-CP and increases to 40–70% in the advanced phases. Progression to a cytogenetically evolved acute leukemia is also frequently seen in CMML cases and has also been reported in aCML (6, 7).
Chromosomal aberrations such as translocations and partial deletions or duplications are usually caused by unfaithful repair of DNA double-strand breaks (DSBs). Human cells may use at least three different repair mechanisms: homologous recombination (HR), non-homologous end-joining (NHEJ) and single-strand annealing (SSA) to deal with DSBs that represent a “clear and present danger” to cell survival and genomic integrity (8). SSA by definition represents a very unfaithful type of repair because it arises from the annealing of complementary single strands formed after extensive resection at a DSB. Thus, when sequence repeats are present near a DSB, they can undergo SSA, resulting in the deletion of sequences between the repeats (e.g., tumor suppressor genes). When present on nearby chromatids the result may be a translocation (9). Because ~50% of the mammalian genome consists of repeat sequences (e.g., Alu), SSA is a potentially important pathway of mutagenesis.
Large submicroscopic intra-chromosomal deletions in regions with high overall density of Alu sequence repeats have been detected in BCR/ABL-positive leukemias implicating SSA activity (10). This finding and our previous report that BCR/ABL-transformed cells contain numerous DSBs prompted us to investigate the potential influence of FTKs on SSA activity (11).
MATERIALS AND METHODS
FTK-transformed cells
Murine growth factor-dependent 32Dcl3 myeloid cells were transfected with pSR〈 or pMSCV retroviral expression plasmids encoding BCR/ABL, TEL/ABL, TEL/JAK2 or TEL/PDGFβR kinases and neomycin resistance as previously described (12). Imatinib-resistant BCR/ABL mutants (P-Loop Y253H and E255K, T315I and Activation Loop H396P) in the pEYK3.1 retroviral vector were obtained from Dr. George Daley (Dana Farber Cancer Institute, Boston, MA, USA). Parental cells were used after being transduced with control plasmids and selection for G418 resistance. Cells were cultured in the presence of pre-tested concentrations of IL-3 necessary to maintain the proliferation of parental cells.
Primary cells
Peripheral blood mononuclear cells or CD34+ cells from healthy donors (n=13) were purchased from StemCell Technologies and those from CP (n=11) and BC (n=6) CML patients were obtained from the Stem Cell and Leukemia Core Facility of the University of Pennsylvania (Philadelphia, PA, USA), the Terry Fox Laboratory of the British Columbia Cancer Agency (Vancouver, BC, Canada), and the 2nd Department of Internal Medicine, Oncology and Hematology, Robert Bosch Hospital, Stuttgart, Germany. All cells were obtained with informed consent according to the practices of the host institutions. Lineage marker-negative (Lin−) CD34+ cells were isolated immunomagnetically immediately after thawing using first the EasySep Negative Selection Human Progenitor Cell Enrichment Cocktail followed by the EasySep Human CD34 Positive Selection Kit (StemCell Technologies, Inc., Vancouver, BC, Canada).
Reactive oxygen species (ROS) assay
Levels of intracellular ROS were analyzed using the redox-sensitive fluorochrome 2’,7’-dichlorofluorescein-diacetate (DCFDA) (Sigma) as previously described (13). The oxidized form of DCFDA, carboxy-DCFDA (Molecular Probes, Eugene, OR, USA), was used as a control for uptake, retention, and decay.
SSA assay
The SA-GFP reporter containing the I-SceI-inducible DSB site was generously provided by Maria Jasin (Sloan-Kettering Cancer Center, New York, N.Y) (14) and integrated into the genome of 32Dcl3 cells, which were then transfected with retroviral expression constructs containing various FTKs. Growth factor-independent cell mixtures were obtained and expression of particular FTK and its tyrosine kinase activity was confirmed by Western analysis (Supplementary Figure 1). Additional cells were transfected with an otherwise empty neomycin resistance-encoding retroviral construct and selected for resistance to G418 to provide control cells. SSA activity was examined as described before for HR (15). Briefly, cells were electroporated with 100 µg of pCβA-Sce expression plasmid encoding I-SceI endonuclease and 20 µg of pDsRed1-Mito (Clontech, Paolo Alto, CA). Expression of I-SceI causes a DSB in the specific restriction site included in the SA-GFP cassette, and pDsRed1-Mito encodes red fluorescent protein with a mitochondrial localization signal to control the efficiency of transfection (approximately 35%). SSA activity was determined as the number of GFP+/Red1+ cells in 105 Red1+ cells. When indicated, cells were incubated with 1 µM imatinib mesylate (Novartis Pharma AG, Basel, Switzerland)) starting from 24 hours before transfection with I-SceI and continuing until the end of experiment. The fragments of the SA-GFP cassette containing DSB repair site were amplified by PCR from GFP+ and GFP− cells using the following primers: 1 5’-ATGGTGAGCAAGGGCGAGGA, 2 5’-AAAGACCCCAACGAGAAGCGCGAT, and 3 5’-TTACTTGTACAGCTCGTCCAT. The products from GFP+ cells were sequenced to confirm restoration of GFP sequence accompanied by the loss of the I-SceI restriction site.
Western analysis
Total cell lysates were prepared as previously described (16). Proteins were examined by Western blotting with the use of antibodies recognizing RAD52 (Cell Signaling Technology, Inc., Beverly, MA, USA), ERCC1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and actin (Santa Cruz).
Immunofluorescence
Nuclear localization of the indicated proteins was detected by immunofluorescence, as previously described (11). Cells were stained first with antibodies against γ-H2AX (Upstate Biotechnology, Lake Placid, NY, USA), or RAD52 (Cell Signaling) and ERCC1 (Santa Cruz) followed by secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes). Negative controls were performed without primary antibodies. DNA was counterstained with 4’,6’diamedino-2-phenylindole (DAPI). Specific staining was visualized using an inverted Olympus IX70 fluorescence microscope equipped with 100x UPlan Apo lens (numeric aperture 1.35), and a Cooke Sensicam QE camera (The Cooke Company, Auburn Hills, MI, USA). At least 50 individual cells were analyzed per experimental group. Images were acquired with Slidebook 3.0 (Intelligent Imaging Innovations, Denver, CO, USA). All graphic adjustments were performed using Adobe Photoshop.
RESULTS AND DISCUSSION
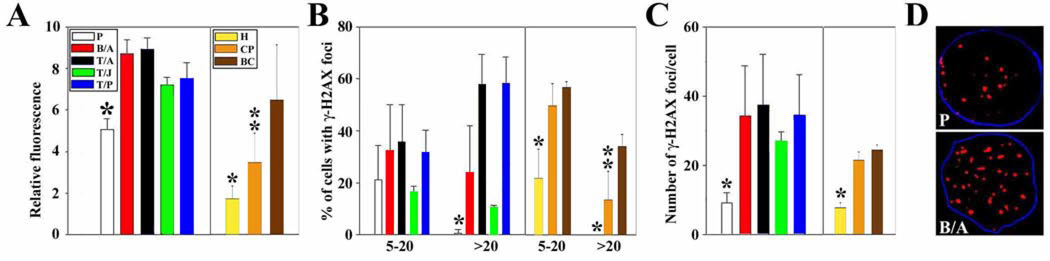
Our previous report showed that BCR/ABL kinase elevated the amount of ROS in leukemia cells, which increased the number of DSBs (11). Here we show that 32Dcl3 cells transformed by BCR/ABL and other FTKs such as TEL/ABL, TEL/JAK2 and TEL/PDGFβR (Supplementary Figure 1) display higher levels of ROS than the parental cells (Figure 1A, left panel). More ROS were also detected in primary Lin−CD34+ CP and BC CML cells in comparison to their normal counterparts (Figure 1A, right panel).
Figure 1. FTKs enhance the production of ROS and DSBs.
Results in BCR/ABL (B/A), TEL/ABL (T/A), TEL/JAK2), and TEL/PDGFβR (T/P) – transformed cells were compared with results in G418-resistant control cells (P). Results in Lin−CD34+ CML CP and BC cells were compared with Lin−CD34+ peripheral blood cells isolated from healthy volunteers (H). (A) ROS levels were measured by fluorescence. (B,C) γ-H2AX nuclear foci were detected by immunofluorescence; the numbers indicate (B) the percentage of cells with 4–20 and >20 γ-H2AX foci/cell and (C) the mean number of foci/nucleus in cells containing ≥ 4 foci. (D) Representative nuclei containing γ-H2AX foci; nuclei borders are marked in blue. p<0.05 in comparison to other groups (*) and to BC (**).
By comparison to their respective controls, BCR/ABL and other FTK-transformed 32Dcl3 cells as well as primary Lin−CD34+ CP and BC CML cells also showed an increase in γ-H2AX nuclear foci (Figure 1B and C), which paint DSBs (Figure 1D) (11). This was seen both as a significant increase in the percentage of leukemic cells containing more than 20 γ-H2AX foci (Figure 1B, left panel) as well as in the average number of γ-H2AX foci per cell (Figure 1C, left panel). Based on our previous reports (11, 13) and this work, we postulate that FTK-mediated enhancement of ROS may be responsible for the generation of an excess of DSBs in leukemic cells.
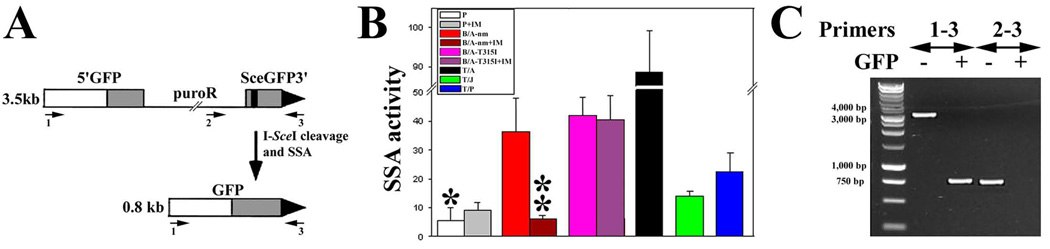
DSBs are usually repaired by HR and NHEJ, however, a relatively rare and extremely unfaithful SSA pathway can occasionally be used (8). To examine the potential influence of FTK expression on SSA activity, cells carrying an integrated SA-GFP reporter cassette (Figure 2A, upper diagram) were generated as described in the Methods. Cells transformed by BCR/ABL or other FTKs displayed a 6 to 16-fold enhanced SSA activity by comparison to control cells (Figure 2B). Imatinib completely abolished the non-mutated BCR/ABL kinase-mediated activation of SSA without exerting any effect on enhanced SSA activity stimulated by the imatinib-resistant BCR/ABL-T315I mutant kinase or on the basal SSA activity measured in parental cells. This clearly shows that the enhanced SSA activity was dependent on the functionality of the BCR/ABL kinase. The various degrees of SSA stimulation obtained by different FTKs may reflect quantitative and qualitative differences in their specific kinase activities.
Figure 2. FTKs stimulate SSA.
(A) The structure of SA-GFP reporter cassette is shown before (upper panel, GFP− cells) and after (lower panel, GFP+ cells) I-SceI cleavage and SSA. The cassette consists of the 5’GFP and SceGFP3’ fragments, which have 266 bp of homology and intervening sequence encoding puromycin-resistance (puroR). The black strip represents the I-SceI site in the SceGFP3’ and the large black triangle depicts the 3’ end of the cassette. Repair of the I-SceI generated DSB in SceGFP3’ by SSA results in a functional GFP gene when a DNA strand from SceGFP3’ is annealed to the complementary strand of 5’GFP, followed by appropriate DNA-processing steps. As a result, SSA between the homologous sequences in the GFP gene fragments produces a 2.7-kb deletion in the chromosome. The SA-GFP reporter can also be repaired by HR and NHEJ, but without restoration of a functional GFP gene (14). (B) I-SceI and Red1-Mito were expressed in parental (P) and FTK-transformed (BCR/ABL non-mutated = B/A-nm, BCR/ABL-T315I mutant = B/A-T315I, TEL/ABL = T/A, TEL/JAK2 = T/J, and TEL/PDGFβR = T/P) cells containing SA-GFP reporter cassette and cultured in the presence of IL3 and imatinib (IM) when indicated. SSA activity was determined as the number of GFP+/Red1+ cells in 105 Red1+ cells. * p<10−8, <10−8, <10−8, 10−2 and <10−7 in comparison to B/A-nm, B/A-T315I, T/A, T/J and T/P, respectively; and ** p<0.03 in comparison to B/A. (C) PCR products from genomic DNA of GFP+ and GFP− cells.
PCR reactions were performed on genomic DNA from GFP− and GFP+ cells using the primers shown in Figure 2A. The results confirmed that functional GFP gene in GFP+ cells was recovered by SSA. As expected, PCR reactions with primers 1–3 generated ~3.5 kb and ~0.8 kb bands from GFP− and GFP+ cells, respectively, detecting intact SA-GFP reporter cassette and SSA-restored GFP gene (Figure 2C). Thus, SSA-mediated DSB repair resulted in genomic instability associated with a loss of ~2.7 kb chromosome fragment. In contrast, PCR reactions with primers 2–3 amplified ~0.8 kb band from GFP− cells and no band from GFP+ cells confirming that a segment containing puroR had been lost during SSA.
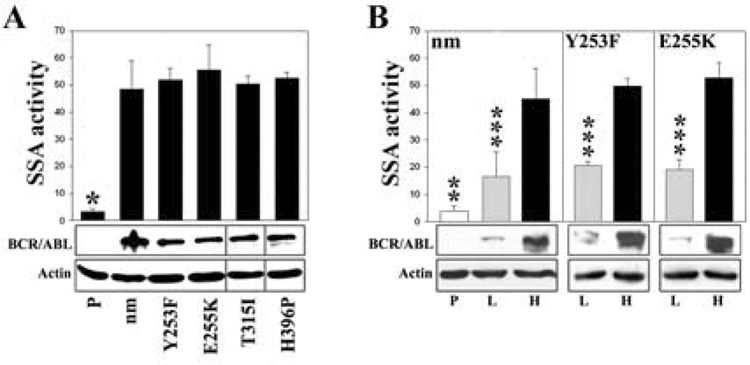
Imatinib-resistant BCR/ABL kinase mutants may promote malignant progression in CML patients being treated with imatinib due to the different kinase activities and transforming properties they endow on hematopoietic cells (17, 18). Therefore, it was of interest to use the SA-GFP reporter system in a similar fashion to compare the SSA activity in 32Dcl3 cells expressing non-mutated and mutated BCR/ABL known to confer imatinib resistance (e.g.; P-Loop Y253H and E255K, T315I, and Activation Loop H396P). Accordingly, additional lines carrying the reporter construct and these mutant forms of BCR/ABL were constructed and analyzed (Figure 3A, bottom panel). The results showed that imatinib-resistant BCR/ABL kinase mutants stimulated SSA in a similar manner as the non-mutated kinase (Figure 3A, upper panel).
Figure 3. Non-mutated and imatinib-resistant BCR/ABL kinase mutants stimulate SSA in a dose-dependent manner.
(A) Similar high levels of non-mutated (nm), Y253F, E255K, T315I and H396P BCR/ABL kinase proteins, and (B) low (L) and high (H) levels of nm, Y253F, and E255K BCR/ABL kinase proteins were expressed in parental cells (P) containing the SA-GFP reporter cassette (lower panels). Cells were transfected with I-SceI and Red1-Mito and maintained in the presence of IL-3. SSA activity was determined as the number of GFP+/Red1+ cells in 105 Red1+ cells (upper panels). * p<10−7 in comparison to other groups, ** p<10−2, <10−4, and <10−3 in comparison to L groups of nm, Y253F, and E255K, respectively; and *** p=0.02, 0.006, and 0.02 in comparison to corresponding H groups.
Enhanced levels of BCR/ABL kinase expression were noted in comparisons of the cells circulating in the blood of CML patients with BC disease as compared to CP (5). In addition, increased levels of BCR/ABL kinase have been found in the CD34+ CML cells which contain the leukemic stem and progenitor cells responsible for propagation of the malignant clone (19). Therefore, we also investigated the impact of the level of BCR/ABL gene expression on SSA in 32Dcl3 cells expressing high and low levels of BCR/ABL (Figure 3B, bottom panel). Interestingly, the results showed a 5-fold stimulation of SSA in cells expressing lower levels of BCR/ABL and a 15-fold stimulation of SSA activity in cells expressing higher levels of BCR/ABL (Figure 3B, upper panel). The evidence of a BCR/ABL “dose-dependent” stimulation of SSA activity was also seen in cells expressing imatinib-resistant BCR/ABL mutants, for example Y253H and E255K (Figure 3B, upper panel).
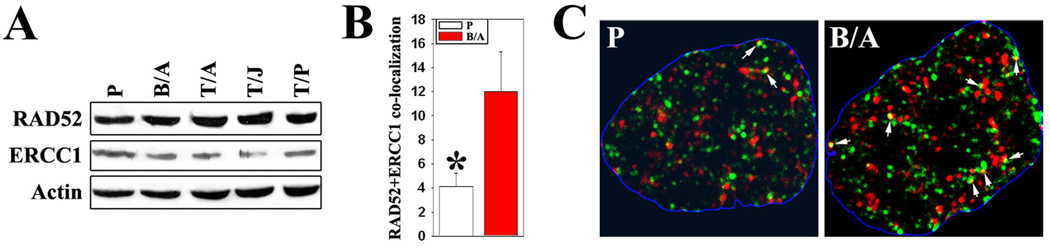
A current model of SSA assumes that the RAD52 protein directs the annealing of complementary single strands of DNA and ERCC1/XPF endonuclease is involved in strand-processing steps (20). FTKs and IL-3 were independently able to stimulate the expression of RAD52 and ERCC1 in 32Dcl3 cells (data not shown). FTK-transformed cells did not manifest significant changes in expression of RAD52 and ERCC1 proteins in comparison to control cells incubated under conditions where FTK-transformed cells showed enhanced SSA (Figure 4A). Interestingly BCR/ABL-positive cells also displayed enhanced co-localization of these proteins in the nucleus in comparison to control cells (Figure 4B and C) suggesting functional modifications of RAD52 and/or ERCC1 in the leukemic cells.
Figure 4. BCR/ABL facilitates RAD52-ERCC1 co-localization.
(A) Western analysis of RAD52 and ERCC1 expression in parental and FTK-transformed cells cultured in the presence of IL-3. Actin served as loading control. (B) Detection of RAD52 (green), ERCC1 (red) and co-localizing (yellow) foci in parental and BCR/ABL-positive cells presented as percentage of RAD52 + ERCC1 staining/RAD52 staining. * p<10−5 in comparison to B/A. (C) Representative nuclear staining for RAD52 and ERCC1; white arrows indicate co-localization sites; nuclei borders are marked in blue.
In summary, we have shown that BCR/ABL, TEL/ABL, TEL/JAK2 and TEL/PDGFβR oncogenic tyrosine kinases stimulate SSA, a rare and unfaithful mechanism of DSB repair. Enhanced SSA may have a significant impact on accumulation of additional chromosomal abnormalities such as intrachromosomal deletions and translocations contributing to malignant progression and further treatment resistance of MPDs expressing FTKs (3–7). This speculation is further supported by the consistent demonstration of a “dose–dependent” effect of BCR/ABL on SSA activity, given the highly elevated levels of BCR/ABL expression found in CML stem and progenitor-enriched populations even prior to the emergence of BC (19). Taken together, these findings implicate FTKs as playing an important role in contributing to the generation of treatment resistant and progressed subclones in CML and other MPDs by induction of SSA.
Supplementary Material
ACKNOWLEDGEMENTS
T.S was supported by the grants from NIH/NCI 1R01CA89052 and the Department of Defense W81XWH-05-1-0214. E.T.P.P. was sponsored by the Physician Scientist Training Program 5R25DK059644-05 from NIH/NIDDK. C.J.E was supported by the Department of Defense CM064020 and the National Cancer Institute of Canada. We would like to thank Akhil Reddy and Jonathan Wosen for excellent technical assistance.
REFERENCES
- 1.Tefferi A, Gilliland DG. Oncogenes in myeloproliferative disorders. Cell Cycle. 2007;6:550–566. doi: 10.4161/cc.6.5.3919. [DOI] [PubMed] [Google Scholar]
- 2.Penserga ET, Skorski T. Fusion tyrosine kinases: a result and cause of genomic instability. Oncogene. 2007;26:11–20. doi: 10.1038/sj.onc.1209756. [DOI] [PubMed] [Google Scholar]
- 3.Bacher U, Haferlach T, Hiddemann W, et al. Additional clonal abnormalities in Philadelphia-positive ALL and CML demonstrate a different cytogenetic pattern at diagnosis and follow different pathways at progression. Cancer Genet Cytogenet. 2005;157:53–61. doi: 10.1016/j.cancergencyto.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Soverini S, Colarossi S, Gnani A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin Cancer Res. 2006;12:7374–7379. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 5.Neviani P, Santhanam R, Trotta R, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8:355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Fenaux P, Beuscart R, Lai JL, Jouet JP, Bauters F. Prognostic factors in adult chronic myelomonocytic leukemia: an analysis of 107 cases. J Clin Oncol. 1988;6:1417–1424. doi: 10.1200/JCO.1988.6.9.1417. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez JM, del Canizo MC, Cuneo A, et al. Clinical, hematological and cytogenetic characteristics of atypical chronic myeloid leukemia. Ann Oncol. 2000;11:441–444. doi: 10.1023/a:1008393002748. [DOI] [PubMed] [Google Scholar]
- 8.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res. 2001:480–481. 37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 9.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 10.Kolomietz E, Al-Maghrabi J, Brennan S, et al. Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood. 2001;97:3581–3588. doi: 10.1182/blood.v97.11.3581. [DOI] [PubMed] [Google Scholar]
- 11.Nowicki MO, Falinski R, Koptyra M, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104:3746–3753. doi: 10.1182/blood-2004-05-1941. [DOI] [PubMed] [Google Scholar]
- 12.Slupianek A, Hoser G, Majsterek I, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–4201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koptyra M, Falinski R, Nowicki MO, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108:319–327. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slupianek A, Nowicki MO, Koptyra M, Skorski T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair (Amst) 2006;5:243–250. doi: 10.1016/j.dnarep.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slupianek A, Schmutte C, Tombline G, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 17.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 18.Griswold IJ, MacPartlin M, Bumm T, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copland M, Hamilton A, Elrick LJ, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–4539. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 20.Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem. 2004;279:13634–13639. doi: 10.1074/jbc.M313779200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.