Abstract
The Arp2/3 complex was first purified from Acanthamoeba castellanii by profilin affinity chromatography. The mechanism of interaction with profilin was unknown but was hypothesized to be mediated by either Arp2 or Arp3. Here we show that the Arp2 subunit of the complex can be chemically cross-linked to the actin-binding site of profilin. By analytical ultracentrifugation, rhodamine-labeled profilin binds Arp2/3 complex with a Kd of 7 μM, an affinity intermediate between the low affinity of profilin for barbed ends of actin filaments and its high affinity for actin monomers. These data suggest the barbed end of Arp2 is exposed, but Arp2 and Arp3 are not packed together in the complex exactly like two actin monomers in a filament. Arp2/3 complex also cross-links actin filaments into small bundles and isotropic networks, which are mechanically stiffer than solutions of actin filaments alone. Arp2/3 complex is concentrated at the leading edge of motile Acanthamoeba, and its localization is distinct from that of α-actinin, another filament cross-linking protein. Based on localization and actin filament nucleation and cross-linking activities, we propose a role for Arp2/3 in determining the structure of the actin filament network at the leading edge of motile cells.
INTRODUCTION
Active motility and passive mechanical properties of eukaryotic cells depend on the actin cytoskeleton. Motile cells move across a substrate using actin polymerization to drive membrane protrusion at the leading edge (Tilney et al., 1981; Wang, 1985). Adherent cells transmit forces between sites of attachment via bundles and orthogonal networks of cross-linked actin filaments. Even nonmotile cells such as Saccharomyces cerevisiae and Schizosaccharomyces pombe rely on actin filaments to transport cellular components and to complete cell division. Despite our detailed understanding of actin biochemistry and the impressive catalog of known actin-binding proteins, a fundamental question remains unanswered: How exactly do eukaryotic cells control the organization of the actin cytoskeleton? To move or even to change shape, cells must somehow regulate, both spatially and temporally, the de novo nucleation of actin filaments as well as their organization into higher order structures such as bundles or isotropic networks.
One potentially important regulator of cytoskeletal organization is the Arp2/3 complex, a seven-subunit protein complex first identified by profilin affinity chromatography (Machesky et al., 1994). The complex is composed of two actin-related proteins, Arp2 and Arp3, and five novel proteins, p40, p35, p19, p18, and p14.
Five lines of evidence implicate Arp2/3 complex in regulation of the actin cytoskeleton. (1) The complex is ubiquitous and essential. The genes for Arp2 and Arp3 are highly conserved across eukaryotic phyla, apparently as ancient as actin and essential in both budding and fission yeast (for recent reviews see Frankel and Mooseker, 1996; Mullins et al., 1996; Machesky, 1997). This is not true for many proteins associated with the actin cytoskeleton, which appear to have overlapping or nonessential functions. (2) The Arp2/3 complex is directly associated with the actin cytoskeleton. In Acanthamoeba, Arp2/3 complex binds to and decorates the sides of actin filaments (Mullins et al., 1997), and in sea urchin egg extracts the complex binds F-actin affinity columns and cosediments with actin filaments (Terasaki et al., 1997). The complex also sediments with the cytoskeletal fraction after lysis of human platelets (Welch et al., 1997a). In vivo Arp2/3 complex is localized to actin-rich regions, especially the leading edges of motile cells and the actin-rich comet tails of Listeria monocytogenes (Machesky et al., 1994; Kelleher et al., 1995; Mullins et al., 1997, Welch et al., 1997a, 1997b) as well as to actin-rich cortical patches in S. pombe and S. cerevisiae (Balasubramanian et al., 1996; Moreau et al., 1996; Winter et al., 1997). (3) Arp2/3 complex interacts with the actin-regulatory protein, profilin. In vitro the complex binds profilin affinity columns and in S. pombe, homologs of Arp3 and p40 interact genetically with profilin (Machesky et al., 1994; Balasubramanian et al., 1996; McCollum et al., 1996). (4) In S. cerevisiae Arp3 function is required for proper organization and motility of cortical actin patches (Moreau et al., 1996; Winter et al., 1997) and for the internalization step of endocytosis (Moreau et al., 1997). (5) Along with partially purified mammalian cytoskeletal extracts, Arp2/3 complex is sufficient to reconstitute actin-based Listeria motility (Welch et al., 1997a).
Based on structural arguments, Kelleher et al. (1995) suggested that the Arp2/3 complex might nucleate de novo assembly of actin filaments. Welch et al. (1997a) invoked such a nucleation activity to account for the ability of Arp2/3 complex to induce the formation of actin clouds around Listeria cells. And recently we demonstrated that Arp2/3 complex from Acanthamoeba tightly caps the pointed ends of actin filaments and nucleates formation of filaments that grow from their barbed ends (Mullins and Pollard, unpublished data). These data suggest that the Arp2/3 complex plays an important role in initiating polymerization of actin at the leading edge of motile cells. Here we show that profilin binds the Arp2/3 complex by direct interaction with the Arp2 subunit. This is the first demonstration of direct binding between an actin-related protein and a conventional actin-binding protein, and it supports the idea of Kelleher et al. (1995) that surfaces on the barbed end of conventional actin are conserved on Arp2 and exposed at the surface of the Arp2/3 complex. We also show that in vitro Arp2/3 organizes actin filaments into small bundles and orthogonal networks of the type seen at the leading edges of motile cells, suggesting that the Arp2/3 complex not only regulates actin polymerization but also filament organization at the leading edge.
MATERIALS AND METHODS
Protein Purification
We purified Arp2/3 complex from Acanthamoeba: extract by both poly-L-proline affinity chromatography and by conventional column chromatography essentially as previously described (Machesky et al., 1994; Mullins et al., 1997) with some modifications. Cells were homogenized in either pyrophosphate extraction buffer as described before (Lynch et al., 1991; Machesky et al., 1994) or sucrose extraction buffer (10 mM Tris-HCl, pH 8.0, 11.6% sucrose, 1 mM EGTA, 1 mM ATP, 1 mM dithiothreitol (DTT), 0.1 mM benzamidine, 2 mg/l leupeptin, 20 mg/l soybean trypsin inhibitor, and 10 mg/l pepstatin A). For preparations using sucrose extraction, the extract was not dialyzed against a low-salt pyrophosphate buffer but was loaded directly onto diethylaminoethyl (DEAE)-cellulose equilibrated with 10 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.5 mM DTT, 0.5 mM ATP, 0.5 mM phenylmethylsulfonylfluoride, and 0.1 mM benzamidine (Tseng et al., 1984). For both sucrose and phosphate extraction procedures, flow-through from the DEAE-cellulose column was collected and loaded onto poly-L-proline Sepharose (Kaiser et al., 1989). Arp2/3 complex was eluted from the poly-L-proline Sepharose with 0.4 M MgCl2 in 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 100 mM glycine, and 1 mM DTT. Profilin was subsequently eluted with 8 M urea.
Purification of Arp2/3 complex by ion exchange chromatography was modified from the method originally used by Machesky et al. (1994). Briefly, the DEAE flow-through was adjusted to pH 6.0 by the addition of solid 2-[N-morpholino]ethanesulfonic acid (MES) and loaded onto P-11 phosphocellulose (PC) (Whatman, Maidstone, England) equilibrated with 25 mM MES, pH 6.0. The PC column was washed with 300 mM NaCl and eluted with 500 mM NaCl. Peak fractions were pooled, concentrated, and chromatographed in 300 mM NaCl on a Sephacryl S-300 gel filtration column. The Arp-containing pool from this column was then diluted to a final NaCl concentration of 100 mM and loaded onto a Mono-S FPLC column (Pharmacia Biotech, Piscataway, NJ). The flow-through from this column, containing the majority of the Arp2/3 complex, was loaded onto hydroxyapatite Bio-Gel HT (Bio-Rad, Hercules, CA), and the complex was eluted with 100 mM sodium pyrophosphate and 100 mM NaCl.
For storage, purified Arp2/3 complex was dialyzed into 10 mM imidazole (pH 7.5), 150 mM NaCl, 0.2 mM MgCl2, 0.2 mM ATP, and 1.0 mM dithiothreitol, concentrated by dialysis against solid sucrose, and stored on ice.
Acanthamoeba actin was purified from DEAE column fractions by polymerization-depolymerization and gel filtration (Pollard, 1984). Rabbit skeletal muscle actin was purified from acetone powder by the method of Spudich and Watt (1971) followed by gel filtration (MacLean-Fletcher and Pollard, 1980).
Chemical Cross-linking
The zero-length chemical cross-linker 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), along with N-hydroxysuccinimide (NHS) were obtained from Pierce (Rockford, IL). Stock solutions (20×) were made fresh in dry dimethylsulfoxide (DMSO) immediately before use. Final concentration of DMSO in all reactions was ≤10%. Reactions were carried out for 1 h at room temperature.
Preparation of Fluorescently-labeled Proteins
Acanthamoeba actin was labeled with pyrene iodoacetamide (Pollard, 1984). Acanthamoeba profilin-II mutants N58C and S38C were labeled with rhodamine maleimide (Vinson, De La Cruz, Kaiser, Higgs, Pollard, unpublished data).
Profilin Binding by Equilibrium Ultracentrifugation
We did equilibrium analytical ultracentrifugation in a Beckman model XL-A ultracentrifuge. We loaded samples containing rhodamine-labeled profilin-II into six-hole, charcoal-filled Epon centerpieces and centrifuged them to equilibrium (at least 20 h at each speed) in a Beckman model An60ti rotor. We monitored absorption at 550 nm, a wavelength specific for the rhodamine-label and collected data sets every hour. To monitor the approach to equilibrium we calculated total root mean square (RMS) deviation of each data set from the final data set collected using the program MATCH (Jeff Lary, National Analytical Ultracentrifuge Facility, Storrs CT). We considered experiments to have reached equilibrium when there was no change in RMS deviation of consecutive data sets. We parsed the data from individual samples into separate data sets using the program REEDIT (Jeff Lary, National Analytical Ultracentrifuge Facility).
Absorption optics allow us to selectively monitor labeled profilin in the presence of other proteins, to determine directly the distributions of free and bound profilin, and to calculate equilibrium binding constants (Schachman, 1960; Schachman et al., 1962). A similar method has been used previously to measure equilibrium binding of pyrene-labeled actobindin to actin (Bubb et al., 1994). First we fit each data set to a sum of two exponential functions by linear least squares methods
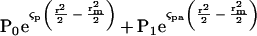 |
where Po and P1 are the fitting constants to be determined, r is radial position, rm is the radius of the meniscus, and ςp and ςpa are the effective reduced molecular weights of profilin alone and profilin bound to Arp2/3 complex or to actin monomers. The effective reduced molecular weight is defined as
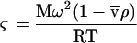 |
where M is molecular mass (g/mol), ω is the angular velocity (rad/sec), 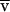 is the partial specific volume (cm3/gm), ρ is the solvent density (gm/cm3), R is the gas constant, and T is the absolute temperature (K). From these fits we determined the total amounts of free and bound profilin by
is the partial specific volume (cm3/gm), ρ is the solvent density (gm/cm3), R is the gas constant, and T is the absolute temperature (K). From these fits we determined the total amounts of free and bound profilin by
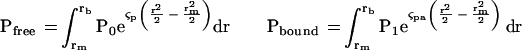 |
Where rm and rb are the radii of the meniscus and the bottom of the sample cell. The total amount of profilin is simply the sum of the bound and free. The total amount of Arp2/3 complex or actin can be calculated from the relative loading concentration.
Ptotal = Pfree + Pbound
Atotal = CrPtotal
Where Cr is the ratio of the initial Arp2/3 complex or actin concentration to the initial profilin concentration. For a stoichiometry of 1:1, the amount of bound profilin will equal the amount of bound Arp2/3 complex or actin and we calculate the distribution of free Arp2/3 or actin
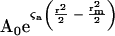 |
from
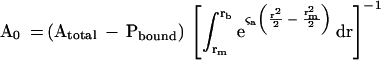 |
The dissociation equilibrium constant is then given by
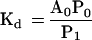 |
It is apparent from the formulas above that the accuracy of this method depends on accurate determination of both the fitting parameters P0 and P1 as well as the positions of the meniscus and base of the sample cell. The position of the meniscus is marked by a spike in the data set and can be assigned with confidence. We added a small amount of a dense imiscible liquid (usually carbon tetrachloride) to each sample chamber to form a flat base, perpendicular to the direction of centrifugation, so the exact position of the bottom of the sample column was less obvious. For our calculation we chose the base of each sample chamber to satisfy conservation of mass according to the following equation
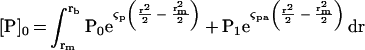 |
Where [P]0 is the initial baseline concentration of labeled profilin in the sample cell at time t = 0.
Rheometry
Rheological measurements were made with a cone and plate Rheometrics rheometer (Rheometrics Inc., Piscataway, NJ) in the small amplitude(strain ≤ 2%), forced oscillation mode (Sato et al., 1984). Arp2/3 complex in KME buffer (50 mM KCl, 10 mM imidazole, pH 7.0, 1 mM EGTA, and 1 mM MgCl2) was mixed with actin in Buffer G [2 mM Tris-HCl (pH 8.0), 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, and 1 mM NaN3], polymerized by adding one-tenth volume of 10 × KME and immediately placed between the metal plates of the rheometer to polymerize at 25°C. The plates were kept in a water-saturated chamber to prevent sample dehydration. Measurements of G′ and G" were made every 30 s using time sweep mode to observe the gel formation. After G′ and G" reached a plateau, frequency sweep mode was used to measure the rheological parameters: the value of complex modulus ‖G*‖, where ‖G*‖=(G’2+G“2)1/2; and the phase shift δ, where δ=tan–1(G”/G’) (Ferry, 1980).
Electron Microscopy
Acanthamoeba actin filaments in the presence and absence of Arp2/3 complex were negatively stained with freshly prepared 1% uranyl formate (Aebi et al., 1981). Other samples were fixed, embedded, and sectioned (Maciver et al., 1991). Acanthamoeba actin (10 μM) was polymerized in the presence or absence of 1 μM Arp2/3 complex in a 50 μl volume on a porcelain tray in a humid chamber for 1 h at 24°C. Samples were overlaid with 1% glutaraldehyde and 2 mg/ml tannic acid in 100 mM sodium phosphate, pH 7.0, 50 mM KCl, and 5 mM MgCl2 for 30 min at 24°C, washed with 100 mM sodium phosphate, pH 7.0, and then with 100 mM sodium phosphate, pH 6.0, fixed again with 0.1% OsO4 in 100 mM sodium phosphate, pH 6.0, at 24°C for 30 min, and then dehydrated with an ethanol series and embedded in Epon for thin sectioning. All micrographs were made at 80 kV with a JEOL JEM100CX or a Zeiss 10A electron microscope (Carl Zeiss, Thornwood, NY) at nominal magnifications between 23,000× and 46,000×. Actual magnification was determined by calibration with tropomyosin paracrystals. Negative images were scanned digitally and prepared for presentation using Adobe Photoshop.
Monoclonal Antibodies
Mouse monoclonal antibodies against Acanthamoeba profilin (Kaiser and Pollard, 1996) and Acanthamoeba α-actinin (Kaiser, Sato, Karaki, and Pollard, unpublished) were prepared by standard methods (Kiehart et al., 1984).
Fluorescence Microscopy
Acanthamoeba were maintained and fixed as previously described (Yonemura and Pollard, 1992). Cells were incubated with rabbit polyclonal antisera specific for Acanthamoeba Arp2, Arp3 (Kelleher, et al., 1995), p40, or p35 (Mullins et al., 1997) or mouse monoclonal antibodies specific for Acanthamoeba α-actinin, for 1 h at 24°C. Preabsorbed (Yonemura and Pollard, 1992) Cy-3 conjugated goat anti-rabbit IgG secondary antibody (Amersham, Arlington Heights, IL) was applied for 30 min. BODIPY-FL phallicidin (Molecular Probes, Sunnyvale, CA) was used to label filamentous actin. Cells were mounted in 50% glycerol containing 20 mM DTT and 5.6 mM p-phenylenediamine. We observed and photographed labeled cells using both phase-contrast and fluorescence microscopy as described previously (Fujiwara and Pollard, 1976). Figures were prepared using Adobe Photoshop from digitally scanned 35-mm negatives.
RESULTS
Profilin Binds the Arp2 Subunit of the Arp2/3 Complex
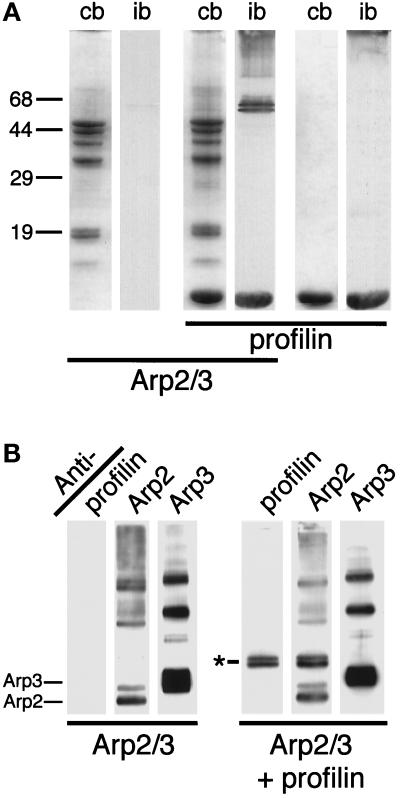
By optimizing conditions we were able to cross-link both profilin-I and profilin-II to the Arp2/3 complex using the zero-length cross-linker EDC with NHS. At 20 μM profilin and 3 μM Arp2/3, EDC/NHS produces two new bands on SDS-PAGE of 56 kDa and 59 kDa that react with antibodies against Acanthamoeba profilin (Figure 1A). Profilin-I and profilin-II produced identical sets of bands. Both bands react with anti-Arp2 antibodies but not with antibodies to Arp3 or any other member of the complex (Figure 1B). We conclude that the two bands are two different heterodimers of Arp2 and profilin, cross-linked at different residues and with slightly different mobilities on SDS- PAGE gels. Under other conditions, chemical cross-linking agents that covalently cross-link actin to profilin fail to cross-link profilin to any subunit of Arp2/3 complex (Mullins et al., 1997).
Figure 1.
EDC with NHS cross-links Acanthamoeba profilin to the Arp2 subunit of the Arp2/3 complex. (A) SDS-PAGE of proteins cross-linked with EDC/NHS and stained with Coomassie blue (cb) or immunoblotted (ib) with monoclonal antiprofilin antibodies. Samples, as indicated by the horizontal bars: 3 μM Arp2/3 complex, Arp2/3 plus 20 μM profilin-II, profilin-II alone. (B) Immunoblot of Arp2/3 complex and profilin-II cross-linked with EDC/NHS and probed with antibodies against profilin (lanes 1 and 4), Arp2 (lanes 2 and 5), and Arp3 (lanes 3 and 6). Lanes 1–3, 3 μM Arp2/3 complex; lanes 4–6, 3 μM Arp2/3 and 20 μM profilin-II. Asterisk marks the two profilin/Arp2 cross-linked products. Identical results were obtained with Acanthamoeba profilin-I. Conditions: Cross-linking was carried out for 1 h at 24°C with 5 mM EDC and 5 mM NHS. Buffer: 150 mM NaCl, 0.2 mM MgCl2, 0.1 mM EGTA, 0.2 mM ATP, 1 mM DTT, 10 mM imidazole, pH 7.5. Arp2/3 complex was purified by profilin:poly-L-proline affinity chromatography.
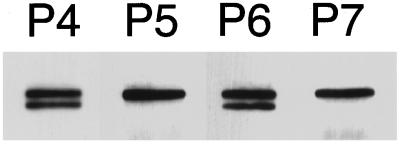
A panel of monoclonal antibodies to profilin reveals that the 56-kDa product is similar to the profilin-actin complex cross-linked by EDC/NHS. EDC/NHS cross-links E364 of Acanthamoeba actin to K115 of Acanthamoeba profilin-I or profilin-II (Vandekerchove et al., 1989). This binding site has been studied with the aid of four epitope-mapped monoclonal anti-profilin antibodies, P4, P5, P6 and P7 (Kaiser and Pollard, 1996). P5 and P7 bind profilin near K115, and cross-linking profilin to actin by EDC/NHS blocks binding of both antibodies (Kaiser and Pollard, 1996). Antibodies P4 and P6 bind to different regions of profilin and still recognize profilin cross-linked to actin. All four antibodies recognize the upper band, but only P4 and P6 react with the 56-kDa band (Figure 2). P5 and P7 both fail to recognize this band. We conclude that the lower band is Arp2 cross-linked to profilin at or near K115, in the actin-binding site of profilin. The upper band is a distinct cross-linked product of Arp2 and profilin with a different mobility on SDS-PAGE in which Arp2 is probably cross-linked to a residue of profilin other than K115. The presence of this product does not necessarily suggest two binding sites for profilin on Arp2 but rather the proximity of another cross-linkable residue of profilin to a residue on Arp2 that is not conserved on conventional actin.
Figure 2.
Arp2 binds at or near the actin-binding site on profilin. Immunoblot of SDS-PAGE of profilin-II cross-linked to Arp2/3 complex with EDC/NHS and probed with four epitope-mapped, monoclonal anti-profilin antibodies, P4, P5, P6, and P7. Antibodies P4 and P6 bind free profilin and profilin cross-linked to actin by EDC/NHS. Both also react with the two Arp2-profilin cross-linked products. P5 and P7 bind free profilin but not profilin cross-linked to actin and do not bind the smaller of the two Arp2-profilin cross-linked products. Conditions as in Figure 1.
Profilin Binds Actin More Strongly than the Arp2/3 Complex
We used sedimentation equilibrium ultracentrifugation to determine the affinity of profilin-II for actin and Arp2/3 complex. To measure the concentration of profilin in the presence of actin or Arp2/3 complex, we labeled profilin-II mutants in which either serine-38 or asparagine-58 was replaced by cysteine (S38C and N58C profilin-II) with rhodamine maleimide. Actin and poly-L-proline binding of these rhodamine-labeled constructs is indistinguishable from that of unlabeled profilin (Vinson, De La Cruz, Kaiser, Higgs, and Pollard, unpublished data). At equilibrium, both rhodamine-labeled mutants form exponential gradients characteristic of single, homogeneous, and thermodynamically ideal species. For N58C we calculated a native molecular weight of 12,930 Da, close to the value of 13,045 Da deduced from amino acid sequence.
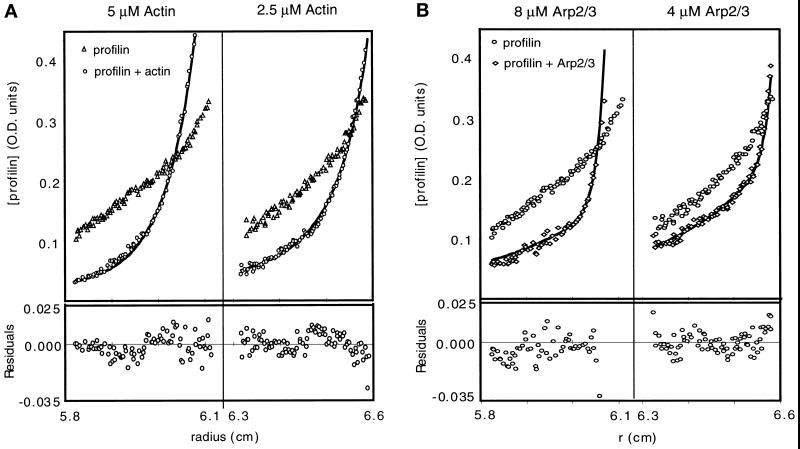
Measuring the distribution of labeled profilin in the presence of actin or Arp2/3 complex by absorbance at 550 nm gave a signal equal to the sum of the absorbances of bound and free profilin. We determined equilibrium dissociation constants for profilin binding actin and Arp2/3 complex by least-squares fitting of the equilibrium distribution of labeled profilin to a sum of two distributions, one determined by the effective reduced molecular weight of profilin alone (ςp) and the other by that of profilin bound to the larger protein (ςpa). We measured the effective reduced molecular weights of profilin, actin, and Arp2/3 complex individually, at the same speeds and in the same buffers used in the binding experiments and used these values in curve fitting. We independently calculated and averaged equilibrium dissociation constants from six data sets at three different protein concentrations spun at two speeds.
By this method N58C profilin-II binds Ca2+-ATP Acanthamoeba actin monomers with a Kd of 0.92 ± 1.0 μM (n = 6, Figure 3A) and Arp2/3 complex with a Kd of 7.4 ± 0.5 μM (n = 6, Figure 3B). The conditions differed in these two experiments. A low-ionic strength buffer was used in the actin binding experiments to keep actin monomers from polymerizing, while 150 mM NaCl was used in the Arp2/3 binding experiment to keep the complex from aggregating. The large uncertainty in the actin experiment arises from the fact that the concentration of profilin needed for detection (5 μM) is well above the Kd of the interaction. Similar results were obtained with the S38C mutant, indicating that the specific cysteine mutations and rhodamine labeling do not alter the binding to Arp2/3 complex.
Figure 3.
Analytical ultracentrifugation analysis of profilin-II binding to (A) actin and (B) Arp2/3 complex. Samples were centrifuged to equilibrium at 20,000 rpm at 24°C in an An60ti rotor in a Beckman model XL-A analytical ultracentrifuge. (A) Sedimentation equilibrium distributions of 5.3 μM rhodamine-labeled N58C Acanthamoeba profilin-II in the absence (triangles) and presence (circles) of Acanthamoeba actin. Left panel, 5 μM monomeric actin. Right panel, 2.5 μM monomeric actin. Solid lines are fits to the data assuming a Kd of 0.9 μM. Residuals of the fit are shown in the lower panels. (B) Sedimentation equilibrium distributions of 5.3 μM rhodamine-labeled profilin with (diamonds) and without (circles) Arp2/3 complex. Left panel, 8 μM Arp2/3 complex; right panel, 4 μM Arp2/3 complex. Solid lines are least squares fits of a two-component model with a Kd of 7.4 μM. Residuals are shown in the lower panels. Conditions were as follows. Buffer: 150 mM NaCl, 0.2 mM MgCl2, 0.1 mM ATP, 10 mM imidazole, pH 7.5. Temperature: 25°C. Arp2/3 complex was purified by profilin:poly-L-proline affinity chromatography.
Arp2/3 Complex Cross-links and Bundles Actin Filaments
Previously we found that Arp2/3 complex binds to the sides of actin filaments (Mullins et al., 1997) and hypothesized that it could cross-link filaments into networks or bundles. Preliminary experiments showed that Arp2/3 complex pellets with actin filaments under centrifugal forces where actin filaments alone do not pellet (our unpublished results). We characterized these higher order structures by quantitative rheometry and electron microscopy.
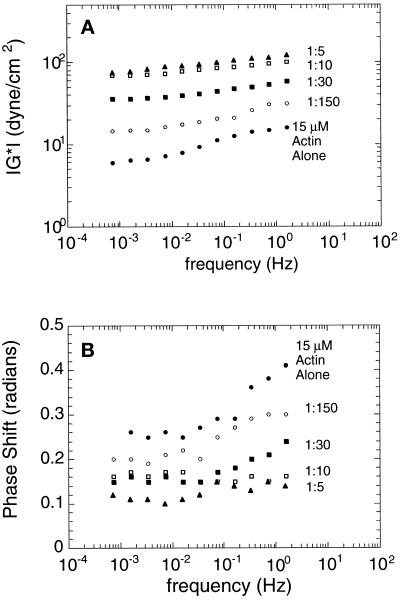
By quantitative rheometry, Arp2/3 complex increases the stiffness of solutions of actin filaments in a concentration-dependent manner (Figure 4). Addition of Arp2/3 complex increases the complex modulus, ‖G*‖—a measure of stiffness, and decreases the phase shift, δ—a measure of fluid- or solid-like behavior (Figure 4B). In these assays, δ = 0 radians for a solid and δ = 1.6 radians for a Newtonian fluid. Viscoelastic materials like actin filaments have intermediate values of δ.
Figure 4.
Rheological properties of actin filaments in the presence of Arp2/3 complex. (A) Frequency dependence of complex modulus, ‖G*‖. (B) Frequency dependence of the phase shift. Samples contained 15 μM rabbit skeletal muscle actin in the absence (•) or presence of Arp2/3 complex at 0.1 μM (○), 0.5 μM (▪), 1.5 μM (□), or 3 μM (▴). Conditions were as follows. Cone and plate rheometer in the small amplitude (strain ≤ 2%), forced oscillation mode. Buffer: 50 mM KCl, 10 mM imidazole, pH 7.0, 1 mM EGTA, and 1 mM MgCl2. Arp2/3 complex was purified by conventional ion exchange chromatography.
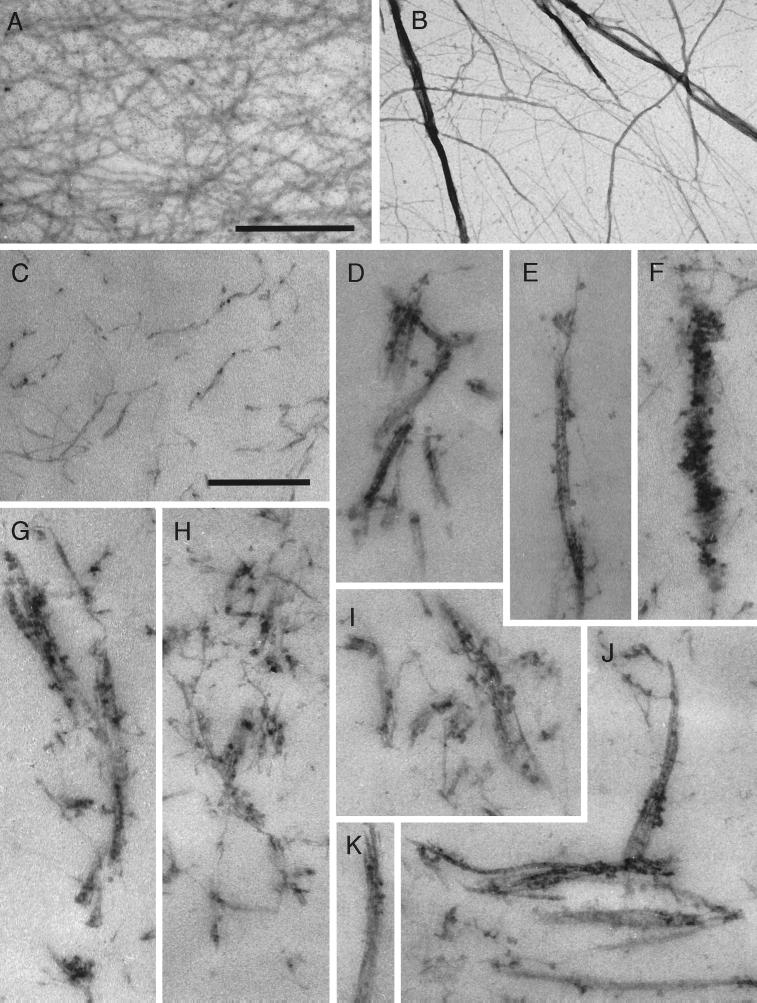
In the presence of Arp2/3 complex, actin filaments form networks of individual filaments and bundles of filaments that are visible in electron micrographs of thin sections of samples imbedded in Epon and of samples adsorbed to EM grids and negatively stained with 1% uranyl formate (Figure 5). Pure actin filaments have a low tendency to associate, and the density of individual filaments in thin sections of actin filaments alone is quite uniform. Actin filament networks in the presence of Arp2/3 complex are quite heterogeneous in thin sections cut from a single sample. Individual filaments are visible, but many are aggregated into bundles containing (within the thickness of the section) two to eight filaments. Individual filaments appear to fray off the ends of these bundles, and some appear to run from one bundle to another. Bundles tend to associate over a wide range of angles from nearly parallel to perpendicular and are studded with 10-nm particles not seen in samples of pure actin filaments or in bundles induced by α-actinin (Wachsstock et al., 1993b). The size of these particles is consistent with the low resolution structure of the complex determined by electron microscopy of rotary shadowed samples (Mullins et al., 1997). There was no obvious periodicity in the association of Arp2/3 complex with the filament bundles.
Figure 5.
Electron micrographs of negatively stained or thin-sectioned actin filaments and Arp2/3-induced filament bundles. (A and B) Negative stained samples. (C–K) Thin sectioned samples. Actin alone (A, C). Actin with Arp2/3 complex (B, D–K). Bar in panels A and B is 395 nm. Bar in panels C–K is 395 nm. Conditions were as follows. Negative stained samples: samples of either 3 μM Acanthamoeba actin alone or 3 μM actin and 0.5 μM Arp2/3 complex were adsorbed to carbon-coated electron microscope grids and negatively stained with 1% uranyl formate (see MATERIALS AND METHODS). Thin sectioned samples: 10 μM Acanthamoeba actin was polymerized in the absence or presence of 1 μM Arp2/3 complex. Samples were fixed, stained, embedded in Epon, and thin-sectioned (see MATERIALS AND METHODS). Arp2/3 complex in the negatively stained samples was purified by profilin:poly-L-proline affinity chromatography and in the thin-sectioned samples by conventional ion exchange chromatography.
The Distributions of Arp2/3 Complex and α-Actinin Differ in Motile Cells
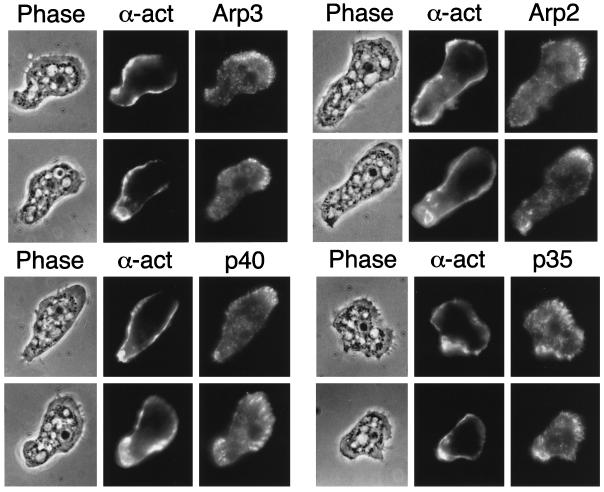
Arp2/3 and α-actinin are both concentrated in cortical regions of Acanthamoeba castellanii, but the leading edge is enriched in Arp2/3 complex compared with α-actinin (Figure 6). As noted before (Kelleher et al., 1995; Mullins et al., 1997), the staining for Arp2/3 complex members in Acanthamoeba is concentrated at the leading edge of the cells where it has a distinctly fibrillar character. In contrast, α-actinin is enriched in peripheral, cortical regions of the cell that do not stain well for Arp2/3 complex subunits. In the regions where it is most enriched, α-actinin staining extends out to the plasma membrane but lacks the fibrillar character of Arp2/3 staining. At the leading edge, α-actinin staining becomes weak and diffuse (as in the panels on the lower right of Figure 6, costained for p35), or is undetectable (panels on the upper left of Figure 6, costained for Arp3). Even in cells with diffuse staining near the leading edge, α-actinin does not extend to the tip of the leading edge like Arp2/3 complex. The localizations of Arp2/3 and α-actinin overlap most frequently at the rear of the cell, directly opposite the leading edge (Figure 6, especially lower right panel). Arp2/3 subunits, Arp3, Arp2, p40, and p35, are also more abundant in the central cytoplasm than α-actinin.
Figure 6.
Localization of Arp2/3 complex and α-actinin in Acanthamoeba castellanii by indirect immunofluorescence. Phase: phase contrast micrographs of individual adherent amoebas oriented with their leading edges pointing toward the upper right. α-act: distribution of α-actinin. Arp3/Arp2/p40/p35: individual distributions of four different subunits of the Arp2/3 complex.
DISCUSSION
Profilin Binding
From its discovery, Arp2/3 complex has been assumed to bind profilin, since it can be purified by affinity chromatography directly on profilin-agarose and indirectly on poly-L-proline Sepharose (Machesky et al., 1994). By analogy with actin, profilin was assumed to bind one of the actin-related proteins. The profilin-binding site of actin is more conserved in structural models of Arp2 than of Arp3 (Kelleher et al., 1995). In line with these predictions, we find that EDC/NHS cross-links both profilin-I and profilin-II to Arp2 but not to Arp3.
With either profilin-I or profilin-II EDC/NHS produces two cross-linked profilin-Arp2 conjugates with slightly different mobilities on SDS-PAGE gels. Reaction of these cross-linked products with a panel of epitope-mapped, anti-profilin monoclonal antibodies shows that the 56-kDa product is similar to profilin cross-linked to actin-E364 by EDC, because cross-linked Arp2 blocks binding of two antibodies with epitopes in the actin-binding site, just like cross-linked actin (Kaiser and Pollard, 1996). We conclude that Arp2 contacts the actin-binding site of profilin. In cross-linking experiments there was no detectable difference between profilin-I and profilin-II. Arp1 filaments are thought to bind spectrin (Holleran et al., 1996), but ours is the first direct experimental evidence that an Arp binds to the actin-binding site of a conventional actin-binding protein. Since the profilin binding site on actin is located at the barbed end of the subunit, the barbed end of Arp2 is probably exposed on the surface of the complex.
By sedimentation equilibrium ultracentrifugation, rhodamine-profilin-II binds Arp2/3 with a Kd of 7.4 μM. This method yielded a Kd for profilin binding to actin of 0.9 μM. Under similar conditions, using fluorescence anisotropy, Vinson and Pollard (unpublished data) measured a Kd of 0.2 μM for the same rhodamine-labeled profilin mutant binding to Acanthamoeba Ca2+-ATP-actin. By both measurements the affinity of profilin for conventional actin monomers is at least 10 times higher than for Arp2/3 complex. The dissociation rate constant of profilin from actin is 4 s−1 (Vinson, DeLaCruz, Kaiser, Higgs, and Pollard, unpublished data), so we predict that profilin is in a rapid equilibrium with Arp2 at a rate of at least 50 s−1.
Arp2/3 Complex Cross-links Actin Filaments
Filament cross-linking proteins are traditionally classed as either bundling or network-forming proteins, depending on their tendency to form either parallel bundles or randomly oriented isotropic networks of filaments. However, many actin-binding proteins, such as α-actinin, induce the formation of both bundles and isotropic networks. Wachsstock et al. (1993a) proposed a model to describe the dependence of the mechanical properties of cross-linked actin filament networks on the concentrations of actin and cross-linker. At low concentrations of cross-linker, filaments are bound in a random isotropic network that behaves like a single elastic solid. In this region, the complex modulus ‖G*‖ increases and the phase shift δ decreases as a function of cross-linker concentration. Above a threshold concentration that depends on the affinity of the cross-linker, actin filaments become organized into bundles. These bundles, although long and stiff, can slip past each other in solution and behave more like a very viscous fluid. In this region ‖G*‖ begins to decrease, and δ begins to increase as a function of cross-linker concentration.
To determine whether Arp2/3 complex preferentially forms bundles or isotropic networks in solution we performed quantitative rheometry on actin filaments cross-linked with Arp2/3 complex. With 15 μM filamentous actin, concentrations of Arp2/3 complex from 0.1–3.0 μM produced monotonically increasing values of ‖G*‖ and decreasing values of δ, indicating that this range of concentrations and cross-linker:actin ratios produces primarily isotropic networks (Figure 4). At the same actin concentration, α-actinin from Acanthamoeba undergoes a transition from network to bundle formation at 0.1 μM, and α-actinin from chicken gizzard smooth muscle undergoes the same transition at 1.0 μM. Arp2/3 complex, therefore, has a greater tendency to form isotropic networks than α-actinin. In the electron microscope, however, we also observe filament bundles induced by Arp2/3 complex. Therefore, Arp2/3 complex probably belongs, along with α-actinin, to that class of actin-binding proteins that can, under varying conditions of concentration and shear, form both bundles and isotropic networks of filaments.
Arp2/3 complex decorates the sides of actin filaments (Mullins et al., 1997) and caps pointed ends (Mullins et al., 1998). This combination is sufficient to link filaments into orthogonal networks. The concentration of filament pointed ends in our experiments, however, is in the nanomolar or subnanomolar range so the question becomes whether a nanomolar concentration of filament-filament cross-links is sufficient to explain the large changes in viscosity we observe in our experiments. Wachsstock et al. (1993b) demonstrated that the stiffness of a network is related in a complicated way to both the number and dynamics of cross-links. We can estimate the number of possible end-side cross-links in our rheometry experiments from the affinity of Arp2/3 for pointed ends, approximately 20 nM (Mullins and Pollard, unpublished data). With 5 nM filament ends in a solution of 15 μM filamentous actin, 0.5 μM Arp2/3 will cap 4.8 nM of the pointed ends, and at 1.5 μM it will cap 4.9 nM. Over the same concentration range, however, the stiffness of the filament network increases by more than a factor of 2, suggesting that the network formation activity cannot be completely explained by end-side associations but is also produced by side-side cross-links. The existence of two filament side-binding sites in addition to the pointed end binding site is also consistent with the fact that three subunits can be chemically cross-linked to actin filaments (Mullins et al., 1997).
Subcellular Localizations of Arp2/3 and α-Actinin
Arp2/3 complex cross-links actin filaments in a manner similar to α-actinin, so we wondered if the cellular functions of these proteins overlap. One way to address this question is to determine the extent to which the subcellular distributions of the two proteins overlap. By immunofluorescence microscopy, we find both Arp2/3 and α-actinin enriched in peripheral regions of cells, but the staining patterns do not overlap completely, particularly at the leading edge (Figure 10). The staining of Arp2/3 subunits is enriched at the leading edge of adherent Acanthamoeba and fibrillar in character, while α-actinin staining is diffuse or undetectable at the leading edge (Figure 6). We consistently observe that, even in cells with α-actinin staining near the leading edge, the staining does not reach the plasma membrane while Arp3, Arp2, p40, and p35 staining always extends to the tip of the leading edge.
The fact that Arp2/3 localizes to the tip of the leading edge suggests that it does not passively diffuse into the region but is involved in the initial events of leading edge formation. This pattern is consistent with the recent observation that pseudopodia produced by rapidly polymerizing actin filaments in the tails of motile Listeria monocytogenes also do not contain α-actinin (Sechi et al., 1997). One explanation for the absence of α-actinin in such actin-rich structures is that they form more rapidly than α-actinin can diffuse into them. The cellular concentrations of filamentous actin and α-actinin in Acanthamoeba are approximately 100 and 5 μM (Gordon et al., 1976; Pollard et al., 1986) and the Kd of α-actinin binding to actin filaments is 5 μM so at a given time more than 95% of the cellular α-actinin is bound to existing filaments. The dissociation rate constant of Acanthamoeba α-actinin is 5 s−1 (Wachsstock et al., 1993a) while the rate of addition of actin monomers to existing filaments in vivo can be as high 1000 s−1, so it is possible in a cell for filament formation to outpace α-actinin redistribution, especially in regions where actin polymerization is driving membrane protrusion.
Role of Profilin in Arp2/3 Function
How does profilin, which interacts with both actin and Arp2, affect the interaction of actin with Arp2/3 complex? Arp2/3 complex nucleates barbed end growth of filaments and remains tightly attached to the pointed ends of filaments. Profilin-bound actin monomers add to the barbed end of an actin filament with the same kinetics as free monomers so we would not expect profilin to affect the ability of actin to bind Arp2/3 complex. Arp2/3 binds filament ends with nanomolar affinity (Mullins et al., 1998) and profilin with micromolar affinity so we would also not expect profilin to significantly inhibit binding of Arp2/3 to actin filament ends.
The affinity of profilin for its ligands varies considerably: submicromolar for actin monomers, an order of magnitude weaker for Arp2/3 complex, and yet another order of magnitude weaker for both actin dimers and the ends of actin filaments. For barbed ends the Kd is estimated at 50 μM (Cooper and Pollard, 1985; Kaiser et al., 1986), and for cross-linked actin dimers it is estimated at 70 μM (Mockrin and Korn, 1983). Cooper and Pollard (1985) speculated that steric hindrance from adjacent subunits in a filament reduces the affinity for profilin. The low affinity of profilin for cross-linked actin dimers (Mockrin and Korn, 1983) indicates that a single additional monomer could produce such hindrances. If steric constraints are indeed responsible for the low affinity of actin filaments and dimers for profilin, then these constraints are not as severe in the Arp2/3 complex, suggesting that Arp2 and Arp3 are not packed exactly like actin subunits at the end of a filament.
Given a Kd of 7 μM, is there enough free profilin in Acanthamoeba to bind a substantial fraction of Arp2/3 complex? The concentrations of profilin (Tseng et al., 1984) and unpolymerized actin (Gordon et al., 1976) are about 100 μM each and that of Arp2/3 complex is 2 μM (Kelleher et al., 1995). Calculations based on equilibrium binding constants and results from cell fractionation studies (Perelroizen et al., 1994; Vinson, DeLaCruz, Kaiser, Higgs, and Pollard, in preparation; Kaiser, unpublished observations) indicate that only 70–80% of cellular profilin is bound to actin. Thus, in the absence of other competing ligands, a cellular pool of 20–30 μM profilin is available to bind Arp2/3 complex. At these concentrations about 75% of the available Arp2/3 complex would be saturated with profilin. So, despite its relative weakness, the interaction may have physiological significance. Arp2 and Arp3 bind and hydrolyze ATP, but the exchange of nucleotide from the Arp2/3 complex is very slow (Mullins and Kelleher, unpublished observations) so one possible function of profilin binding is to facilitate nucleotide exchange in the complex.
Biological Implications of Filament Cross-linking Activity
At the leading edge of motile cells we expect Arp2/3 complex to nucleate filament formation, cap the filament pointed ends (Mullins and Pollard, unpublished data), and bind the sides of filaments (Mullins et al., 1997). These activities are sufficient to form orthogonal networks of actin filaments in which the pointed ends are attached to the sides of other filaments in T junctions of the type observed at the leading edges of fish keratocytes (Small, et al., 1995; Svitkina et al., 1997). As discussed above, one side binding and one pointed end binding site are probably insufficient to produce the stiff orthogonal networks observed in our rheometric experiments, so there may be an additional filament binding site.
Arp2/3 complex is thus capable of promoting polymerization at the leading edge and attaching newly formed filaments to a rigid structure, a requirement for the “Elastic Brownian ratchet” model of polymerization-driven motility (Mogilner and Oster, 1996). As the actin filaments grow at their membrane-proximal ends and drive forward motility (Tilney et al., 1981; Wang, 1985), the bound complex will be translocated away from the membrane. This would explain the uniform localization of the complex throughout the actin-rich zone of the leading edge and all along the actin filament tails of motile Listeria (Welch, et al., 1997a). The partitioning of Arp2/3 complex to the leading edge and α-actinin to other parts of the amoeba cortex suggests that other cross-linking proteins (with different properties) replace the complex at the lateral margins of protrusive zones.
Our observations raise several questions for future study: Are the nucleation and filament cross-linking activities separable? If so, are they regulated separately in the cell? Which activity of the complex is essential for cell polarity and motility—nucleation, filament organization, or both? The answers to these questions will provide us with a more complete picture of how living cells regulate motility and shape changes.
ACKNOWLEDGMENTS
We thank members of the Pollard laboratory for technical assistance, advice, and helpful discussion, particularly Pam Maupin who prepared thin sections, Don Kaiser who supplied us with monoclonal antibodies, and Mike Ostap who participated in almost every amoeba preparation. We thank Cynthia Wolberger for use of her analytical ultracentrifuge and Tom Laue for checking our math. This work was supported by NIH research grant GM-26338 to T.D.P., a fellowship to R.D.M. from the Jane Coffin Childs Fund for Cancer Research, and a Jenkins Fellowship to J.X.
REFERENCES
- Aebi U, Fowler WE, Isenberg G, Pollard TD, Smith PR. Crystalline actin sheets: their structure and polymorphism. J Cell Biol. 1981;91:340–351. doi: 10.1083/jcb.91.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Feoktistova A, McCollum D, Gould KL. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Lewis MS, Korn ED. Actobindin binds with high affinity to a covalently cross-linked actin dimer. J Biol Chem. 1994;269:25587–25591. [PubMed] [Google Scholar]
- Cooper JA, Pollard TD. Effects of capping protein on the kinetics of actin polymerization. Biochemistry. 1985;24:793–799. doi: 10.1021/bi00324a039. [DOI] [PubMed] [Google Scholar]
- Ferry JD. Viscoelestic Properties of Polymers. 3rd ed. New York: John Wiley and Sons; 1980. [Google Scholar]
- Frankel S, Mooseker MS. The actin-related proteins. Curr Opin Cell Biol. 1996;8:30–37. doi: 10.1016/s0955-0674(96)80045-7. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Pollard TD. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J Cell Biol. 1976;71:848–875. doi: 10.1083/jcb.71.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Eisenberg E, Korn ED. Characterization of a cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J Biol Chem. 1976;251:4778–4786. [PubMed] [Google Scholar]
- Holleran EA, Tokito MK, Karki S, Holzbaur ELF. Centractin (Apr1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J Cell Biol. 1996;135:1815–1830. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser DA, Goldschmidt-Clermont PJ, Levine BA, Pollard TD. Characterization of renatured profilin purified by urea elution from poly-L-proline agarose columns. Cell Motil. 1989;14:251–262. doi: 10.1002/cm.970140211. [DOI] [PubMed] [Google Scholar]
- Kaiser DA, Pollard TD. Characterization of actin and poly-L-proline binding sites of Acanthamoeba profilin with monoclonal antibodies and by mutagenesis. J Mol Biol. 1996;255:89–107. doi: 10.1006/jmbi.1996.0070. [DOI] [PubMed] [Google Scholar]
- Kaiser DA, Sato M, Ebert R, Pollard TD. Purification and characterization of two isoforms of Acanthamoeba profilin. J Cell Biol. 1986;102:221–226. doi: 10.1083/jcb.102.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard. TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Kaiser DA, Pollard TD. Monoclonal antibodies demonstrate limited structural homology between myosin isozymes from Acanthamoeba. J Cell Biol. 1984;99:1002–1014. doi: 10.1083/jcb.99.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Brzeska H, Baines IC, Korn ED. Purification of myosin-I and myosin-I heavy chain kinase. Methods Enzymol. 1991;196:12–23. doi: 10.1016/0076-6879(91)96004-b. [DOI] [PubMed] [Google Scholar]
- Machesky LM. Cell motility: complex dynamics at the leading edge. Curr Biol. 1997;7:R164–R167. doi: 10.1016/s0960-9822(97)70079-4. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Wachsstock D, Schwarz WH, Pollard TD. The actin filament severing protein actophorin catalyzes the formation of rigid bundles of actin filaments crosslinked with alpha-actinin. J Cell Biol. 1991;115:1621–1628. doi: 10.1083/jcb.115.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean-Fletcher S, Pollard TD. Identification of a factor in conventional muscle actin preparation which inhibits actin filament self-association. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Mockrin SC, Korn ED. Kinetics of polymerization and ATP hydrolysis by covalently cross-linked actin dimers. J Biol Chem. 1983;258:3215–3221. [PubMed] [Google Scholar]
- Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Madania A, Martin RP, Winsor B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Kelleher JF, Pollard TD. Actin’ like actin? Trends Cell Biol. 1996;6:208–212. doi: 10.1016/0962-8924(96)20017-0. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, R.D., Heuser, J.A., Pollard, T.D. (1998). The interaction of Arp2/3 complex with actin: nucleation, high-affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA (in press). [DOI] [PMC free article] [PubMed]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Purification of a high molecular-weight actin filament gelation protein from Acanthamoeba that shares antigenic determinants with vertebate spectrins. J Cell Biol. 1984;99:1970–1980. doi: 10.1083/jcb.99.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Tseng PC-H, Rimm DL, Bichell DP, Williams RC, Sinard J. Characterization of alpha-actinin from Acanthamoeba. Cell Motil. 1986;6:649–661. doi: 10.1002/cm.970060613. [DOI] [PubMed] [Google Scholar]
- Sato M, Wong TZ, Brown D, Allen RD. Rheological properties of living cytoplasm: a preliminary investigation of squid axoplasm. Cell Motil. 1984;4:7–23. doi: 10.1002/cm.970040103. [DOI] [PubMed] [Google Scholar]
- Schachman HK. Deductions from hydrodynamic and thermodynamic measurements. Brookhaven Symp Biol. 1960;13:49–70. [PubMed] [Google Scholar]
- Schachman HK, Gropper L, Hanlon S, Putney F. Ultracentrifuge studies with absorption optics. II. Incorporation of a monochrometer and its application to the study of proteins and interacting systems. Arch Biochem Biophys. 1962;99:175–190. doi: 10.1016/0003-9861(62)90259-x. [DOI] [PubMed] [Google Scholar]
- Sechi AS, Wehland J, Small JV. The isolated comet tail pseudopodium of Listeria monocytogenes: a tail of two actin filament populations, long and axial and short and random. J Cell Biol. 1997;137:155–167. doi: 10.1083/jcb.137.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Herzog M, Anderson K. Actin filament organization in the fish keratocyte lamellipodium. J Cell Biol. 1995;129:1275–1286. doi: 10.1083/jcb.129.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Terasaki AG, Ohnuma M, Mabuchi I. Identification of actin-binding proteins from sea urchin eggs by F-actin affinity column chromatography. J Biochem. 1997;122:226–236. doi: 10.1093/oxfordjournals.jbchem.a021733. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Bonder EM, DeRosier DJ. Actin filaments elongate from their membrane-associated ends. J Cell Biol. 1981;90:485–494. doi: 10.1083/jcb.90.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng PC H, Runge MS, Cooper JA, Williams RC, Jr, Pollard TD. Physical, immunochemical, and functional properties of Acanthamoeba profilin. J Cell Biol. 1984;98:214–221. doi: 10.1083/jcb.98.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerchove J, Kaiser DA, Pollard TD. Acanthamoeba actin and profilin can be crosslinked between glutamic acid 364 of actin and lysine 115 of profilin. J Cell Biol. 1989;109:619–626. doi: 10.1083/jcb.109.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsstock DH, Schwarz WH, Pollard TD. Affinity of alpha-actinin for actin filaments determines the structure and mechanical properties of actin filament gels. Biophys J. 1993a;65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsstock DH, Schwarz WH, Pollard TD. Structure and mechanical properties of actin filament gels. Biophys J. 1993b;65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Exchange of actin subunits at the leading edge of living fibroblasts: Possible role of treadmilling. J Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997a;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997b;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Podtelejnikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Yonemura SY, Pollard TD. Localization of myosin-I and myosin-II in Acanthamoeba. J Cell Sci. 1992;102:629–642. doi: 10.1242/jcs.102.3.629. [DOI] [PubMed] [Google Scholar]