FIGURE 10.
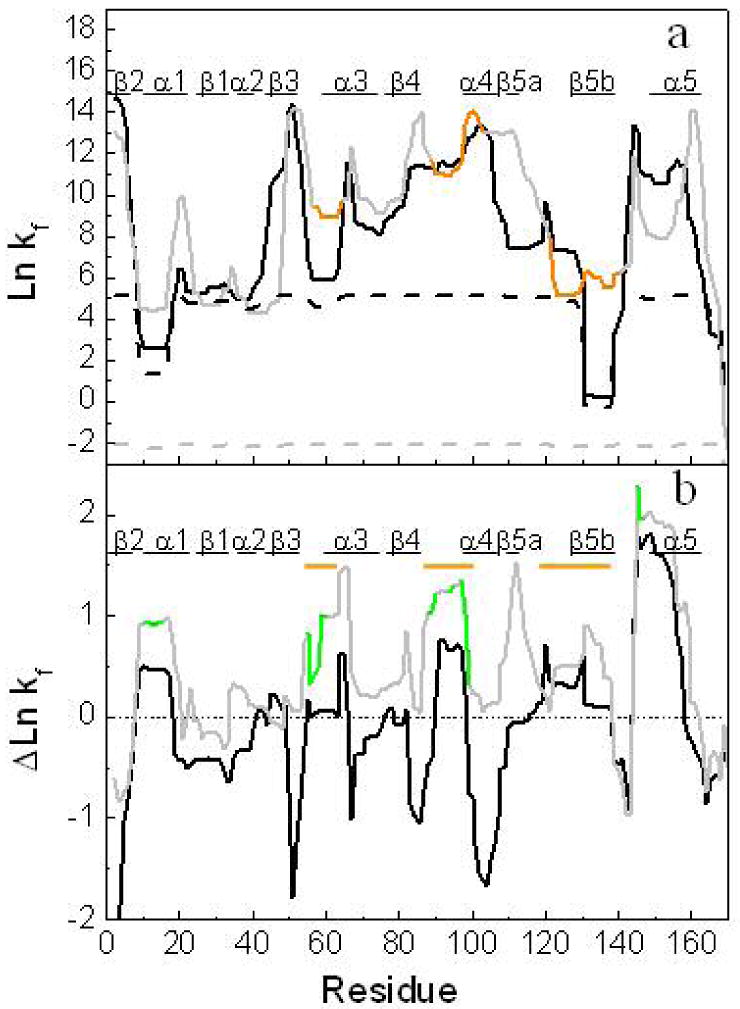
Local stability of H. pylori apoflavodoxin compared to that of Anabaena apoflavodoxin. (a) Natural logarithms of the predicted folding constant with COREX algorithm (ln kf) for each amino acid of H. pylori apoflavodoxin at 4 °C (black line) and Anabaena apoflavodoxin (gray line) and at 65 °C (in dashed line). The regions determined to be unfolded by equilibrium Φ-analysis(23) in the thermal intermediate of Anabaena apoflavodoxin are colored in orange. The elements of secondary structure of the proteins are shown at the top. (b) Difference in folding constant values of H. pylori (black line) and Anabaena (gray line) between 4 °C and 25 °C. The FMN binding regions in Anabaena flavodoxin are colored in green, the proposed unfolded regions in the thermal intermediate are marked as orange lines. For a better comparison between proteins and its structural elements, a sequence alignment was performed and the predicted folding constant per residue were plotted in the resulted sequences in both figures.
