Abstract
Vitamin E is a dietary lipid that is essential for vertebrate health and fertility. The biological activity of vitamin E is thought to reflect its ability to quench oxygen- and carbon- based free radicals, and thus to protect the organism from oxidative damage. However, recent reports suggest that vitamin E may also display other biological activities. Here, to examine possible mechanisms that may underlie such non-classical activities of vitamin E, we investigated the possibility that it functions as a specific modulator of gene expression. We show that treatment of cultured hepatocytes with RRR-α-tocopherol alters the expression of multiple genes and that these effects are distinct from those elicited by another antioxidant. Genes modulated by vitamin E include those that encode key enzymes in the cholesterol biosynthetic pathway. Correspondingly, vitamin E caused a pronounced inhibition of de novo cholesterol biosynthesis. The transcriptional activities of vitamin E were mediated by attenuating the post-translational processing of the transcription factor SREBP-2 that, in turn, led to a decreased transcriptional activity of sterol responsive elements in the promoters of target genes. These observations indicate that vitamin E possesses novel transcriptional activities that affect fundamental biological processes. Cross talk between tocopherol levels and cholesterol status may be an important facet of the biological activities of vitamin E.
The term vitamin E refers to a family of structurally related neutral plant lipids that are critical for vertebrate health and fertility. Numerous studies established that members of the vitamin E family are efficient chain-breaking radical scavengers both in vitro and in vivo, and led to the definition of vitamin E as the most important lipid-soluble antioxidant (1). While all isoforms of vitamin E possess comparable radical trapping activity in vitro (2), RRR-α-tocopherol (denoted herein as αTOH) exhibits the highest potency in biological assays (3). This vitamer preference stems from the combined activities of the hepatic α-tocopherol transfer protein (TTP) that selectively retains αTOH (3–5), and the catabolic actions of hepatic enzymes that degrade other forms of vitamin E to water-soluble products (6).
The selectivity for αTOH raises the possibility that this vitamer has unique biological roles, in addition to its anti-oxidant function. Indeed, novel regulatory properties have been ascribed in recent years to αTOH, including the modulation of apoptosis, cell adhesion, and specific enzymatic activities (cf. 7, 8). Moreover, αTOH was shown to regulate the expression levels of several mRNAs and proteins, such as collagen α1 (9), the scavenger receptors SR-BI (10) and CD36 (11, 12), α-tropomyosin (13), the nuclear receptor PPARγ (14), and the adhesion molecule VCAM-I (15). These anecdotal observations were further supported by several studies that documented genomic responses to vitamin E using expression-profiling approaches (16–23). However, interpretation of such analyses is complicated by the known genomic responses to changes in cellular redox status (24). Indeed, treatment with vitamin E elicits pronounced changes in the expression of known antioxidant-responsive genes, such as glutathione-S-transferase, cytochrome P450-1A2, catalase, superoxide dismutase, and metallothioprotein (18, 19, 22). Thus, at present, only very limited information is available regarding the molecular mechanisms that mediate genomic responses to vitamin E. Here, we report the identification of specific genomic activities of αTOH that are distinct from those induced by another anti-oxidant, and provide insights into the molecular mechanisms that mediate these activities.
EXPERIMENTAL PROCEDURES
Cell culture
HepG2 C3A cells were cultured at 37 °C and 5% CO2 with Dulbecco’s modified Eagle’s medium (CellGro) supplemented with 10% FBS (Atlanta Biologicals) or LPDS (Sigma Chemical Co.). Antioxidants were added as either pre-formed serum complexes (25–27) or from ethanol stocks directly to the media. Both methods resulted in comparable uptake efficiencies as determined by accumulation of radioactively labeled tocopherol. Final ethanol concentrations did not exceed 0.2% (v/v). Unless otherwise indicated, treatments lasted 48 hours, with fresh media change after 24 hours. Cell viability was evaluated using an MTT reduction conversion assay (28, Sigma Chemical Co.).
Micro-array Analyses
Total RNA was isolated using the TRIzol reagent (Invitrogen) and RNA quality was evaluated using the 2100 Bioanalyzer (Agilent Technologies). Double-stranded cDNA was synthesized using a SuperScript II Reverse Transcriptase Kit (Invitrogen). Phase Lock Gel, phenol/chloroform extraction, and ethanol precipitation were employed to purify the resultant cDNA. cRNA was synthesized using the GeneChip IVT Labeling Kit (Affymetrix). In vitro transcription was carried out for four hours at 37°C, using biotinylated ribonucleotides (Enzo Diagnostics) for labeling. Labeled cRNA was purified with an RNeasy kit (Qiagen), and fragmented in fragmentation buffer for 30 minutes at 94 °C. Fragmented cRNA was hybridized to human U133A GeneChip micro-arrays (Affymetrix) in a rotating hybridization oven for 16 hours at 45°C. Staining was performed on Affymetrix fluidics station utilizing streptavidin/phycoerythrin conjugate (Molecular Probes), followed by biotinylated antibody to streptavidin (Vector Laboratories), and finally via a second streptavidin/phycoerythrin conjugate. Stained micro-arrays were scanned on a Hewlett Packard’s GeneArray Scanner, and data was compiled with Affymetrix Microarray Suite 5.0 software. Each treatment condition was repeated in three independent cultures, each of which was subjected to three independent hybridization reactions, for a total of nine micro-arrays per experimental condition.
Data Analyses
Intensity data from each of the nine micro-arrays was uploaded to GeneTraffic version 2.8 (Iobion). Raw data were normalized using Robust Multichip Analysis, and p-values were calculated using an F-class ratio with variance stabilization, which served as a measure of the difference between the normalized intensity-value means of various treatment groups relative to the variability of intensity values of each replicate within a given treatment group. Significantly regulated gene lists (p < 0.05, absolute fold change greater than 1.2) were then manually clustered based upon known cellular function.
Measurements of Intracellular ROS
Triplicate wells of HepG2 C3A cells in 96-well plates were serum-starved for 72 hours, washed twice with minimum essential medium lacking phenol red (CellGro), and supplemented with various concentrations of NAC or αTOH (delivered from ethanolic stocks) for 24 hours. The cell-permeable dye DCFDA (Molecular Probes) was then added to 5 μg/mL in Hank’s buffered saline solution lacking phenol red (Sigma Chemical Co.) for two hours at 37°C. DCFDA rapidly diffuses across cell membranes, where the diacetate moiety is cleaved by intracellular esterases to yield 2′,7′-dichlorodihydrofluorescein that oxidizes upon interaction with ROS to the highly fluorescent DCF that serves as a sensitive quantitative read-out for ROS levels in live cells (29, 30). DCF fluorescence was measured with a Tecan SpectraFluor Plus microplate reader (excitation = 485 nm; emission = 535 nm). Assay range and linearity were ensured using known concentrations of hydrogen peroxide.
Validation of Micro-array Results
Total RNA was isolated from treated cells, and cDNA was prepared as described above. The expression profiles of selected genes were quantified, in triplicate, on a 7500 Real Time RT-PCR System instrument (Applied Biosystems). FAM dye-labeled Taqman MGB probes (Applied Biosystems) were utilized to assess mRNA levels of the genes encoding HMG-CoA reductase, the low-density lipoprotein (LDL)-receptor, and A-kinase anchor protein 12 (AKAP12). Threshold cycle (Ct) values were determined for each replicate and normalized to β-actin or 18S mRNA levels detected by the same procedure. These measurements were then converted to fold change values. Where indicated, actinomycin D (Sigma) was added from DMSO stock to 1 μM.
Reporter Gene Assays
HepG2 C3A cells grown in MEM/10% LPDS were transiently transfected with a pGL3-basic vector encoding firefly luciferase under the control of the SREBP2 promoter (generous gift of Dr. Timothy Osborne, University of California Irvine; see (31) for details) using the Fugene6 reagent (Roche). Two hours after transfection, triplicate wells were treated as indicated for 36 hours, lysed and luciferase expression was assayed using a commercial kit (Promega) on a Berthold luminometer. Where indicated, the single sterol response element (SRE) sequence in this promoter was mutated to change the wild-type SRE (5′ATCACCCCAT3′) to a non-functional response element (5′ATAAAAAAAT3′) using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). All constructs were verified by sequencing.
Measurements of SREBP-2 Processing
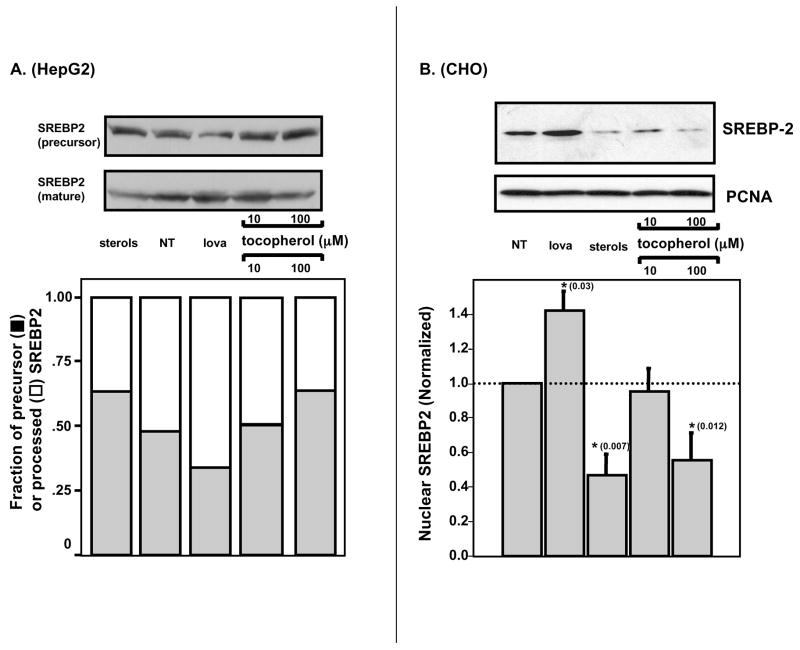
Chinese hamster ovary (CHO) cells were propagated in Ham’s F-12 media supplemented with 10% FBS, then adapted to LPDS for two days. Mevalonic acid (50 μM; Sigma Chemical Co.) and other treatments were added as indicated, after which the cells were treated with 10 μM MG132 for 5 additional hours. The cells were then harvested and nuclear extracts prepared as described previously (32). Briefly, scraped cells were washed with cold PBS and resuspended in 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 M EDTA, 0.1 mM EGTA, 1 mM DTT, and 0.5 mM PMSF). After swelling for 15 minutes on ice, NP-40 was added to 0.58%, the sampled were mixed vigorously and microfuged at 3000 × g for 30 seconds. The pellet was resuspended in 20 mM HEPES pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF, rocked at 4°C for 15 minutes, and the nuclear extract were obtained after microcentrifugation at 14,000 rpm for 5 minutes at 4° C. The presence of processed SREBP-2 in the nuclear fraction was measured by and anti-SREBP-2 immunoblotting (Caymen Chemicals, antibody 10007663). Equal loading was ensured by immunoblotting with antibodies for proliferating cell nuclear antigen (PCNA, Cell Signaling clone PC10). Fractionation of HepG2 cells into nuclear and membranous fractions (Figure 4A) was done according to the method of Goldstein and Brown (33). Immunoblots were quantified by densitometry using Scion Image software (Scion Corp.).
Figure 4. Vitamin E inhibits proteolytic activation of SREBP-2.
HepG2 cells (panel A) or CHO cells (panel B) were cultured in media containing 10% LPDS and treated with 50 μM lovastatin, 10 μg/μl cholesterol + 0.1 μg/μl 25-hydroxy cholesterol, or the indicated concentration of αTOH for 24 hours. Cells were then treated with 10 μM MG132 for five hours, washed, and SREBP-2 levels assessed by anti-SREBP-2 immunoblotting. A. Representative analysis of nuclear and membrane fractions from treated HepG2 cells. B. Representative analyses of nuclear fractions from treated CHO cells. Shown are averages and standard deviations. Asterisks denote statistically significant changes from control samples, as determined by a student’s t-test. P-values obtained from statistical analyses are shown in parentheses. NT – non-treated.
Measurements of de novo Cholesterol Biosynthesis
De novo biosynthesis of cholesterol was measured by following the incorporation of radio-labeled acetate into cholesterol following the protocol of Goldstein and Brown (34). HepG2 C3A cells were cultured in 6-well plates for 18 hours, washed with PBS and media was changed to DMEM + 10% lipoprotein-deficient serum (LPDS, Sigma Chemical Co.). Twenty-four hours later, triplicate wells were treated with either of the following: 1–100 μM αTOH, 10 μg/μl cholesterol + 0.1 μg/μl 25-hydroxycholesterol, 1 mM NAC, or vehicle control (ethanol). After 24 hours, cells were pulsed with 2 μCi of [1-14C]acetic acid (2.11 GBq/mmol, 57.0 mCi/mmol, GE Amersham) for 6 hours. The cells were washed with PBS and lipids were extracted by incubation with 2 mL of 3:2 hexane:isopropanol at 25°C for 30 minutes. Solvent was then evaporated under N2 stream, and the lipids were resuspended in 60 μL hexane and resolved by thin layer chromatography (TLC) developed with 80:20:1 petroleum ether:diethyl ether:acetic acid. Resolved lipids were visualized with iodine vapor, and radioactive incorporation into cholesterol was quantified by scintillation counting.
RESULTS
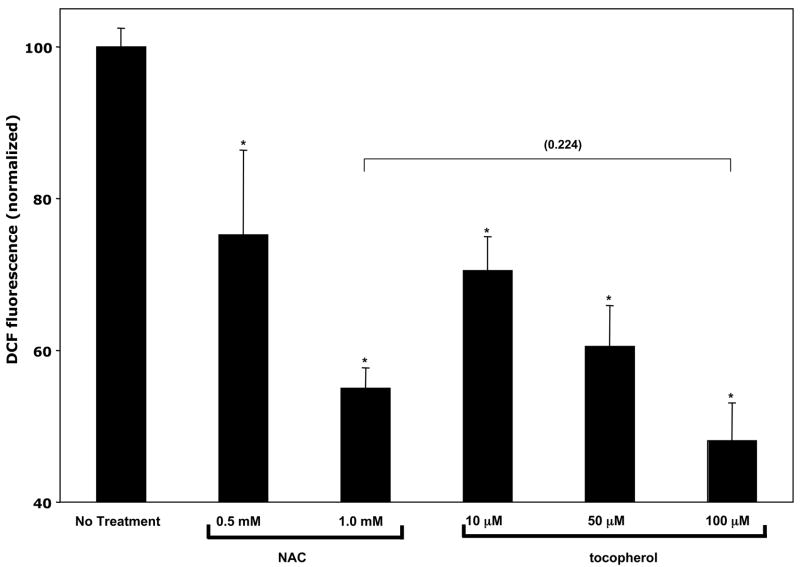
The main goal of the studies presented here was to evaluate to impact of vitamin E (and, specifically, αTOH) on gene expression in human hepatocytes. A possible confounding factor in such experiments stems from the non-specific effects that antioxidants have on multiple redox-responsive cellular targets. To examine the unique activity of αTOH, we compared its transcriptional activities effects to those of another established antioxidant, NAC (35). We hypothesized that specific targets of vitamin E actions would respond to vitamin E treatment, but not to NAC treatment. To compare the efficacy of αTOH and NAC in quenching reactive oxygen species in our cell system, we employed DCF, a cell permeable fluorescent probe that quantitatively reports on intracellular ROS levels (29, 36–38). Data in Figure 1 show that treatment of HepG2 cells with either NAC or αTOH caused a dose-dependent decrease in cellular ROS levels. Importantly, 100 μM αTOH elicited a ca. 50% reduction in cellular ROS levels, similar to the change induced by NAC. These results show that treatment of HepG2 cells with 1 mM NAC is an appropriate control for the antioxidant effects of 100 μM αTOH.
Figure 1. Quenching of intracellular reactive oxygen species by N-acetyl L-cysteine (NAC) and RRR-α-tocopherol (αTOH).
Confluent HepG2 cells were serum-starved for 72 hours, washed twice with minimum essential medium lacking phenol red, and treated with the indicated concentrations of NAC or αTOH for 24 hours. Reactive oxygen species levels were measured using DCFDA fluorescence as described in Materials and Methods. Shown are means and standard deviations from three independent experiments. Fluorescence values were normalized to those obtained from untreated cells (36,011 fluorescence units).
To evaluate the specific effects of vitamin E on hepatic gene expression, we determined the global expression profile of HepG2 cells that were treated with 100 μM αTOH, and compared it with the expression profile of untreated cells, or those treated with 1 mM NAC. RNA extracted from the different experimental groups was processed as described in Materials and Methods, and hybridized to Affymetrix U133A expression micro-arrays (Affymetrix) on which ca. 22,000 transcripts and ESTs are displayed, representing approximately 14,500 unique human genes.
In three independent array experiments, 57 genes displayed statistically significant differences in expression levels between the αTOH treatment group and untreated controls (p < 0.05). Of these, 29 were up-regulated by αTOH, while the expression of 28 genes was repressed by this treatment. Changes in the expression levels of these genes were in the 1.2- to 2.0-fold range. The affected genes can be grouped into four major categories when clustered on the basis of known cellular functions: cell proliferation, lipid status, protein stability and metabolism, and transcriptional regulation (Table 1). Treatment with αTOH caused specific and significant reduction in the expression levels of 17 genes that play central roles in regulating cellular lipid status (Table 2). Interestingly, 10 of these αTOH-responsive gene products catalyze key steps in the de novo biosynthesis of cholesterol: dehydrocholesterol reductase, farnesyl diphosphate synthase, HMG-CoA synthase, isopentenyl-diphosphate delta isomerase, lanosterol synthase, lathosterol oxidase, squalene monooxygenase, squalene synthase and sterol-C4-methyl oxidase (Table 2). Notably, mRNA levels of the HMG-CoA reductase gene, encoding the rate-limiting enzyme in cholesterol biosynthesis (39), and of the low-density lipoprotein (LDL) receptor, a key regulator of cholesterol transport (40), were also significantly reduced by TOH treatment (Table II). Importantly, we observed that none of these transcripts were affected by NAC treatment.
Table 1.
Functional clustering of TOH-responsive genes
| Cellular function | Total Number of TOH- responsive genes1 | Number of genes up- regulated by TOH treatment | Number of genes down- regulated by TOH treatment |
|---|---|---|---|
| Cell Proliferation | 23 | 19 | 4 |
| Lipid Status | 17 | 0 | 17 |
| Protein Stability and Metabolism | 9 | 6 | 3 |
| Transcriptional
Regulation |
5 | 3 | 2 |
| Other | 3 | 1 | 2 |
| Total | 57 | 29 | 28 |
p < 0.05, absolute fold change > 1.2.
Table 2.
Effect of αTOH on the expression of lipid homeostatic genes
| Fold Change1 | Gene Identity | Accession Number |
|---|---|---|
| −1.37 | 7-dehydrocholesterol reductase2 | NM_001360 |
| −1.33 | acetoacetyl Coenzyme A thiolase | NM_005891 |
| −1.28 | cytochrome P450, family 51, A1 | NM_000786 |
| −1.34 | farnesyl diphosphate synthase2 | NM_002004 |
| −1.41 | HMG-CoA reductase2 | NM_000859 |
| −1.55 | HMG-CoA synthase2 | NM_002130 |
| −1.42 | hydroxysteroid (17-beta) dehydrogenase | NM_016371 |
| −1.47 | isopentenyl-diphosphate delta isomerase2 | NM_004508 |
| −1.27 | lanosterol synthase2 | NM_002340 |
| −1.29 | lathosterol oxidase2 | NM_006918 |
| −1.21 | low density lipoprotein receptor | NM_000527 |
| −1.22 | phosphate cytidylyltransferase 2, ethanolamine | NM_002861 |
| −1.32 | squalene monooxygenase2 | NM_003129 |
| −1.27 | squalene synthase2 | NM_004462 |
| −1.28 | stearoyl-CoA desaturase (delta-9-desaturase) | NM_005063 |
| −1.58 | sterol-C4-methyl oxidase2 | NM_006745 |
p < 0.05.
genes products that catalyze reactions in the de novo cholesterol biosynthetic pathway.
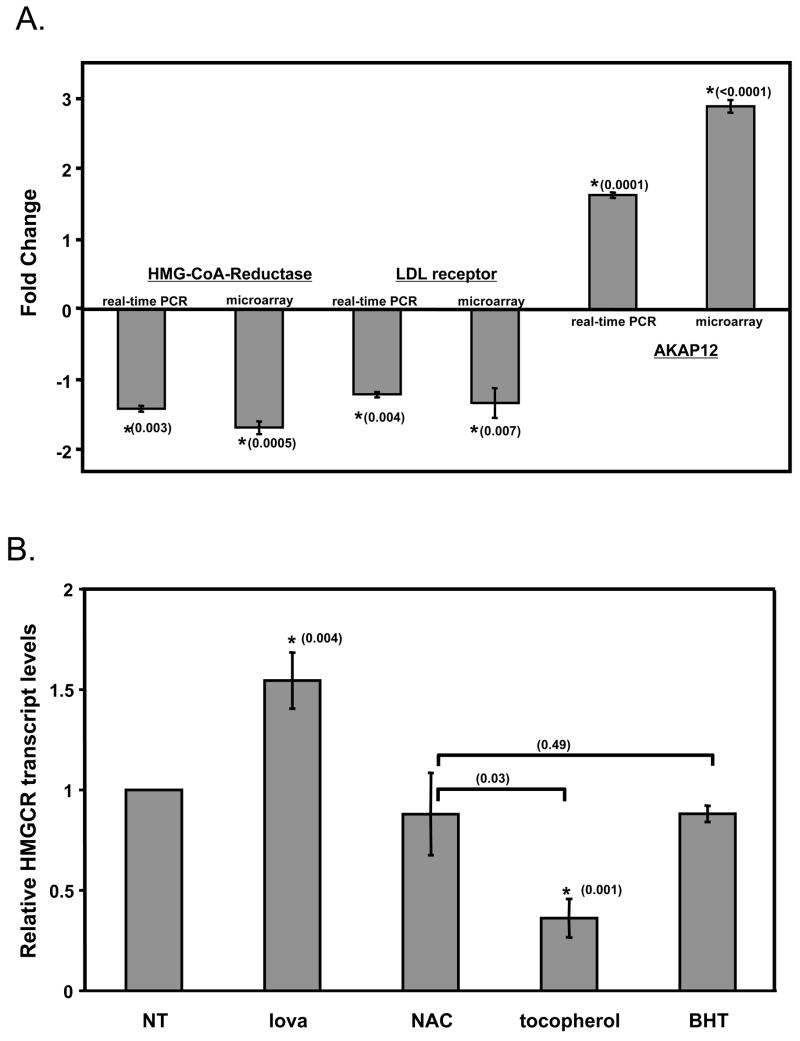
To verify the results obtained from micro-array analyses we employed quantitative, real-time RT-PCR. The data show that αTOH treatment caused a significant decrease in the levels of mRNAs encoding the HMG-CoA reductase and the LDL-receptor, and that the magnitude of the effect was similar to that observed in expression arrays (Figure 2). Real-time RT-PCR of the signal transducer AKAP12 transcript revealed that αTOH treatment caused a similar effect to that observed in the expression arrays (i.e. 2–3-fold increase in mRNA levels, Figure 2A). These results indicate that the effects of αTOH on gene expression observed in the micro-array experiment were both significant and reproducible. Additional experiments revealed that the vitamin E-induced changes in the HMG CoA Reductase transcript were specific, and did not occur following treatments with the soluble antioxidant NAC, or the lipid soluble antioxidant BHT (Figure 2B). These observations raise the possibility that the regulation of HMG CoA reductase transcript stems from specific, possibly antioxidant-independent, actions of vitamin E.
Figure 2. Validation of micro-array data.
A. Comparison of real-time RT-PCR and microarray data. B. The effect of different antioxidants on HMG CoA Reductase gene expression. HepG2 cells were treated for 24 hours with 100 μM αTOH (or ethanol as vehicle control), 50 μM lovastatin, 1 mM NAC or 200 μM BHT, and total RNA was isolated and utilized for cDNA synthesis. Expression levels of the indicated genes were quantified by real time RT-PCR, as described in Materials and Methods. Shown are averages and standard deviations of three independent experiments. Asterisks denote statistically significant changes from control samples, as determined by a student’s t-test. P-values obtained from statistical analyses are shown in parentheses. NT- not treated.
The vitamin E-induced decreases in transcript levels observed in cholesterol homeostatic genes could in principle result from inhibition of transcription, or, alternatively, from facilitation of mRNA degradation. To discriminate between these possibilities, we measured the effect of vitamin E on gene expression in the presence of a actinomycin D, a selective inhibitor of DNA-directed RNA synthesis (41). Cells were treated with 1 μM actinomycin D together with 100 μM αTOH (or ethanol), and the levels of mRNAs encoding HMG-CoA reductase and the LDL-receptor were measured using real-time RT-PCR. We observed that in the presence of the transcriptional inhibitor actinomycin D, vitamin E did not affect the expression of these transcripts (data not shown), demonstrating that αTOH attenuates the mRNA levels of cholesterol homeostatic genes by inhibiting transcription, and not by affecting mRNA stability.
Molecular Mechanisms of αTOH Action
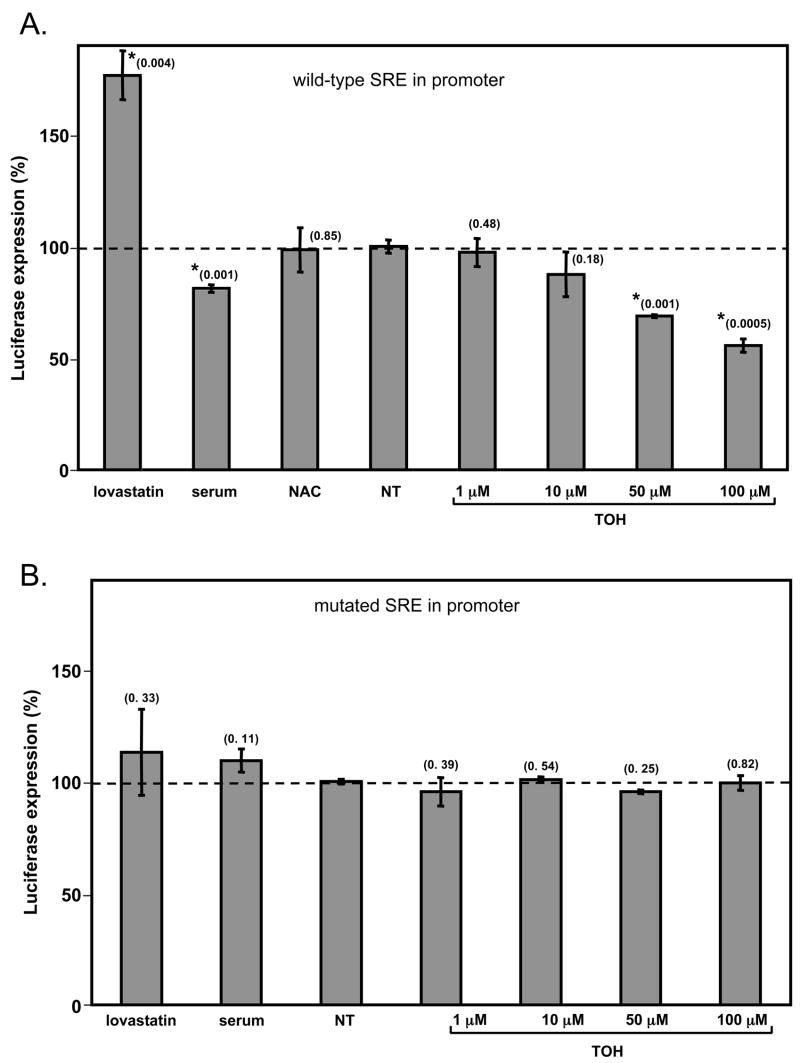
The 10 cholesterol homeostatic genes that are affected by vitamin E treatment share common regulatory features in their promoter regions. Specifically, the sequence 5′ATCACCCCAT3′ is found upstream of the transcription start sites of all of these genes. This conserved regulatory sequence, known as the sterol response element (SRE), is the binding site for the transcription factor SREBP-2, which mediates many established responses to sterols (42). To examine whether SREBP-2 mediates the transcriptional activities of vitamin E, we employed a reporter construct in which the luciferase gene is under the regulation of an SRE-containing promoter (the SREBP-2 promoter, possessing a single SRE at position –293, (31)). We transfected the reporter construct into HepG2 cells, and monitored the effect of αTOH treatment on luciferase expression. Inhibiting cholesterol biosynthesis by treatment of the cells with the HMG-CoA-reductase inhibitor lovastatin caused a ca. 2-fold increase in luciferase expression (Figure 3A), as anticipated from the established response of the SREBP-2 promoter to depletion of cellular sterol pools (43). Conversely, increasing cholesterol levels by the addition of exogenous (cholesterol rich) serum lipoproteins caused a ca. 20% repression of promoter activity (Figure 3A). This feedback inhibition of SRE-containing promoters by sterols is an established feature of cholesterol homeostatic regulation (31, 44–46). Importantly, we observed that treatment of the cells with vitamin E lead to a dose-dependent attenuation of reporter gene expression, reaching approximately 50% inhibition at 100 μM αTOH. The inhibitory effect of αTOH on promoter activity does not reflect a general cell response to antioxidants or redox state, as NAC did not affect promoter activity (Figure 3A). To implicate specific promoter regions in the transcriptional responses to vitamin E, we ablated the single SRE sequence in the luciferase reporter construct using site-directed mutagenesis (see Materials and Methods). As expected, the SRE-defective promoter no longer responded to alterations in cholesterol status induced by lovastatin and serum (Figure 3B). Importantly, mutating the SRE sequence also completely abolished transcriptional responses to αTOH (Figure 3B). As the experimental treatments used in these experiments did not affect cell viability (data not shown), we conclude that αTOH inhibits the expression of these cholesterol homeostatic genes by attenuating SRE-mediated transcriptional responses.
Figure 3. Vitamin E inhibits transcription of a sterol-responsive element (SRE) -driven reporter gene.
HepG2 cells were transiently transfected with 0.5 μg of a pGL3-basic plasmid encoding the structural gene for firefly luciferase driven by the SREBP-2 promoter containing either the wild-type (panel A) or a mutated (panel B) SRE sequences. Two hours after transfection, cells were treated with 10% FBS, 1 mM NAC, 1 μM lovastatin or the indicated tocopherol concentration for 36 hours. Luciferase expression was measured in clarified lysates as described in Materials and Methods. Shown are means and standard deviations of three independent experiments. Luminescence readings were normalized to those measured in untreated lysates (1.2 × 106 luminescence units). Asterisks denote statistically significant changes from control samples, as determined by a student’s t-test. P-values obtained from statistical analyses are shown in parentheses. NT – non-treated.
Physiological regulation of SREBP-2 by sterols occurs through post-translational proteolytic activation of the SREBP-2 transcription factor that controls its translocation from the endoplasmic reticulum to its site of action in the nucleus (47). We examined whether the transcriptional effects of vitamin E, like those of sterols, are mediated by post-translational processing of SREBP-2. To this end, we fractionated treated HepG2 and CHO cells, and assayed the levels of proteolytically-activated SREBP-2 by immunoblotting. As expected, processing of SREBP-2 was enhanced when cholesterol levels were depleted (i.e. in the presence of lovastatin), and reduced upon the addition of exogenous sterols, as shown in Figure 4. Treatment with vitamin E elicited a dose-responsive decrease in the amount of activated (nuclear) SREBP-2, reaching ca. 50% inhibition at 100 μM αTOH, similar to the magnitude of inhibition elicited by sterols (Figure 4).
These results indicate that vitamin E attenuates the transcriptional responses of sterol response elements in the promoter regions of cholesterol homeostatic genes, and that these events are mediated by the actions of SREBP-2.
Attenuation of de novo Cholesterol Biosynthesis by αTOH in vivo
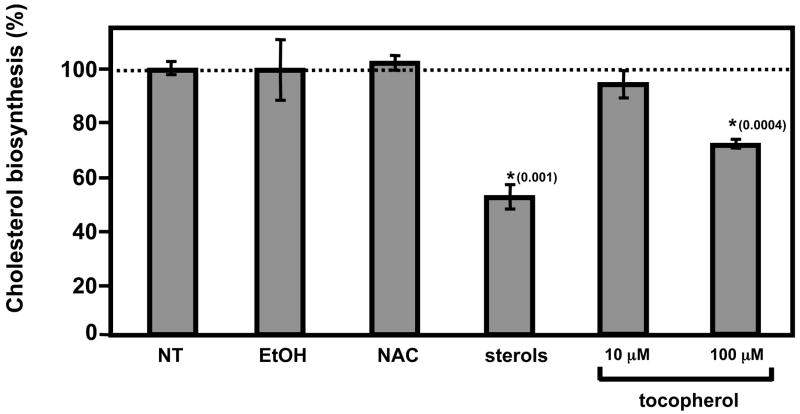
The observations that tocopherol specifically attenuates the expression of key regulators of cholesterol homeostasis raise the possibility that vitamin E levels impact on cellular cholesterol status. To evaluate the physiological significance of these findings, we examined whether treatment with vitamin E influences the rate of de novo cholesterol biosynthesis in cultured cells. HepG2 cells were incubated with the radio-labeled precursor [14C]-acetate for six hours, and incorporation of the label into cholesterol was measured after lipid extraction and thin layer chromatography. Treatment with vitamin E resulted in a dose-dependent inhibition of cholesterol biosynthesis, reaching ~30% inhibition at 100 μM αTOH (Figure 5). The magnitude of inhibition by αTOH was comparable to that induced by established physiological attenuators of the cholesterol biosynthetic pathway, namely high sterols (10 μg/μL cholesterol + 0.1 μg/μL 25-hydroxycholesterol). Importantly, suppression of cholesterol biosynthesis was a specific attribute of vitamin E, as treatment with vehicle control (ethanol) or the general antioxidant NAC did not affect incorporation of acetate into cholesterol (Figure 5). These findings indicate that the specific effects of αTOH on the cholesterol biosynthetic pathway are significant at the physiological end-point of the pathway, namely, the production of cholesterol.
Figure 5. Vitamin E attenuates de novo cholesterol biosynthesis.
HepG2 cells were cultured in 10% LPDS and treated for 24 hours as indicated in the text. Two μCi of [1-14C]acetic acid were added to the culture media for six hours, prior to lipid extraction and chromatographic analysis of labeled cholesterol. Shown are averages and standard deviations of three independent experiments, after normalization to non-treated samples (12,104 CPM).
Discussion
It is commonly believed that the biological requirement for vitamin E stems from the ability of the vitamin to scavenge intracellular free radicals. By counteracting the harmful actions of reactive oxygen species and lipid radicals, the vitamin is thought to prevent cellular damage and the onset of related pathologies. In support of this notion, vitamin E deficient humans and animals display pathologies that are commonly attributed to elevated oxidative stress, such as atherosclerosis and neuronal degeneration. However, recent observations suggest that members of the vitamin E family possess biological activities in addition to those elicited by other antioxidants. Thus, tocopherol was shown to influence apoptosis (48, 49, 50) and proliferation (51), and to modulate the activity of some enzymes (52–55). An additional striking ‘non-classical’ effect of vitamin E is its documented ability to modulate the expression of certain mRNAs and proteins. Recently, a number of groups utilized cDNA micro-array approaches to determine the effect of vitamin E on mRNA expression profiles. Gohil et al (16, 21–23) compared the gene expression profiles in brains and livers from TTP−/− (vitamin E- deficient) mice with those from wild-type mice and reported pronounced differences (2- to 20-fold) in the levels of multiple transcripts that play important roles in myriad biological functions. Similarly, Barella and colleagues compared the expression profile of vitamin E- depleted rats with those obtained from normal animals (17–19, 56). A major confounding factor in interpreting the data from these studies stems from the established genomic responses to changes in redox status. Vertebrate cells have evolved sensitive mechanisms to detect changes in the levels of oxygen, oxygen radicals and reducing agents, and to relay this information to specific transcription factors, such as the hypoxia responsive HIF-1 (24), the redox responsive NF-kB (57) and the anti-oxidant responsive Nrf2 (58, 59). Thus, treatment of cells with vitamin E can (and is likely to) lead to transcriptional responses that originate from changes in cellular redox status, and are not vitamin E–specific. Indeed, the aforementioned expression profiling experiments documented pronounced changes in the expression of multiple known redox-sensitive genes.
To isolate genomic responses that are specific to vitamin E, we focused our attention to αTOH-responsive mRNAs that were not affected by treatment with another antioxidant, NAC. The data revealed a limited number of transcripts that are attenuated specifically by vitamin E. Of special interest are the observations that genes involved in maintaining lipid homeostasis are down-regulated by vitamin E. Of these, ten encode enzymes that catalyze important steps in the de-novo biosynthesis of cholesterol, including the rate-limiting enzyme of this pathway, HMG-CoA reductase. The data further demonstrate that these specific genomic responses to vitamin E are mediated by the sterol-responsive transcription factor SREBP-2. Moreover, αTOH appears to elicit its regulatory action on SREBP-2 through a mechanism similar to that employed by sterols, namely by attenuating the proteolytic activation of SREBP-2 and its translocation into the nucleus. The transcriptional activities of vitamin E have a marked physiological impact in that treatment with αTOH caused a pronounced inhibition of de novo cholesterol biosynthesis.
The precise molecular mechanisms by which vitamin E influences SREBP-2 activity are not completely understood. It is possible that αTOH shares the sensing mechanism utilized for sterols, i.e. binding to sterol-sensing domains such as that found in the sterol sensor SCAP (60, 61). Alternatively, novel and specific mechanisms may exist that convey information about tocopherol levels to the transcriptional machinery.
Our observations suggest that a ‘cross-talk’ exists between the physiological levels of vitamin E and the mechanisms that regulate cholesterol homeostasis. Thus, one would expect that, in whole animals, vitamin E levels will impact overall cholesterol status. Indeed, it was reported that increased dietary intake of vitamin E is accompanied by a dose-dependent reduction in total serum cholesterol in the rat, quail and rabbit. The impact of vitamin E on the health risk in human populations still awaits complete resolution (62–73). The observations described in this study reinforce the notion of physiologically relevant ‘non-classical’ activities of vitamin E, and reveal that key roles of the vitamin in supporting human health may be mediated by its ability to regulate the rates of transcription of specific target genes.
Acknowledgments
This work was supported by award DK067494 from the National Institutes of Health.
We thank Timothy Osborne (University of California Irvine) for reagents, the University of Rochester’s Micro-array Core Facility for their assistance with the profiling experiments, and Noa Noy, Samantha Morley and Julie Beckenstein for critical readings of the manuscript.
ABBREVIATIONS
- PMSF
phenylmethylsulfonyl fluoride
- αTOH
RRR-α-tocopherol
- FBS
fetal bovine serum
- LPDS
lipoproein-deficient serum
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- NAC
N-acetyl cysteine
- DCFDA
2′,7′-dichlorodihydrofluorescin diacetate
- DCF
2′,7′-dichlorofluorescein
- ROS
reactive oxygen species
- HMG-CoA
3-hydroxy-3-methylglutaryl coenzyme A, SREBP-2, Sterol-responsive Element-binding Protein 2
- BHT
butylated hydroxytoluene
References
- 1.Ingold KU, Webb AC, Witter D, Burton GW, Metcalfe TA, Muller DP. Vitamin E remains the major lipid-soluble, chain-breaking antioxidant in human plasma even in individuals suffering severe vitamin E deficiency. Arch Biochem Biophys. 1987;259:224–225. doi: 10.1016/0003-9861(87)90489-9. [DOI] [PubMed] [Google Scholar]
- 2.Burton GW, Cheeseman KH, Doba T, Ingold KU, Slater TF. Vitamin E as an antioxidant in vitro and in vivo. Ciba Found Symp. 1983;101:4–18. doi: 10.1002/9780470720820.ch2. [DOI] [PubMed] [Google Scholar]
- 3.Traber MG, Arai H. Molecular mechanisms of vitamin E transport. Annu Rev Nutr. 1999;19:343–355. doi: 10.1146/annurev.nutr.19.1.343. [DOI] [PubMed] [Google Scholar]
- 4.Blatt DH, Leonard SW, Traber MG. Vitamin E kinetics and the function of tocopherol regulatory proteins. Nutrition. 2001;17:799–805. doi: 10.1016/s0899-9007(01)00637-2. [DOI] [PubMed] [Google Scholar]
- 5.Kaempf-Rotzoll DE, Traber MG, Arai H. Vitamin E and transfer proteins. Curr Opin Lipidol. 2003;14:249–254. doi: 10.1097/00041433-200306000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Sontag TJ, Parker RS. Cytochrome P450 omega-hydroxylase pathway of tocopherol catabolism. Novel mechanism of regulation of vitamin E status. J Biol Chem. 2002;277:25290–25296. doi: 10.1074/jbc.M201466200. [DOI] [PubMed] [Google Scholar]
- 7.Azzi A, Ricciarelli R, Zingg JM. Non-antioxidant molecular functions of alpha-tocopherol (vitamin E) FEBS Lett. 2002;519:8–10. doi: 10.1016/s0014-5793(02)02706-0. [DOI] [PubMed] [Google Scholar]
- 8.Zingg JM, Azzi A. Non-antioxidant activities of vitamin E. Curr Med Chem. 2004;11:1113–1133. doi: 10.2174/0929867043365332. [DOI] [PubMed] [Google Scholar]
- 9.Chojkier M, Houglum K, Lee KS, Buck M. Long- and short-term D-alpha-tocopherol supplementation inhibits liver collagen alpha1(I) gene expression. Am J Physiol. 1998;275:G1480–1485. doi: 10.1152/ajpgi.1998.275.6.G1480. [DOI] [PubMed] [Google Scholar]
- 10.Witt W, Kolleck I, Fechner H, Sinha P, Rustow B. Regulation by vitamin E of the scavenger receptor BI in rat liver and HepG2 cells. J Lipid Res. 2000;41:2009–2016. [PubMed] [Google Scholar]
- 11.Devaraj S, Hugou I, Jialal I. Alpha-tocopherol decreases CD36 expression in human monocyte-derived macrophages. J Lipid Res. 2001;42:521–527. [PubMed] [Google Scholar]
- 12.Ricciarelli R, Zingg JM, Azzi A. Vitamin E reduces the uptake of oxidized LDL by inhibiting CD36 scavenger receptor expression in cultured aortic smooth muscle cells. Circulation. 2000;102:82–87. doi: 10.1161/01.cir.102.1.82. [DOI] [PubMed] [Google Scholar]
- 13.Aratri E, Spycher SE, Breyer I, Azzi A. Modulation of alpha-tropomyosin expression by alpha-tocopherol in rat vascular smooth muscle cells. FEBS Lett. 1999;447:91–94. doi: 10.1016/s0014-5793(99)00277-x. [DOI] [PubMed] [Google Scholar]
- 14.Davies GF, McFie PJ, Khandelwal RL, Roesler WJ. Unique ability of troglitazone to up-regulate peroxisome proliferator- activated receptor-gamma expression in hepatocytes. J Pharmacol Exp Ther. 2002;300:72–77. doi: 10.1124/jpet.300.1.72. [DOI] [PubMed] [Google Scholar]
- 15.Zapolska-Downar D, Zapolski-Downar A, Markiewski M, Ciechanowicz A, Kaczmarczyk M, Naruszewicz M. Selective inhibition by alpha-tocopherol of vascular cell adhesion molecule-1 expression in human vascular endothelial cells. Biochem Biophys Res Commun. 2000;274:609–615. doi: 10.1006/bbrc.2000.3197. [DOI] [PubMed] [Google Scholar]
- 16.Gohil K, Schock BC, Chakraborty AA, Terasawa Y, Raber J, Farese RV, Jr, Packer L, Cross CE, Traber MG. Gene expression profile of oxidant stress and neurodegeneration in transgenic mice deficient in alpha-tocopherol transfer protein. Free Radic Biol Med. 2003;35:1343–1354. doi: 10.1016/s0891-5849(03)00509-4. [DOI] [PubMed] [Google Scholar]
- 17.Barella L, Muller PY, Schlachter M, Hunziker W, Stocklin E, Spitzer V, Meier N, de Pascual-Teresa S, Minihane AM, Rimbach G. Identification of hepatic molecular mechanisms of action of alpha-tocopherol using global gene expression profile analysis in rats. Biochim Biophys Acta. 2004;1689:66–74. doi: 10.1016/j.bbadis.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Hyland S, Muller D, Hayton S, Stoecklin E, Barella L. Cortical gene expression in the vitamin E-deficient rat: possible mechanisms for the electrophysiological abnormalities of visual and neural function. Annals of nutrition & metabolism. 2006;50:433–441. doi: 10.1159/000094635. [DOI] [PubMed] [Google Scholar]
- 19.Rota C, Barella L, Minihane AM, Stocklin E, Rimbach G. Dietary alpha-tocopherol affects differential gene expression in rat testes. IUBMB Life. 2004;56:277–280. doi: 10.1080/15216540410001724133. [DOI] [PubMed] [Google Scholar]
- 20.Rota C, Rimbach G, Minihane AM, Stoecklin E, Barella L. Dietary vitamin E modulates differential gene expression in the rat hippocampus: potential implications for its neuroprotective properties. Nutritional neuroscience. 2005;8:21–29. doi: 10.1080/10284150400027123. [DOI] [PubMed] [Google Scholar]
- 21.Oommen S, Vasu VT, Leonard SW, Traber MG, Cross CE, Gohil K. Genome wide responses of murine lungs to dietary alpha-tocopherol. Free Radic Res. 2007;41:98–109. doi: 10.1080/10715760600935567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasu VT, Hobson B, Gohil K, Cross CE. Genome-wide screening of alpha-tocopherol sensitive genes in heart tissue from alpha-tocopherol transfer protein null mice (ATTP(−/−)) FEBS Lett. 2007;581:1572–1578. doi: 10.1016/j.febslet.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gohil K, Godzdanker R, O’Roark E, Schock BC, Kaini RR, Packer L, Cross CE, Traber MG. Alpha-tocopherol transfer protein deficiency in mice causes multi-organ deregulation of gene networks and behavioral deficits with age. Ann N Y Acad Sci. 2004;1031:109–126. doi: 10.1196/annals.1331.012. [DOI] [PubMed] [Google Scholar]
- 24.Haddad JJ. Oxygen sensing and oxidant/redox-related pathways. Biochem Biophys Res Commun. 2004;316:969–977. doi: 10.1016/j.bbrc.2004.02.162. [DOI] [PubMed] [Google Scholar]
- 25.Asmis R. Physical partitioning is the main mechanism of alpha-tocopherol and cholesterol transfer between lipoproteins and P388D1 macrophage-like cells. Eur J Biochem. 1997;250:600–607. doi: 10.1111/j.1432-1033.1997.0600a.x. [DOI] [PubMed] [Google Scholar]
- 26.Qian J, Atkinson J, Manor D. Biochemical Consequences of Heritable Mutations in the alpha-Tocopherol Transfer Protein. Biochemistry. 2006;45:8236–8242. doi: 10.1021/bi060522c. [DOI] [PubMed] [Google Scholar]
- 27.Qian J, Morley S, Wilson K, Nava P, Atkinson J, Manor D. Intracellular trafficking of vitamin E in hepatocytes: the role of tocopherol transfer protein. J Lipid Res. 2005;46:2072–2082. doi: 10.1194/jlr.M500143-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Sladowski D, Steer SJ, Clothier RH, Balls M. An improved MTT assay. Journal of immunological methods. 1993;157:203–207. doi: 10.1016/0022-1759(93)90088-o. [DOI] [PubMed] [Google Scholar]
- 29.Ellerby LM, Bredesen DE. Measurement of cellular oxidation, reactive oxygen species, and antioxidant enzymes during apoptosis. Methods Enzymol. 2000;322:413–421. doi: 10.1016/s0076-6879(00)22040-5. [DOI] [PubMed] [Google Scholar]
- 30.Amoroso S, Gioielli A, Cataldi M, Di Renzo G, Annunziato L. In the neuronal cell line SH-SY5Y, oxidative stress-induced free radical overproduction causes cell death without any participation of intracellular Ca(2+) increase. Biochim Biophys Acta. 1999;1452:151–160. doi: 10.1016/s0167-4889(99)00110-x. [DOI] [PubMed] [Google Scholar]
- 31.Shin DJ, Osborne TF. Thyroid hormone regulation and cholesterol metabolism are connected through Sterol Regulatory Element-Binding Protein-2 (SREBP-2) J Biol Chem. 2003;278:34114–34118. doi: 10.1074/jbc.M305417200. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 35.Lee JE, Kim KM, Cho JW, Suh SI, Suh MH, Kwon TK, Park JW, Bae JH, Song DK, Cho CH, Bae I, Baek WK. Pyrrolidine dithiocarbamate induces cyclooxygenase-2 expression in NIH 3T3 fibroblast cells. Biochem Biophys Res Commun. 2002;298:230–234. doi: 10.1016/s0006-291x(02)02430-0. [DOI] [PubMed] [Google Scholar]
- 36.Liou JS, Chen CY, Chen JS, Faller DV. Oncogenic ras mediates apoptosis in response to protein kinase C inhibition through the generation of reactive oxygen species. J Biol Chem. 2000;275:39001–39011. doi: 10.1074/jbc.M007154200. [DOI] [PubMed] [Google Scholar]
- 37.Pani G, Colavitti R, Bedogni B, Anzevino R, Borrello S, Galeotti T. A redox signaling mechanism for density-dependent inhibition of cell growth. J Biol Chem. 2000;275:38891–38899. doi: 10.1074/jbc.M007319200. [DOI] [PubMed] [Google Scholar]
- 38.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, Goldschmidt-Clermont PJ, Finkel T. Regulation of reactive-oxygen-species generation in fibroblasts by Rac1. Biochem J. 1996;318:379–382. doi: 10.1042/bj3180379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodwell VW, Nordstrom JL, Mitschelen JJ. Regulation of HMG-CoA reductase. Advances in lipid research. 1976;14:1–74. doi: 10.1016/b978-0-12-024914-5.50008-5. [DOI] [PubMed] [Google Scholar]
- 40.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 41.Wadkins RM, Jares-Erijman EA, Klement R, Rudiger A, Jovin TM. Actinomycin D binding to single-stranded DNA: sequence specificity and hemi-intercalation model from fluorescence and 1H NMR spectroscopy. J Mol Biol. 1996;262:53–68. doi: 10.1006/jmbi.1996.0498. [DOI] [PubMed] [Google Scholar]
- 42.Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Zhang F, Li C, Lin M, Briggs MR. Synergistic activation of human LDL receptor expression by SCAP ligand and cytokine oncostatin M. Arterioscler Thromb Vasc Biol. 2003;23:90–96. doi: 10.1161/01.atv.0000046229.77566.e5. [DOI] [PubMed] [Google Scholar]
- 44.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 45.Amemiya-Kudo M, Shimano H, Hasty AH, Yahagi N, Yoshikawa T, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga J, Harada K, Gotoda T, Sato R, Kimura S, Ishibashi S, Yamada N. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res. 2002;43:1220–1235. [PubMed] [Google Scholar]
- 46.Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 47.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azzi A, Boscoboinik D, Fazzio A, Marilley D, Maroni P, Ozer NK, Spycher S, Tasinato A. RRR-alpha-tocopherol regulation of gene transcription in response to the cell oxidant status. Z Ernahrungswiss. 1998;37:21–28. [PubMed] [Google Scholar]
- 49.de Nigris F, Franconi F, Maida I, Palumbo G, Anania V, Napoli C. Modulation by alpha- and gamma-tocopherol and oxidized low-density lipoprotein of apoptotic signaling in human coronary smooth muscle cells. Biochem Pharmacol. 2000;59:1477–1487. doi: 10.1016/s0006-2952(00)00275-6. [DOI] [PubMed] [Google Scholar]
- 50.Gunawardena K, Murray DK, Meikle AW. Vitamin E and other antioxidants inhibit human prostate cancer cells through apoptosis. The Prostate. 2000;44:287–295. doi: 10.1002/1097-0045(20000901)44:4<287::aid-pros5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 51.Venkateswaran V, Fleshner NE, Klotz LH. Modulation of cell proliferation and cell cycle regulators by vitamin E in human prostate carcinoma cell lines. The Journal of urology. 2002;168:1578–1582. doi: 10.1016/S0022-5347(05)64524-7. [DOI] [PubMed] [Google Scholar]
- 52.Clement S, Tasinato A, Boscoboinik D, Azzi A. The effect of alpha-tocopherol on the synthesis, phosphorylation and activity of protein kinase C in smooth muscle cells after phorbol 12- myristate 13-acetate down-regulation. Eur J Biochem. 1997;246:745–749. doi: 10.1111/j.1432-1033.1997.t01-2-00745.x. [DOI] [PubMed] [Google Scholar]
- 53.Ozer NK, Palozza P, Boscoboinik D, Azzi A. d-alpha-Tocopherol inhibits low density lipoprotein induced proliferation and protein kinase C activity in vascular smooth muscle cells. FEBS Lett. 1993;322:307–310. doi: 10.1016/0014-5793(93)81592-n. [DOI] [PubMed] [Google Scholar]
- 54.Mahoney CW, Azzi A. Vitamin E inhibits protein kinase C activity. Biochem Biophys Res Commun. 1988;154:694–697. doi: 10.1016/0006-291x(88)90195-7. [DOI] [PubMed] [Google Scholar]
- 55.Pentland AP, Morrison AR, Jacobs SC, Hruza LL, Hebert JS, Packer L. Tocopherol analogs suppress arachidonic acid metabolism via phospholipase inhibition. J Biol Chem. 1992;267:15578–15584. [PubMed] [Google Scholar]
- 56.Muller PY, Netscher T, Frank J, Stoecklin E, Rimbach G, Barella L. Comparative quantification of pharmacodynamic parameters of chiral compounds (RRR- vs. all-rac-alpha tocopherol) by global gene expression profiling. Journal of plant physiology. 2005;162:811–817. doi: 10.1016/j.jplph.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 57.Gius D, Botero A, Shah S, Curry HA. Intracellular oxidation/reduction status in the regulation of transcription factors NF-kappaB and AP-1. Toxicol Lett. 1999;106:93–106. doi: 10.1016/s0378-4274(99)00024-7. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 60.Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 61.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 62.Chen LH, Liao S, Packett LV. Interaction of dietary vitamin E and protein level of lipid source with serum cholesterol level in rats. J Nutr. 1972;102:729–732. doi: 10.1093/jn/102.6.729. [DOI] [PubMed] [Google Scholar]
- 63.Jack Yang NY, Desai ID. Effect of high levels of dietary vitamin E on liver and plasma lipids and fat soluble vitamins in rats. J Nutr. 1977;107:1418–1426. doi: 10.1093/jn/107.8.1418. [DOI] [PubMed] [Google Scholar]
- 64.Morrissey RB, Donaldson WE. Cholesteremia in Japanese quail: response to a mixture of vitamins C and E and choline chloride. Artery. 1979;5:182–192. [PubMed] [Google Scholar]
- 65.Komaratat P, Chupukcharoen N, Wilairat P. Effect of vitamin E on cholesterol plasma lipoprotein distribution and metabolism in rabbit. Int J Vitam Nutr Res. 1985;55:167–171. [PubMed] [Google Scholar]
- 66.Phonpanichrasamee C, Komaratat P, Wilairat P. Hypocholesterolemic effect of vitamin E on cholesterol-fed rabbit. Int J Vitam Nutr Res. 1990;60:240–244. [PubMed] [Google Scholar]
- 67.Wilson RB, Middleton CC, Sun GY. Vitamin E, antioxidants and lipid peroxidation in experimental atherosclerosis of rabbits. J Nutr. 1978;108:1858–1867. doi: 10.1093/jn/108.11.1858. [DOI] [PubMed] [Google Scholar]
- 68.Stephens NG, Parsons A, Schofield PM, Kelly F, Cheeseman K, Mitchinson MJ. Randomised controlled trial of vitamin E in patients with coronary disease: Cambridge Heart Antioxidant Study (CHAOS) Lancet. 1996;347:781–786. doi: 10.1016/s0140-6736(96)90866-1. [DOI] [PubMed] [Google Scholar]
- 69.de Gaetano G. Low-dose aspirin and vitamin E in people at cardiovascular risk: a randomised trial in general practice. Collaborative Group of the Primary Prevention Project. Lancet. 2001;357:89–95. doi: 10.1016/s0140-6736(00)03539-x. [DOI] [PubMed] [Google Scholar]
- 70.Boaz M, Smetana S, Weinstein T, Matas Z, Gafter U, Iaina A, Knecht A, Weissgarten Y, Brunner D, Fainaru M, Green MS. Secondary prevention with antioxidants of cardiovascular disease in endstage renal disease (SPACE): randomised placebo-controlled trial. Lancet. 2000;356:1213–1218. doi: 10.1016/s0140-6736(00)02783-5. [DOI] [PubMed] [Google Scholar]
- 71.Rapola JM, Virtamo J, Haukka JK, Heinonen OP, Albanes D, Taylor PR, Huttunen JK. Effect of vitamin E and beta carotene on the incidence of angina pectoris. A randomized, double-blind, controlled trial. Jama. 1996;275:693–698. doi: 10.1001/jama.1996.03530330037026. [DOI] [PubMed] [Google Scholar]
- 72.Marchioli R. Dietary supplementation with n-3 polyunsaturated fatty acids and vitamin E after myocardial infarction: results of the GISSI-Prevenzione trial. Gruppo Italiano per lo Studio della Sopravvivenza nell’Infarto miocardico. Lancet. 1999;354:447–455. [PubMed] [Google Scholar]
- 73.Miller ER, 3rd, Pastor-Barriuso R, Dalal D, Riemersma RA, Appel LJ, Guallar E. Meta-analysis: high-dosage vitamin E supplementation may increase all-cause mortality. Ann Intern Med. 2005;142:37–46. doi: 10.7326/0003-4819-142-1-200501040-00110. [DOI] [PubMed] [Google Scholar]