Abstract
Background
Bile acid reclamation following ileo-cecal resection (ICR) may prevent colonic mucosa from chronic injury. In this study, we hypothesized that in a murine model of ICR the remnant colon would upregulate the cellular machinery necessary for BA reclamation and would do so in an FXR- and bacteria-dependent manner.
Methods
Conventional (WT), conventional FXR knockout (FXR null) and germ free (GF) mice were randomized to undergo either ICR or sham operation. The ascending colon was harvested for histology & immunohistochemistry and changes in bile acid homeostatic gene expression determined by RT-PCR 7d following surgery.
Results
Following ICR WT mice showed significant increases in the expression of genes regulating bile acid transport including IBABP, Asbt, Ostβ and FGF 15. Increased expression of IBABP and Asbt was confirmed by immunohistochemistry. Induction of bile acid transport genes was absent or attenuated in FXR null and GF mice.
Conclusion
Bacterial dependent up regulation of IBABP is FXR mediated in the colon following ICR. Mice lacking microbiota (GF) or FXR are unable to increase the expression of IBABP or FGF 15.
Introduction
Resection of the ileum, cecum and proximal colon is commonly required for the management of various surgical problems, including advanced necrotizing enterocolitis and Crohn’s disease. One clinical concern which arises from such a procedure is the potential for diminished capacity of the remaining bowel to reclaim luminal bile acids (BA). Another concern is the potential for exposure of colonocytes to bile acids to cause DNA damage and lead to carcinogenesis.
In the normal intestine, the terminal ileum functions as the conduit by which luminal BA are reclaimed into the enterohepatic circulation.1 BA are taken up by ileal enterocytes and transported to the liver via the portal vein for recycling. BA circulation is a tightly regulated process, particularly by BA themselves. By binding to the primary intracellular bile acid receptor, farnesoid X receptor (FXR),2,3 BA are able to influence the expression of numerous genes involved in their synthesis and transport. In ileal enterocytes this includes genes such as Asbt (luminal BA uptake),4 IBABP (cytosolic transport),5 Ost α/β (BA export into circulation),6 and FGF15 (intestinal signal to hepatocytes).7 At present, the ability of the colon to aid in bile acid reclamation following the loss of the ileum is not understood.
Under normal conditions the colon sees very low concentrations (approximately 3% of total luminal bile) of luminal BA due to reclamation by the terminal ileum. Colonic bacteria play a key role in deconjugation and 7α/β-hydroxylation of remaining luminal BA causing an increase in hydrophobicity and allowing for their passive absorption by colonocytes.8 Increasing BA hydrophobicity also introduces the potential for BA to enter colonocytes and cause DNA damage.9 Indeed, in experimental models, chronic exposure of the colon to bile acids has lead to carcinogenesis,10,11 and bile acids have been linked to incidence of colon cancer in humans.9 In turn, this calls into question the role of colonic bacteria in modulating colonocyte proliferation.
Using our recently developed murine model of ileocecal resection (ICR)12 we investigated the changes in expression of genes associated with BA circulation, including IBABP, Asbt, FGF15 and Ostβ, in colon following resection. These genes were selected because they represent the ability of an ileal enterocytes to take up bile acids (Asbt), shuttle them within the cell (IBABP), extrude them into the circulation (OSTβ) and signal to the liver regarding bile acid concentration (FGF-15). We hypothesized that in response to loss of the ileum the remnant colon would adapt by upregulating the molecular machinery needed for bile acid reclamation. We also hypothesized that upregulation of this machinery is FXR-mediated and bacteria-dependent.
Materials and Methods
Animals and Experimental Design
The University of North Carolina-Chapel Hill Institutional Animal Care and Use Committee approved the protocol for this study. Male C57BL/6J mice, 8–12 weeks old (weight range 25–31 g) were used for this study. Wild type mice were obtained from the Jackson Laboratories (Bar Harbor, ME). FXR null mice were obtained from Dr. Saul Karpen (Baylor College of Medicine, Houston, Texas) with the permission of Dr. Frank J. Gonzalez (National Cancer Institute, NIH, Bethesda, MD). Germ free (GF) mice were housed in the gnotobiotic facility at the University of North Carolina-Chapel Hill. The mice were housed in groups of 5 and randomly assigned to undergo either sham operation (intestinal transection only, n = 3–4/group) or ileocecal resection (ICR, n = 3–4/group). All mice were switched to a liquid diet (Micro-Stabilized Rodent Liquid Diet LAD 101/101A, Purina Mills, St. Louis, MO) 2 days before the operation and maintained on this diet for the duration of the experiment to avoid intestinal obstruction, as previously described.12
Operative Procedure
Mice were weighed, ear tagged and clipped the day of the procedure. All operations were performed under sterile conditions and with the aid of an operating microscope (7X magnification) using inhaled 2% isofluorane and oxygen for anesthesia. Surgeries done under gnotobiotic conditions were performed in a custom built surgical isolator developed by the National Gnotobiotic Rodent Resource Center, Department of Laboratory Medicine, University of North Carolina at Chapel Hill. This allowed animals to be maintained in a germ free environment throughout the experiment. Detailed description of the ICR model has been published 12. Briefly, 12 cm of ileum, cecum and 1–2 cm of proximal colon are removed and a primary anastomosis performed. Mice undergoing sham resections undergo division and re-anastomosis of the intestine 12 cm proximal to the cecum. All mice received a single IP dose of Pipercillin and Tazobactam (100 mg/kg) at the start of the procedure. To verify GF status after surgery, fresh fecal samples were collected from animals 4 and 7 days after surgery (and from co-housed NoTx controls). Feces were sealed in a plastic cup before removal from isolators. Aerobic and anaerobic cultures as well as Gram stains verified lack of bacterial contamination. Homogenized fecal samples were also held for one week at room temperature and examined for molds. Enlarged cecum in T and NoTx animals, co-housed with ICR animals, also verified GF status.
Tissue harvest
At 7d following either sham-operation or ICR, mice were weighed and then sacrificed between 6 am and 9 am by cervical dislocation while under isofluorane anesthesia. The entire bowel was removed, and the intestinal contents gently expressed by cotton swabs and saline flush. For animals that underwent ICR, we measured 1 cm distal from the anastomosis and collected the next 1 cm of colon for fixation in 10% normal buffered formalin. We collected the next 2 cm for RNA isolation. For sham-operated animals, we measured 2 cm distal from the cecum to collect the 1 cm piece of colon for fixation, and collected the next 2 cm for RNA isolation.
Histology and analysis of intestinal morphometrics
Sections were stained with H&E or subjected to immunohistochemistry (as described below). Histological samples were analyzed in a blinded manner.
qRT-PCR
The following genes were selected based on the known function in bile acid reclamation in the ileum13. We hypothesized that similar mechanisms would occur in the colon. Changes in expression of mRNA transcripts following sham or ICR were determined by quantitative RT-PCR. Briefly, total RNA was isolated from colon from both sham and ICR, DNase treated and subjected to real time RT-PCR using Taqman One-Step RT-PCR Master Mix (Applied Biosystems, Foster City, CA) and Taqman Gene Expression assays for Asbt (Mm00488258_m1), IBABP (Mm04434316_m1), Ostβ (Mm00619242_m1), and FGF15 (Mm00433278_m1). β-actin (Mm00607939_s1) expression was used as the endogenous control. Relative changes in expression levels were calculated by the ΔΔCt method utilizing total colonic RNA from wild type unoperated animals as the reference.
Immunofluoresence
Formalin-fixed, paraffin-embedded sections (5 µm) of colon were used to evaluate protein expression of IBABP and Asbt. Sections were deparaffinized, rehydrated, and boiled in 10 mM sodium citrate, pH 6.0. After cooling, sections were washed in PBS and blocked. For IBABP, sections were blocked with blocking solution containing 1% BSA, 0.2% powdered milk, and 0.3% Triton X100 I in PBS for 15 min at room temperature, as described previously.14 For Asbt, sections were blocked with blocking solution containing 10% donkey serum in PBS for 1 h in a humidified chamber at room temperature, as previously described.15 Blocking solution was replaced with primary antibody in respective blocking solution, and the sections were incubated overnight at 4°C in a humidified chamber. The next day, sections were washed in PBS and incubated in respective blocking solutions for 2–4 h at room temperature in a humidified chamber. Nuclei were stained with DAPI (4’,6-diamino-2-phenylindole dihydrocholoride). The primary antibodies used were rabbit anti-IBABP (1:2000)14 and rabbit anti-Asbt (1:500) (a kind gift of P. Dawson, Wake Forest University). The secondary antibodies used were FITC-labeled donkey anti-rabbit antibody (1:500) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and Alexa fluor 488 donkey anti-rabbit antibody (1:500) (Invitrogen Corporation, Carlsbad, CA) for IBABP and Asbt detection, respectively.
Statistical Analysis
All quantitative results are presented as mean values ± standard error of the mean (SEM). Means were compared using analysis of variance (ANOVA) with correction for multiple comparisons using the Fisher’s procedure. For all parameters a p-value of P < 0.05 was considered the level of significance.
Results
Quantification of mRNA expression in segments of adult mouse colon by real time RT-PCR revealed that in wild type animals, colonic IBABP expression significantly increased by 12-fold 7d after ICR when compared to sham operated animals (Fig. 1A). We observed a 5.6-fold increase in Asbt mRNA expression in colon from resected mice compared to sham-operated mice (Fig 1B). Expression of FGF15 mRNA increased 5.9-fold in the colon following ICR compared to sham-operated mice (Fig. 1C). Expression of the basolateral bile acid transporter subunit Ostβ was increased 1.6-fold after ICR compared to sham-operated animals (Fig. 1D).
Figure 1.
ICR stimulates upregulation of mRNA expression of BA reclamation genes. Total RNA from wild type mouse colon 7 d following ICR or sham was subjected to real time RT-PCR to evaluate expression levels of (A) IBABP, (B) Asbt, (C) FGF-15, and (D) Ostβ. Data represent relative fold increases in mRNA expression are expressed as mean±SEM. * indicates statistical significance at P < 0.05.
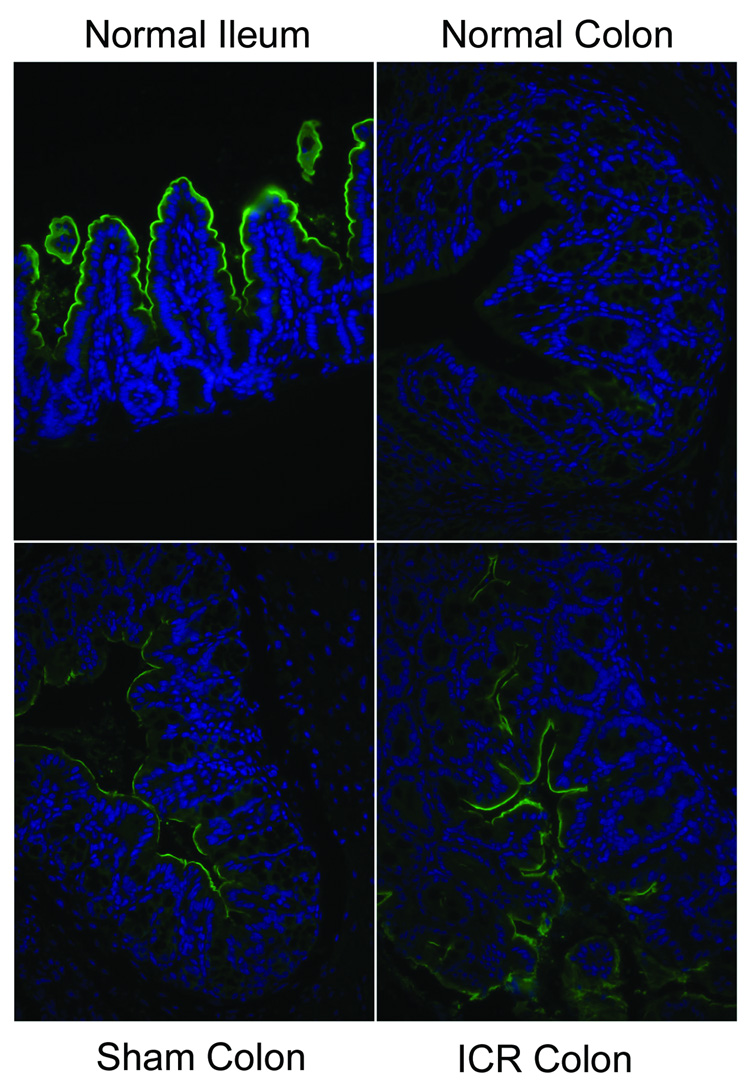
Expression of IBABP protein is normally observed within the cytoplasm of ileal enterocytes (Fig. 2). Following ICR, immunohistochemistry demonstrated IBABP protein expression within colonic epithelium (Fig. 2). Minimal IBABP staining was observed in colonocytes of sham-operated animals. Asbt protein expression is normally found on the apical membrane of ileal enterocytes and is negligible on the apical membrane of colonocytes from unoperated colon (Fig. 3). However, staining of Asbt on the apical membrane of colonocytes was clearly observed following resection and resembled staining patterns seen in the ileum (Fig. 3). Sham operation stimulated some apical expression of Asbt protein.
Figure 2.
Protein expression of IBABP in colonocytes. Immunofluorescence staining was performed on normal ileum, colon from sham operated mice and colon from ICR mice. IBABP expression is clearly cytosolic in ileal enterocytes and is found in all villus enterocytes. Staining in the colon of sham operated mice shows only background staining. IBABP staining in colon of ICR mice shows cytosolic, yet patchy staining in colonocytes.
Figure 3.
Protein expression of Asbt in colonocytes. Immunofluorescence staining was performed on normal ileum, normal colon, colon from sham operated mice and colon from ICR mice. Asbt expression is specifically located on the apical membrane of ileal enterocytes. Little staining is observed on the apical membranes of colonocytes from normal colon. Some Asbt staining is observed on the apical membranes of colonocytes from sham operated mice. Asbt staining appears more extensive on apical membranes of coloncytes from ICR mice.
Others have demonstrated that FXR plays a critical role in regulating the expression of IBABP, Asbt, Ostβ and FGF15 in ileal enterocytes in a bile acid-dependent manner. To determine if a similar mechanism occurs in the colonocytes, we employed FXR null mice to test our hypothesis that FXR mediates upregulation of bile homeostatic genes involved in bile acid reclamation in the colon following ICR. We found that IBABP and FGF15 mRNA expression was negligible in the colon from FXR null mice 7d following ICR (Fig. 4A and C). In contrast neither Asbt nor Ostβ mRNA expression were different in colon from FXR null mice compared to WT mice undergoing ICR (Fig. 4B and D).
Figure 4.
Comparison of mRNA expression of BA reclamation genes in WT, FXR-null and GF mice 7 d following ICR. Total RNA was subjected to real time RT-PCR to evaluate expression levels of (A) IBABP, (B) Asbt, (C) FGF-15, and (D) Ostβ. Expression levels from animals that underwent ICR were compared to levels from sham animals to yield fold change over sham. Data represent fold changes over sham and are expressed as mean±SEM. * indicates statistical significance at P < 0.05.
Others have previously demonstrated that the endogenous bacterial flora in the colon are able to convert taurocholate to deoxycholate, a more hydrophobic dihydroxy bile acid that can be passively absorbed by colonocytes.16 Since intracellular bile acids may be responsible for the upregulation of the bile acid transporters, we evaluated the role of colonic microflora in regulating IBABP, Asbt, Ostβ, and FGF15 gene expression following ICR using GF mice. The results showed that 7 d following ICR the absence of luminal bacteria abolished the expected upregulation of IBABP, Asbt and FGF15, but not Ostβ (Fig. 4).
Discussion
Resection of the terminal ileum, cecum and proximal colon results in loss of the normal pathway for BA reclamation by the small intestine and exposes the remaining colon to potentially damaging concentrations of bile acids. We hypothesized that in a murine model of ICR the remnant colon would upregulate the cellular machinery necessary for BA reclamation and would do so in an FXR- and bacteria-dependent manner. Following ICR, we observed increased mRNA expression of several components of the ileal bile acid transport pathway, including IBABP, Asbt, and FGF15 in colonocytes. This is similar to what was reported by Frankenberg et al. for colon following 0.2% cholic acid diet.17 Immunofluorescence confirms the increases in intracellular IBABP and apical membrane Asbt protein expression in colonocytes following ICR. The reason for Asbt protein expression in colonocytes from sham-operated mice is not clear. It has been our experience that any operative procedure that transects the intestine results in some altered gene expression. In this case, Asbt protein expression may be due to an ileus resulting in increased BA in the colonic lumen or may be due to the single dose of antibiotics given at the time of operation. Using FXR null mice, we found that FXR was essential for the upregulation of IBABP and FGF15 in colonocytes following ICR, but not for Asbt or Ostβ. Furthermore, the use of germ free mice demonstrated that bacteria are also necessary for upregulation of genes involved in BA reclamation in the colon.
The mechanism which drives the upregulation of the genes involved in BA reclamation in the colon is not completely understood, however, our proposed mechanism is outlined in Figure 5. Our data from germ free mice suggest that a critical first step initiating upregulation of BA reclamation genes is deconjugation of luminal BA, such as taurocholate, by colonic microflora which would allow passive diffusion of BA into colonocytes.16,18 Once inside colonocytes, BA, in an FXR-dependent manner, could stimulate the upregulation of IBABP and FGF15, as is seen in the ileum,6,5,7 to allow for intracellular BA shuttling and communication with the liver, respectively. Asbt expression is upregulated, in an FXR-dependent manner, to allow for increased BA uptake by colonocytes. This may occur via PXR, which is expressed in the intestinal tract and has been shown to upregulate Asbt when exposed to agonist.19.
Figure 5.
Potential mechanism for upregulation of BA reclamation genes in colonocytes following ICR. Conjugated BA in the colonic lumen are deconjugated by colonic bacteria. This allows deconjugated BA be passively absorbed by colonocytes. Intracellular binding of BA to FXR or PXR stimulates expression of IBABP, FGF15 and Asbt, which then equips colonocytes with the machinery needed for active BA uptake.
We conclude that the colon is able to adapt to the loss of ileal bile acid uptake in a murine model of ICR by upregulating the machinery necessary to import, transport and export bile acids from the intestinal lumen. This upregulation is partially FXR-dependent and requires colonic bacteria for initiation. This may have clinical implications for the regulation of bile acid uptake in patients following ICR.
Acknowledgements
The authors would like to thank Brooks Scull for help with animal care. We would also like to thank Maureen Bower for assistance with the gnotobiotic animals and germ free surgical preparation. The authors gratefully appreciate the assistance of Dr. Stephen Krasinski with Asbt immunofluoresence.
Financial backing: Center for Gastrointestinal Biology and Disease (P30 DK34987) and The National Gnotobiotic Rodent Resource Center (P40 RR018603)
Abbreviations
- ICR
ileo-cecal resection
- GF
germ free
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zollner G, Marschall HU, Wagner M, Trauner M. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: pathogenetic and therapeutic considerations. Mol Pharm. 2006;3:231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
- 2.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 3.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 4.Chen F, Ma L, Dawson PA, Sinal CJ, Sehayek E, Gonzalez FJ, Breslow J, Ananthanarayanan M, Shneider BL. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem. 2003;278:19909–19916. doi: 10.1074/jbc.M207903200. [DOI] [PubMed] [Google Scholar]
- 5.Hwang ST, Urizar NL, Moore DD, Henning SJ. Bile acids regulate the ontogenic expression of ileal bile acid binding protein in the rat via the farnesoid X receptor. Gastroenterology. 2002;122:1483–1492. doi: 10.1053/gast.2002.32982. [DOI] [PubMed] [Google Scholar]
- 6.Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res. 2006;47:201–214. doi: 10.1194/jlr.M500417-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Flynn C, Montrose DC, Swank DL, Nakanishi M, Ilsley JN, Rosenberg DW. Deoxycholic acid promotes the growth of colonic aberrant crypt foci. Mol Carcinog. 2007;46:60–70. doi: 10.1002/mc.20253. [DOI] [PubMed] [Google Scholar]
- 11.Galloway DJ, Owen RW, Jarrett F, Boyle P, Hill MJ, George WD. Experimental colorectal cancer: the relationship of diet and faecal bile acid concentration to tumour induction. Br J Surg. 1986;73:233–237. doi: 10.1002/bjs.1800730327. [DOI] [PubMed] [Google Scholar]
- 12.Dekaney CM, Fong JJ, Rigby RJ, Lund PK, Henning SJ, Helmrath MA. Expansion of intestinal stem cells associated with long-term adaptation following ileocecal resection in mice. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1013–G1022. doi: 10.1152/ajpgi.00218.2007. [DOI] [PubMed] [Google Scholar]
- 13.Angelin B. Telling the liver (not) to make bile acids: a new voice from the gut? Cell Metab. 2005;2:209–210. doi: 10.1016/j.cmet.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Crossman MW, Hauft SM, Gordon JI. The mouse ileal lipid-binding protein gene: a model for studying axial patterning during gut morphogenesis. J Cell Biol. 1994;126:1547–1564. doi: 10.1083/jcb.126.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006;26:9060–9070. doi: 10.1128/MCB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hylemon PB, Harder J. Biotransformation of monoterpenes, bile acids, and other isoprenoids in anaerobic ecosystems. FEMS Microbiol Rev. 1998;22:475–488. doi: 10.1111/j.1574-6976.1998.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 17.Frankenberg T, Rao A, Chen F, Haywood J, Shneider BL, Dawson PA. Regulation of the mouse organic solute transporter alpha-beta, Ostalpha-Ostbeta, by bile acids. Am J Physiol Gastrointest Liver Physiol. 2006;290:G912–G922. doi: 10.1152/ajpgi.00479.2005. [DOI] [PubMed] [Google Scholar]
- 18.Mekhjian HS, Phillips SF, Hofmann AF. Colonic absorption of unconjugated bile acids: perfusion studies in man. Dig Dis Sci. 1979;24:545–550. doi: 10.1007/BF01489324. [DOI] [PubMed] [Google Scholar]
- 19.Wagner M, Halilbasic E, Marschall HU, Zollner G, Fickert P, Langner C, Zatloukal K, Denk H, Trauner M. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology. 2005;42:420–430. doi: 10.1002/hep.20784. [DOI] [PubMed] [Google Scholar]