Abstract
Human purine nucleoside phosphorylase (PNP) was crystallized with transition state analogue inhibitors Immucillin-H and DADMe-Immucillin-H synthesized with ribosyl mimics of l-stereochemistry. The inhibitors demonstrate that major driving forces for tight binding of these analogues are the leaving group interaction and the cationic mimicry of the transition state, even though large geometric changes occur with d-Immucillins and l-Immucillins bound to human PNP.
Human purine nucleoside phosphorylase (PNP) is required for degradation of 2’-deoxyguanosine and its genetic deficiency is characterized by a lethal T-cell immune deficiency caused by accumulation of dGTP in dividing T-cells.1,2 Human PNP is therefore a target for T-cell disease. The Immucillin family of transition state analogue inhibitors for PNP was developed based on experimentally measured kinetic isotope effects.3 Immucillin-H (ImmH) [1] and DADMe-Immucillin-H (DADMe-ImmH) [3] are picomolar inhibitors of PNP and are in clinical trials for T-cell malignancies and T-cell based autoimmune diseases. 4,5
The Immucillins mimic the ribooxacarbenium ion-like cationic transition state of PNP. ImmH [1] contains an iminoribitol with stereochemistry similar to the β-d-ribose found in purine nucleoside substrates of PNP. DADMe-ImmH [3] mimics the dissociated transition state of human PNP.6 The Immucillins are slow-onset tight-binding inhibitors with initial binding giving Ki values in the low nanomolar range and final equilibrium dissociation constants (Ki* values) in the picomolar range.7,8 Since many inhibitors retain potency in their enantiomeric forms, l-Immucillin-H (l-ImmH) [2] and l-DADMe-Immucillin-H (L-DADMe-ImmH) [4] were synthesized and [4] was a surprisingly potent inhibitor against PNP (Table 1).9 This finding was unexpected because of large losses of binding energy when other geometric or electrostatic features of the Immucillin transition state analogues are changed.8
Table 1.
| Entry | Compound | Ki (pM) | Ki* (pM) | L/D Kd ratio11 |
|---|---|---|---|---|
| 1 |
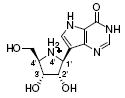
|
Not determined | 58 | 210 |
| 2 |
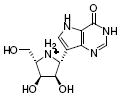
|
190,000 | 12,000 | |
| 3 |
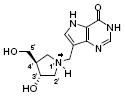
|
1,100 | 11 | 35 |
| 4 |
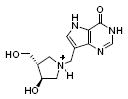
|
380 | Not observed |
We co-crystallized l-ImmH [2] and l-DADMe-ImmH [4] with PNP and phosphate to explore this unusual inhibitor specificity. The structures with l-ImmH [2] and l-DADMe-ImmH [4] were solved to 2.9 and 2.1 Å resolution, respectively, and were refined to Rwork and Rfree values of 21.6% and 26.2% for l-ImmH and to 21.8% and 24.9% for l-DADMe-ImmH, respectively. The Cruickshank DPI14 and Luzzatti15 coordinate errors were 0.14-0.43 for the two complexes; therefore, a 0.5 Å cut off was applied for significant distance differences of interaction. Crystallographic statistics are listed in Table 2, and complete experimental details are reported in the Supporting Information.
Table 2.
Crystallographic statistics for data collection and refinement of human PNP in complex with l-ImmH, l-DADMe-ImmH, and phosphate
| l-ImmH | l-DADMe-ImmH | |
|---|---|---|
| Wavelength (Å) | 0.9791 | 0.97910 |
| space group | H32 | H32 |
| cell: a = b | 143.5 | 142.8 |
| c | 166.2 | 167.3 |
| Resolution (Å) | 2.9 | 2.1 |
| Total Reflections | 129805 | 479977 |
| Unique Reflections | 14741 | 38397 |
| Completeness (%)a | 100(99) | 100(100) |
| Multiplicitya | 6.1(3.2) | 12.5(7.7) |
| Rsym (%)a | 7.3(74) | 7.4(71.4) |
| I/σa | 22(1.4) | 36.1(2.5) |
| No. of protein atoms | 2223 | 2333 |
| No. of water | 20 | 103 |
| Rwork | 21.6 | 21.8 |
| Rfree | 26.2 | 24.9 |
| Average B-factor | 71 | 55 |
| R.m.s. bond (Å) | 0.032 | 0.015 |
| R.m.s. angle (Å) | 2.9 | 1.6 |
| Ramachandran analysis | ||
| Most favored | 96.3 | 94.2% |
| Allowed | 1.7 | 5.4% |
| Disallowed | 2.1 | 0.4% |
| PDB ID | 2Q7O | 3BGS |
Values for the highest resolution shell are given in parentheses.
The PNP protein structures with l-Immucillins bound (Fig. 1) are similar to those previously reported.12,13 The structures of human PNP in complex with ImmH (PDB ID 1RR6) and DADMe-ImmH (PDB ID 1RSZ) allow comparisons of the binding modes for d- and l-Immucillins.
Figure 1.
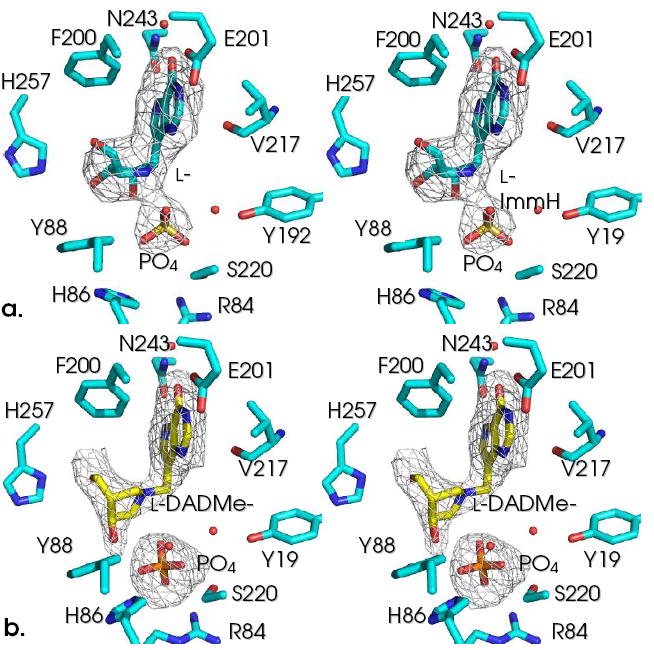
The catalytic sites of PNP•l-Immucillin•PO4 complexes contoured with Fo-Fc maps at 3 σ. l-ImmH [2] is shown in (a) and l-DADMe-ImmH [4] is shown in (b). Note the closer interaction of phosphate with l-ImmH than with l-DADMe-ImmH.
Phosphates in the active sites of PNP containing l-ImmH and l-DADMe-ImmH are in different positions as indicated in side-by-side comparison (Fig. 1). In contrast, the 9-deazahypoxanthine base groups are in similar positions for these same complexes with the l-Immucillins.
l-ImmH [2] binds to PNP with the base in a similar position as ImmH [1], but the plane of the iminoribitol moiety is rotated 180° around the C–C glycosidic bond, thereby preventing the 2’, 3’ and 5’-OH from forming the same favorable H-bond interactions found with ImmH [1] (Fig. 2).12 The 180° rotation of the l-iminoribitol ring causes the inhibitor to be shifted toward the purine binding site and permits phosphate to be translocated along with the inhibitor. Phosphate oxygen interactions with [2] form H-bonds to the 2′-OH and the 4′-NH2+, as well as a favorable (2.6 Å) ion pair between O1 and 4′-NH2+. The 5’-OH in [1] is in H-bond contact with Nδ of His257, and mutational analysis of His257 shows this to be important for binding and catalysis.10 The 180° rotation of l-ImmH with respect to the d-inhibitor prevents this His257 contact to the 5’-hydroxyl even though the 5’-hydroxyl of [2] is rotated away from the iminoribitol ring nitrogen and directed toward His257 (Fig. 2b). Instead, the 3’-OH interacts weakly with Nε of His257 (3.2 Å), and His257 has rotated around the Cβ–Cγ bond relative to the complex with [1]. The 2’-OH interacts in a short hydrogen bond to phosphate O4 (2.3 Å) and to water. The cationic iminoribitol nitrogen interacts with two phosphate oxygens, O1 (2.6 Å) and O4 (3.0 Å). Phosphate O2 interacts with the hydroxyl group of Ser220 (2.3 Å), O1 interacts with the backbone amide nitrogen of Met219, and phosphate O4 interacts with the 2’-OH of l-ImmH [2], as described above (Fig. 3).
Figure 2.
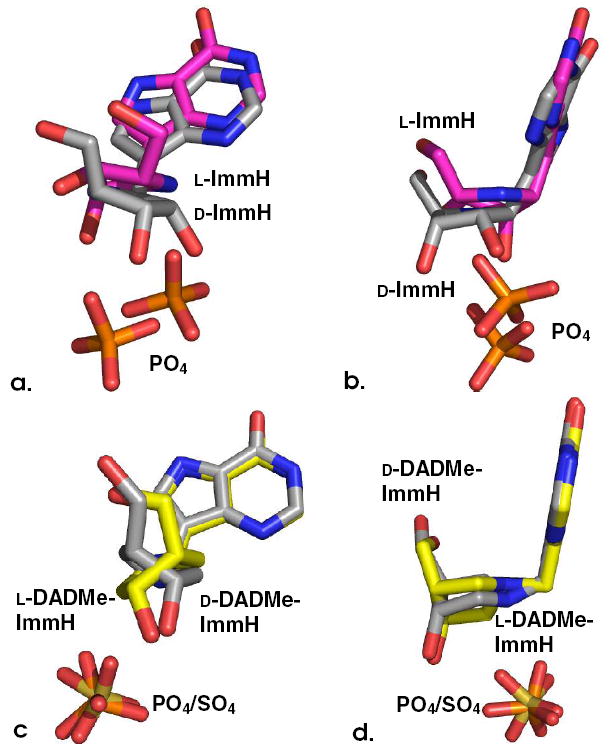
Overlapping views of the ligands from overlayed structures of their respective complexes with human PNP. In (a) and (b), d-ImmH [1] is in (gray) and l-ImmH [2] is in (magenta). In (c) and (d), d-DADMe-ImmH [3] is in (gray) and l-DADMe-ImmH [4] is in (yellow). Phosphates are shown in orange and red, and sulfate (from d-DADMe structure) is shown in tan and red.
Figure 3.
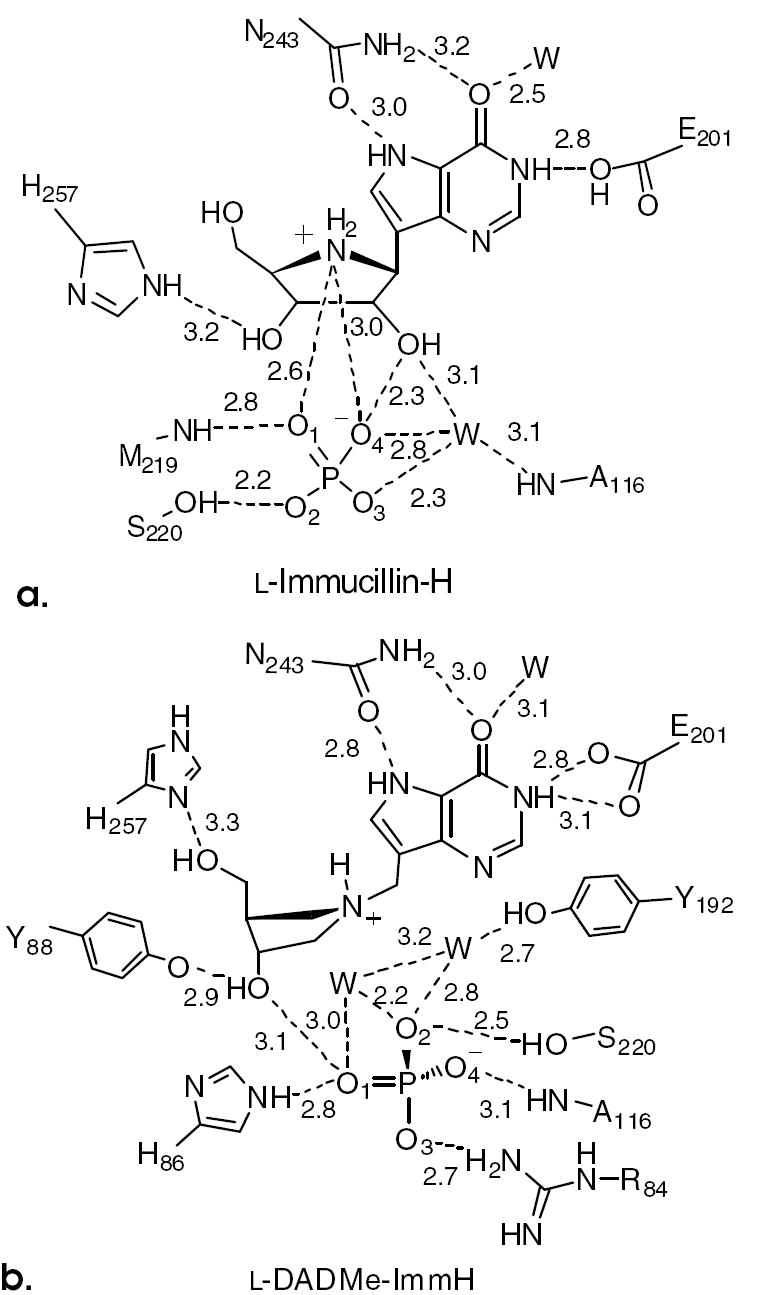
Distance maps for l-Immucillin-H (a) and l-DADMe-Immucillin-H (b) at the catalytic sites of PNP. The proposed nucleophilic oxygens are shown as anions.
Because of the l-ImmH [2] sugar conformation, the base is 0.5 Å closer to Asn243, allowing the formation of favorable leaving group interactions. The Asn243 side chain carbonyl forms a 3.0 Å H-bond with NH7, and its side chain amino group forms a 3.2 Å H-bond to the O6 exocyclic carbonyl oxygen. The 6-oxo group in the l-ImmH [2] complex also interacts with a water (2.5 Å), and Oε1 of Glu201 interacts with NH1 of the purine base (2.8 Å).
To summarize, both l-ImmH [2] and l-DADMe-ImmH [4] have favorable leaving group interactions. The ion pair is short between the iminoribitol cation and phosphate oxygen with [2], and these interactions cause [2] to be a 12 nM inhibitor (Table 1). This factor of 210-fold weaker binding of [2] than the 58 pM Ki* dissociation constant for [1] is readily explained by the altered phosphate position and the weak interactions of the 5’-OH and 3’-OH groups. Surprisingly, displacement of the phosphate group does not abolish the slow-onset binding properties of [2].
His257 anchoring of the 5’-hydroxyls of [1] and [3] occurs from a loop that moves when the catalytic site of PNP becomes occupied. Mutation of His257 affects Km, kcat and slow onset binding of [3].10,16 Although the 5’-hydroxyl group of [4] remains H-bonded to Nδ of His257, a 90° rotation of the pyrrolidine ring is required to place the 5’- and 3’-OH groups near the positions occupied by the 5’-OH of bound [3] (Fig. 2c). With this altered pyrrolidine geometry, the 9-deazahypoxanthine rings of [3] and [4] and their corresponding phosphates remain in the same positions. The 3’-OH of [4] forms a 2.9 Å interaction with Tyr88 (Fig. 3b).
The 5’-OH interaction to His257 is lost in the binding of [2], and instead, His257 interacts weakly (3.2 Å) with the 3’-hydroxyl. This coordination is to the ε-nitrogen, rather than the typical interaction via the δ-nitrogen and requires rotation of the imidazole ring (Fig. 3a). These changes are consistent with the loss of binding energy for [2] and [4]. Structural features causing the slow-onset binding for [2] but not [4] are obscure, but we speculate that slow-onset binding relates to the time-dependent conformational adjustment of the protein to optimize binding of the unusual but tight interactions at the catalytic site.
The phosphate in the l-DADMe-ImmH [4] complex is bound in a similar position as the phosphate of its isomer but is more distant from the pyrrolidine nitrogen with a distance of 3.9 Å in [4] instead of 3.0 Å with [3]. The phosphate has also lost one of the two hydrogen bonds to the 3’-OH of the inhibitor which is formed in the complex with [3]. It is remarkable that these changes in binding interactions, including a 0.9 Å separation of an ion pair in the catalytic site, result in only a 62-fold decrease in binding of [4] and testifies to the adaptability of this catalytic site which is competent for inosine, guanosine, and their 2’-deoxy forms.
A major leaving group interaction for transition state formation in PNP is N7 protonation. The Immucillins capture this interaction through the elevated pKa of NH7 in the 9-deazahypoxanthine rings.17 Interaction with NH7 occurs by a 2.7 Å contact with the side chain carbonyl oxygen of Asn243. This interaction is conserved in the binding of both d- and l-Immucillins. However, in the d-Immucillins, the side chain amino group of Asn243 is also in favorable H-bonding distance to the exocyclic 6-carbonyl oxygen, but this interaction is weak with binding of [2] and [4] (Fig. 3). The Oε1 and Oε2 of Glu201 make weak hydrogen bonds to purine N1 (3.0 Å) in the l-DADMe-ImmH [4] complex.
Structures of l-Immucillins bound to human PNP indicate that one feature responsible for the relatively tight binding of these inhibitors comes from the conserved interaction of the protein to the purine base NH7. Ion pair formation with bound phosphate is similar for the binding of [1] and [2] but differs for [3] and [4]. H-bond interactions are present to both the 5’- and 3’-OH groups of l-DADMe-ImmH [4] but the 5’-OH interaction is lost with l-ImmH. The sum of these interactions causes a 210-fold loss of binding for [2] but only a 35-fold loss for [4] (Table 1), a difference that is attributable to the increased flexibility of the methylene-bridged pyrrolidine compared to the iminoribitol. Considering that relatively simple changes such as conversion of the 5’-OH group to a 5’-F group in [1] cause a 120-fold decrease in binding affinity,8 it is remarkable that the catalytic site of PNP has sufficient adaptability to conform to the l-configuration of the Immucillins.
Stereochemical changes in the binding of transition state analogues are not always so well tolerated. In the interaction of the natural product deoxycoformycin with mammalian adenosine deaminase, the R-isomer is the stereochemical mimic of the transition state and binds with a dissociation constant of 2.5 pM, a factor of 6.4 × 106 more tightly than substrate.18 The S-stereoisomer at the reaction center (the 8-position of the diazepine ring) provides a single stereochemical change in the nucleoside analogue and reduces the binding affinity to 33 μM, a factor of 1.3 × 107 reduced binding.19 Systematic analysis of single atomic changes in the binding of transition state analogues reveals that the loss of each contact characteristic of the transition state interactions also weakens neighboring interactions to give strongly cooperative binding interactions in transition state analogue binding energy.20
Cooperative binding interactions are known to dominate binding of the Immucillins, and the presence of the cationic nature of the transition state undoubtedly facilitates these interactions. Lessons for inhibitor design are that central features of transition state structure, here the elevated pKa at NH7 and the ribocation mimic, are critical features for binding energy and these dominate the interactions compared to those from the exact placement of the ribosyl hydroxyl groups.
Supplementary Material
Crystallographic methods and related information. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by NIH Research Grants GM41916 from the National Institutes of Health and C08X0209 from the New Zealand Foundation for Research, Science and Technology, Wenner-Gren Foundation and the staff at NSLS Brookhaven National Laboratory for the technical assistance at X29A.
References
- 1.Giblett ER, Ammann AJ, Wara DW, Sandman R, Diamond LK. Lancet. 1975;1:1010–1013. doi: 10.1016/s0140-6736(75)91950-9. [DOI] [PubMed] [Google Scholar]
- 2.Bantia S, Kilpatrick JM. Curr Opin Drug Discov Devel. 2004;7:243–247. [PubMed] [Google Scholar]
- 3.Miles RW, Tyler PC, Furneaux RH, Bagdassarian CK, Schramm VL. Biochemistry. 1998;37:8615–8621. doi: 10.1021/bi980658d. [DOI] [PubMed] [Google Scholar]
- 4.Balakrishnan K, Nimmanapalli R, Ravandi F, Keating MJ, Gandhi V. Blood. 2006;108:2392–2398. doi: 10.1182/blood-2006-03-007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.http://www.biocryst.com/bcx_1777.htm; http://www.biocryst.com/bcx_4208.htm
- 6.Lewandowicz A, Schramm VL. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 7.Lewandowicz A, Tyler PC, Evans GB, Furneaux RH, Schramm VL. J Biol Chem. 2003;278:31465–31468. doi: 10.1074/jbc.C300259200. [DOI] [PubMed] [Google Scholar]
- 8.Lewandowicz A, Taylor Ringia EA, Ting L-M, Kim K, Tyler PC, Evans GB, Zubkova OV, Mee S, Painter GP, Lenz DH, Furneaux RH, Schramm VL. J Biol Chem. 2005;280:30320–30328. doi: 10.1074/jbc.M505033200. [DOI] [PubMed] [Google Scholar]
- 9.Clinch K, Evans GB, Fleet GW, Furneaux RH, Johnson SW, Lenz DH, Mee SPH, Rands PR, Schramm VL, Taylor Ringia EA, Tyler PC. Org Biomol Chem. 2006;4:1131–1139. doi: 10.1039/b517883e. Several kinetic values in Table 1. differ significantly from those reported earlier (in ref. 9) and may represent differences in PNP constructs or errors in interpretation of slow-onset inhibition binding constants in the earlier work.
- 10.Murkin AS, Birck MR, Rinaldo-Matthis A, Shi W, Taylor EA, Almo SC, Schramm VL. Biochemistry. 2007;46:5038–5049. doi: 10.1021/bi700147b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.For the [2]/[1] ratio, Ki*/Ki was used and for the [4]/[3] ratio, Ki/Ki* was used. These values represent the final equilibrium dissociation constants (Kd) in every case.
- 12.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 13.Ealick SE, Rule SA, Carter DC, Greehough TJ, Babu YS, Cook WJ, Habash J, Helliwell JR, Stoeckler JD, Parks RE, Chen S-F, Bugg CE. J Biol Chem. 1990;265:1812–1820. doi: 10.2210/pdb2pnp/pdb. [DOI] [PubMed] [Google Scholar]
- 14.Cruickshank DW. Acta Crystallograf D. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 15.Luzzati V. Acta Crystallograf. 1952;5:802–810. [Google Scholar]
- 16.Nunez S, Wing C, Antoniou D, Schramm VL, Schwartz SD. J Phys Chem A. 2006;110:463–472. doi: 10.1021/jp051277u. [DOI] [PubMed] [Google Scholar]
- 17.Sauve AA, Cahill SM, Zech SG, Basso LA, Lewandowicz A, Santos DS, Grubmeyer C, Evans GB, Furneaux RH, Tyler PC, McDermott A, Girvin ME, Schramm VL. Biochemistry. 2003;42:5694–5705. doi: 10.1021/bi034003a. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal RP, Spector T, Parks RE., Jr Biochem Pharmacol. 1977;26:359–367. doi: 10.1016/0006-2952(77)90192-7. [DOI] [PubMed] [Google Scholar]
- 19.Schramm VL, Baker DC. Biochemistry. 1985;24:641–646. doi: 10.1021/bi00324a016. [DOI] [PubMed] [Google Scholar]
- 20.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Shi W, Fedorov A, Lewandowicz A, Cahill SM, Almo SC, Schramm VL. Biochemistry. 2002;41:1448–14498. doi: 10.1021/bi026636f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystallographic methods and related information. This material is available free of charge via the Internet at http://pubs.acs.org.


