The crystal structure of octopine dehydrogenase revealed a specific role of the His5 tag in inducing the crystal contacts required for successful crystallization.
Keywords: His tags, coenzymes, octopine, crystal contacts
Abstract
Over the last decade, protein purification has become more efficient and standardized through the introduction of affinity tags. The choice and position of the tag, however, can directly influence the process of protein crystallization. Octopine dehydrogenase (OcDH) without a His tag and tagged protein constructs such as OcDH-His5 and OcDH-LEHis6 have been investigated for their crystallizability. Only OcDH-His5 yielded crystals; however, they were multiple. To improve crystal quality, the cofactor NADH was added, resulting in single crystals that were suitable for structure determination. As shown by the structure, the His5 tag protrudes into the cleft between the NADH and l-arginine-binding domains and is mainly fixed in place by water molecules. The protein is thereby stabilized to such an extent that the formation of crystal contacts can proceed. Together with NADH, the His5 tag obviously locks the enzyme into a specific conformation which induces crystal growth.
1. Introduction
The NAD(P)H-dependent reductive condensation of the amino group of an amino acid and an α-keto acid is catalyzed by a family of enzymes referred to as opine dehydrogenases (Grieshaber et al., 1994 ▶). The reaction products, the so-called opines (Thompson & Donkersloot, 1992 ▶), have two asymmetric centres and in nature they exhibit either (l,l) or (d,l) stereochemistry (Storey & Dando, 1982 ▶). It was only recently that the determination of the structure of octopine dehydrogenase (OcDH) from the adductor muscle of the great scallop Pecten maximus (Smits et al., 2008 ▶) allowed the elucidation of the reaction mechanism of this enzyme family, although the structure of CENDH from Arthrobacter sp. strain 1C had been solved in the late 1990s (Britton et al., 1998 ▶).
In recent years, His-tagged proteins have been extensively used for rapid and efficient protein purification. However, the role of the His tag in protein crystallization has been under considerable debate ever since (Carson et al., 2007 ▶). On one hand, a His tag can inhibit crystallization, which can be overcome by cleaving off the His tag via an engineered protease site. Conversely, in most cases the His tag does not interfere with crystal formation, although owing to its flexibility the polyhistidine can rarely be detected in the electron-density map. Here, we describe His-tag-induced crystallization of OcDH which was found to be dependent on the length of the His tag.
2. Materials and methods
2.1. Expression and purification of recombinant OcDH
The cloning of tagless OcDH (OcDH-tagless) and His5-tagged OcDH (OcDH-His5) were performed as described previously (Muller et al., 2007 ▶). Additionally, a His6-tagged OcDH variant (OcDH-LEHis6) was cloned. The expression and purification of OcDH-His5 and OcDH-LEHis6 were performed as described previously (Muller et al., 2007 ▶).
2.2. Crystallization, data collection and structure determination of OcDH-His5
Purified OcDH-tagless, OcDH-His5 and OcDH-LEHis6 were dialyzed against 10 mM HEPES pH 7.0, 1 mM EDTA and 1 mM DTT. Protein samples were concentrated to 20 mg ml−1 and optimized crystals of OcDH-His5 were grown as described by Smits et al. (2008 ▶). Data sets for OcDH-His5 were collected on the BW7A or X12 beamlines at the EMBL Outstation, DESY, Hamburg.
3. Results and discussion
3.1. Overexpression and purification of OcDH
Three different constructs were cloned for crystallization purposes (OcDH-tagless, OcDH-His5 and OcDH-LEHis6). The His tags were placed at the C-terminus, since the important NADH-binding site is encoded in the first 20 N-terminal amino acids. In OcDH-LEHis6 two additional amino acids, leucine and glutamate, are located between OcDH and the His tag as encoded on the plasmid.
OcDH-His5 and OcDH-LEHis6 were purified by Ni–NTA chromatography, yielding almost 20 mg homogenous enzyme per litre of cell culture (purity greater than 98%).
The purification of OcDH-tagless required several chromatographic steps and yielded 3–5 mg per litre of cell culture, with a purity of >98%. In terms of activity, the three constructs were indistinguishable (data not shown) and comparable to OcDH purified directly from P. maximus (van Thoai et al., 1969 ▶).
3.2. Crystallization
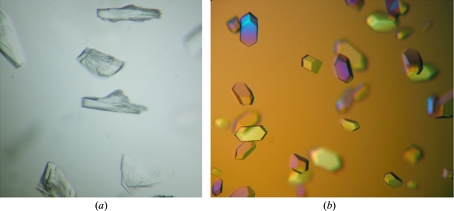
Despite extensive attempts, OcDH-tagless and OcDH-LEHis6 did not yield crystals. It is probable that OcDH can adopt multiple conformations which prevented crystal formation. However, the purified OcDH-His5 yielded small crystals which appeared to be multiple on optical examination (Fig. 1 ▶ a). They diffracted to a resolution of 2.6 Å with multiple lattices in one diffraction image and neither the XDS nor the DENZO (Otwinowski & Minor, 1997 ▶) program packages were able to process the data. All attempts to improve these crystals, using for example seeding, temperature ramping or various crystallization conditions, failed or only produced crystals of mediocre quality.
Figure 1.
Crystals of (a) apo OcDH-His5 and (b) OcDH-His5 with bound NADH coenzyme.
Finally, the coenzyme NADH was added (to a final concentration of 0.8 mM) prior to crystallization. This produced crystals under conditions similar to those in the absence of NADH (Fig. 1 ▶ b). In particular, the incubation temperature appeared to be critical and had to be kept at 285 K. The crystals obtained were single and diffracted to 2.1 Å resolution, which allowed processing of the data and subsequent structure determination (Smits et al., 2008 ▶).
3.3. Structure of OcDH
The three-dimensional structure of OcDH from P. maximus has recently been reported (Smits et al., 2008 ▶). Two distinct domains could be found in OcDH: an NADH-dependent glycerol-3-phosphate dehydrogenase-like domain (domain I) and an octopine dehydrogenase specific domain (domain II; Interpro database). Each domain comprises approximately half of the protein, with domain I containing the classic Rossman fold of dinucleotide-binding proteins (Rossmann et al., 1974 ▶; Schulz, 1992 ▶).
3.4. His tag and crystal contacts
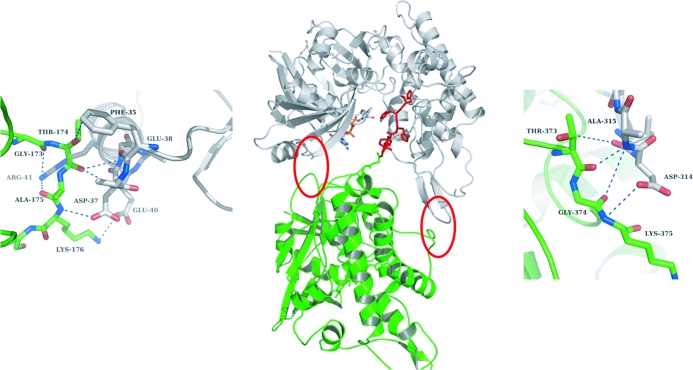
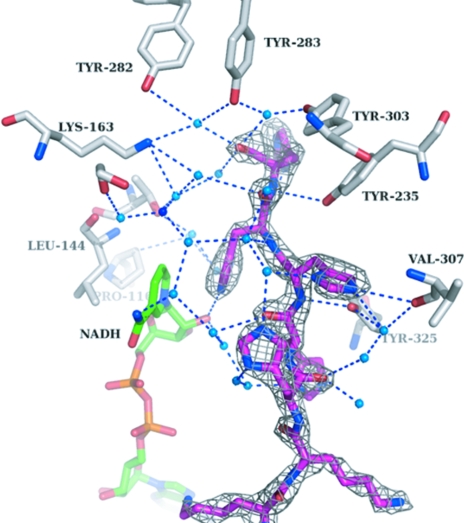
Surprisingly, the His5 tag protrudes into the cleft between domains I and II (Fig. 3). Of the histidines in the His tag, His402 directly interacts with the side chain of Val307, whereas His403 interacts with the 3′-OH moiety of the ribose of NADH. The position and orientation of the other histidines from the His tag are stabilized via a complex water network (Figs. 2 ▶ and 3 ▶). The water molecules were picked automatically using ARP/wARP (Perrakis et al., 1999 ▶) and manually checked for proper density. His400 interacts with Val307 and His401 via two water molecules, His401 interacts with Tyr325 via two water molecules, His402 interacts with Tyr325 via one water molecule, His403 interacts with Thr143, Leu116, Pro116 and Tyr235 via two water molecules and His404 interacts with Lys163, Tyr235, Tyr282, Tyr283 and Tyr303 via one water molecule. The complete His tag thus opens up both domains to a certain extent and the structure is fixed in this conformation. The electron density surrounding the His5 tag is of high quality (Fig. 3 ▶).
Figure 2.
His5-tag-mediated crystal contacts. The central figure shows the overall structure of OcDH (green) with the His5 tag (red) pointing into the cleft between the two domains of a symmetry-related molecule (white). Red circles indicate protein–protein-mediated crystal contacts and enlargements of these regions are shown on the left and right. The top of domain I interacts with the bottom of domain I of a neighbouring molecule. An identical arrangement is observed for domain II.
Figure 3.
Interactions of the His5 tag (magenta) with the surrounding water molecules (blue dots). Highlighted are the side chains (white) which make first-line or second-line interactions with the His5 tag via a water network. The NADH coenzyme is coloured green. The grey mesh shows an F o − F c OMIT map of the His5 tag contoured at 2σ.
The stable conformation of OcDH induced by the His5 tag created crystal contacts which are located at the bottom of both domains (Fig. 2 ▶). Here, Phe35, Glu38, Asp37, Glu40 and Arg41 of domain I of monomer A interact with Gly173, Thr174, Ala175 and Lys176 of domain I of a symmetry-related monomer B. The crystal contacts in domain II are mediated by Val313, Asp314 and Ala315 of monomer A, which interact with Thr373, Gly374 and Lys375 of monomer B. Disruption of any of these interactions by a further opening of the cleft between the two domains would loosen or even diminish crystal formation, as corroborated by the observation that crystals without coenzyme were multiple and of lower quality. Any lengthening of the His5 tag would probably result in a similar orientation of the tag inside the cleft, but would reduce the contacts between the protein molecules in the crystal, thereby inhibiting crystal growth. This might explain why OcDH-LEHis6 did not yield any crystals under the same conditions. The two monomers are probably too far apart to generate similar protein–protein contacts. This specific role of the His5 tag could explain why OcDH-tagless did not crystallize; the two domains are too flexible and the stable conformation suitable for crystallization can only be induced by the insertion of the His5 tag into the cleft in combination with the addition of NADH.
Chimeric proteins are utilized in many applications at the forefront of protein science. In particular, His tags (Smith et al., 1988 ▶) have gained great popularity in recent decades as a purification tool for recombinant proteins. An often unspoken assumption is that these tags have no effect on the structure and function of the protein (Chant et al., 2005 ▶). OcDH is one example where not only the presence but also the length of the His tag is crucial to the crystallization process, as it induces additional and important crystal contacts. A similar observation was made in the structure of a 116-residue protein (PDB code 1v30). Here, a long C-terminal helix, which includes the His6 tag, protrudes outside the molecule and packs with another molecule along the crystallographic twofold axis (Tajika et al., 2004 ▶). The authors noted that they were unable to obtain crystals of the wild-type sequence (no His tag) under the same crystallization conditions.
In summary, the length of the His tag in combination with the cofactor appeared to be crucial in the structure determination of OcDH. Although all three constructs appeared identical in terms of purification and activity, only one construct yielded crystals from which the X-ray structure could be determined. This might also suggest that not only black-and-white answers (His tag or no His tag) should be given for proteins that are ‘difficult to crystallize’, but that variations in the length of the His tag should also be considered in such crystallization efforts.
Acknowledgments
We thank Matthew Groves and Paul Tucker for excellent support at the beamlines of the EMBL Outstation Hamburg (Germany). This work was supported by the DFG (grant GR456/20-4 to MKG).
References
- Britton, K. L., Asano, Y. & Rice, D. W. (1998). Nature Struct. Biol.5, 593–601. [DOI] [PubMed]
- Carson, M., Johnson, D. H., McDonald, H., Brouillette, C. & DeLucas, L. J. (2007). Acta Cryst. D63, 295–301. [DOI] [PubMed]
- Chant, A., Kraemer-Pecore, C. M., Watkin, R. & Kneale, G. G. (2005). Protein Expr. Purif.39, 152–159. [DOI] [PubMed]
- Grieshaber, M. K., Hardewig, I., Kreutzer, U. & Portner, H. O. (1994). Rev. Physiol. Biochem. Pharmacol.125, 43–147. [DOI] [PubMed]
- Muller, A., Janssen, F. & Grieshaber, M. K. (2007). FEBS J.274, 6329–6339. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Rossmann, M. G., Moras, D. & Olsen, K. W. (1974). Nature (London), 250, 194–199. [DOI] [PubMed]
- Schulz, G. E. (1992). Curr. Opin. Struct. Biol.2, 61–67.
- Smith, M. C., Furman, T. C., Ingolia, T. D. & Pidgeon, C. (1988). J. Biol. Chem.263, 7211–7215. [PubMed]
- Smits, S. H. J., Mueller, A., Schmitt, L. & Grieshaber, M. K. (2008). J. Mol. Biol.318, 200-211. [DOI] [PubMed]
- Storey, K. B. & Dando, P. R. (1982). Comput. Biochem. Physiol. B, 73, 521–528.
- Tajika, Y., Sakai, N., Tamura, T., Yao, M., Watanabe, N. & Tanaka, I. (2004). Proteins, 57, 862–865. [DOI] [PubMed]
- Thoai, N. van, Huc, C., Pho, D. B. & Olomucki, A. (1969). Biochim. Biophys. Acta, 191, 46–57. [DOI] [PubMed]
- Thompson, J. & Donkersloot, J. A. (1992). Annu. Rev. Biochem.61, 517–557. [DOI] [PubMed]