The structure of the catalytic subunit of M. jannaschii aspartate transcarbamoylase has been determined in space group P212121 using synchrotron data to a resolution of 3.0 Å and was refined to a final R work and R free of 0.215 and 0.269, respectively.
Keywords: aspartate transcarbamoylase, catalytic subunit, Methanococcus jannaschii
Abstract
Crystals of the catalytic subunit of Methanococcus jannaschii aspartate transcarbamoylase in an orthorhombic crystal form contain four crystallographically independent trimers which associate in pairs to form stable staggered complexes that are similar to each other and to a previously determined monoclinic C2 form. Each subunit has a sulfate in the central channel. The catalytic subunits in these complexes show flexibility, with the elbow angles of the monomers differing by up to 7.4° between crystal forms. Moreover, there is also flexibility in the relative orientation of the trimers around their threefold axis in the complexes, with a difference of 4° between crystal forms.
1. Introduction
Aspartate transcarbamoylase (ATCase; EC 2.1.3.2) catalyzes the second step of de novo pyrimidine biosynthesis, the reaction between carbamoyl phosphate (CP) and aspartate to form N-carbamoyl-l-aspartate and inorganic phosphate (Jones et al., 1955 ▶), and is an important site of regulation of this pathway in many organisms. The structure and properties of the Escherichia coli enzyme have been extensively studied (Herve, 1989 ▶; Allewell, 1989 ▶; Lipscomb, 1992 ▶, 1994 ▶; England et al., 1994 ▶). It has a dodecameric structure (C6R6) composed of two types of subunits. The two larger or catalytic subunits (C3) are each composed of three identical polypeptide chains (M r 33 000), while the three smaller or regulatory subunits (R2) are each composed of two identical polypeptide chains (M r 17 000). The catalytic chains contain two domains: the carbamoyl phosphate (CP) and aspartate-binding (ASP) domains. The regulatory chains also contain two domains: the nucleotide and Zn-binding domains. E. coli ATCase is an allosteric enzyme that exhibits cooperativity for aspartate and heterotropic effects, being activated by ATP and inhibited by CTP. Upon binding the substrate aspartate (in the presence of saturating CP), E. coli ATCase undergoes a conformational change from a low-activity T state to a higher activity R state. The large conformational differences in the X-ray structures of the unliganded ATCase (Stevens et al., 1990a ▶,b ▶) and the N-phosphonacetyl-l-aspartate (PALA) liganded enzyme (Ke et al., 1988 ▶; Jin et al., 1999 ▶) have been proposed to define the structural differences between the T and R states.
Characterization of the ATCase enzyme from the hyperthermophilic and barophilic archaeon Methanococcus jannaschii (Hack et al., 2000 ▶) suggested that it consists of catalytic trimers and regulatory dimers and has a molecular weight similar to that from E. coli. Kinetic analysis of M. jannaschii ATCase from cell-free extracts showed that it has limited homotropic cooperativity and little if any regulatory properties with ATP and CTP. Kinetic analysis of the M. jannaschii catalytic trimer showed hyperbolic kinetics with an activation energy similar to that of the E. coli trimer and with an activity that increases with temperature. It is stable at 358 K.
We have previously determined the structure of the catalytic trimer of M. jannaschii ATCase in a monoclinic crystal form (Vitali et al., 2008 ▶). This study and comparisons with E. coli ATCase and the hyperthermophilic ATCases from Pyrococcus abyssi (Van Boxstael et al., 2003 ▶) and Sulfolobus acidocaldarius (De Vos et al., 2004 ▶) provided insight into the strategies for thermostabilization adopted by the M. jannaschii enzyme. We undertook the structural analysis of the orthorhombic form which is presented in this paper in order to further investigate the structure of the enzyme in different crystalline environments and its thermostabilization strategies. In particular, we were interested in the possible patterns of association of the catalytic subunits in the crystal. An interesting feature of the monoclinic form was the vertical association of two crystallographically independent catalytic subunits into a staggered dimer of trimers.
2. Materials and methods
2.1. Protein preparation and crystallization
The M. jannaschii catalytic trimer was prepared following the procedure of Hack et al. (2000 ▶) from E. coli strain EK1911 which has a deletion in the pyrBI region of the chromosome and contains the plasmids pEK406 coding for the M. jannaschii catalytic chain and pSJS1240 (Kim et al., 1998 ▶) coding for rare archaeal tRNAs. The gene for the M. jannaschii catalytic chain is not associated with any tags in pEK406. The purification involved a 30% ammonium sulfate precipitation step, a heat step at 363 K for 15 min and chromatography using a Q-Sepharose Fast Flow anion-exchange column and a phenyl Sepharose column. The purified protein was concentrated to 15 mg ml−1 in 40 mM KH2PO4, 0.2 mM EDTA and 2 mM 2-mercaptoethanol pH 7.0. Crystals were grown at 295 K by the sitting-drop method from reservoirs containing 2.5 M ammonium sulfate and 0.1 M Tris–HCl pH 8.5. The drops consisted of 5 µl reservoir solution and 5 µl protein solution. Two crystals were used for this study: they belonged to space group P212121, with unit-cell parameters (average for the two crystals) a = 87.13, b = 167.93, c = 319.41 Å, four trimers per asymmetric unit and a V M of 2.80 Å3 Da−1 (Matthews, 1968 ▶).
2.2. X-ray data collection
Diffraction data were measured on the X12B beamline of the National Synchrotron Light Source at Brookhaven National Laboratory using an ADSC Quantum-4 CCD detector. The temperature was 100 K, the wavelength was 0.979 Å and the crystal-to-detector distance was 340.00 mm. The cryoprotectant used was 15% glucose. Two crystals were used. Oscillations were measured for 60 s each in 1.0° intervals of ϕ. A range of 100° in ϕ was measured for the first crystal and a range of 70° for the second. The data were processed with the HKL software package (Otwinowski & Minor, 1997 ▶). The data from the two crystals were reduced and integrated in DENZO and merged and scaled in SCALEPACK. Data statistics are summarized in Table 1 ▶.
Table 1. Data-collection and final refinement statistics.
Values in parentheses are for the highest resolution shell.
| Summary of X-ray diffraction data collection | |
| Space group | P212121 |
| Unit-cell parameters () | a = 87.13, b = 167.93, c = 319.41 |
| Resolution range () | 40.03.0 (3.13.0) |
| Temperature (K) | 100 |
| Wavelength () | 0.979 |
| No. of unique reflections | 92280 |
| Redundancy | 4.4 (2.8) |
| Completeness (%) | 99.5 (96.8) |
| Mean I/(I) | 8.9 (1.4) |
| R merge † | 0.095 (0.67) |
| Final refinement statistics | |
| Resolution range () | 40.03.0 |
| No. of reflections | 86463‡ |
| Non-H protein atoms per ASU | 29609 |
| Waters per ASU | 500 |
| Sulfates per ASU | 4 |
| R work § | 0.215 |
| R free § | 0.269 |
| R.m.s. deviations from ideal geometry | |
| Bond lengths () | 0.01 |
| Bond angles () | 1.3 |
| Impropers () | 0.8 |
| B values (2) | |
| From Wilson plot | 73.3 |
| Mean B over all atoms | 70.0 |
| Ramachandran plot | |
| Most favored regions (%) | 84.5 |
| Additional allowed regions (%) | 13.9 |
| Generously allowed regions (%) | 1.1 |
| Disallowed regions (%) | 0.5¶ |
R
merge = 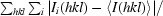
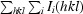 , where I
i(hkl) and I(hkl) are the observed intensity of measurement i and the mean intensity of the reflection with indices hkl, respectively.
, where I
i(hkl) and I(hkl) are the observed intensity of measurement i and the mean intensity of the reflection with indices hkl, respectively.
The number of reflections used in the refinement differs from the number scaled because reflections with |F o(hkl)| = 0 were excluded.
R
work and R
free = 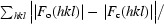
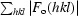 , where |F
o(hkl)| and |F
c(hkl)| are the observed and calculated structure-factor amplitudes, respectively. R
work is calculated for the reflections used in the refinement (90% of data) and R
free for the reflections used in the test set (10% of data).
, where |F
o(hkl)| and |F
c(hkl)| are the observed and calculated structure-factor amplitudes, respectively. R
work is calculated for the reflections used in the refinement (90% of data) and R
free for the reflections used in the test set (10% of data).
Except for Leu263, all residues in this region, namely Ser128 of C6 (complex 1) and C1 (complex 2), Asn129 of C1 (complex 1), C5 (complex 1), C1 (complex 2) and C4 (complex 2) and Glu82 of C4 (complex 1), correspond to poor electron density.
2.3. Structure determination and refinement
The structure was solved using molecular replacement by evolutionary search with EPMR (Kissinger et al., 1999 ▶) and data in the 15.0–4.0 Å resolution range. The search model was the complex of the two trimers in the asymmetric unit of the monoclinic form (Vitali et al., 2008 ▶). Refinement was carried out with CNS (Brünger et al., 1998 ▶) using simulated annealing with torsion-angle dynamics. During the refinement NCS restraints were imposed for both the main chain and side chains for most of the four regions 1–131, 132–146, 147–280 and 281–303 among the 12 monomers. Residues that appeared to differ between monomers were refined independently. The refinement was alternated with manual model building and rebuilding using O (Jones et al., 1991 ▶) and Coot (Emsley & Cowtan, 2004 ▶). Model building was facilitated by electron-density maps computed with phases improved by solvent flipping, density truncation and NCS averaging over all 12 monomers. For each amino acid, two group temperature factors were refined: one for the main chain and one for the side chain. Each trimer had a sulfate in its central tunnel on the noncrystallographic threefold. The sulfates were harmonically restrained to their initial positions as well as with respect to each other together with residues 55–67 of the monomers in each trimer. At the end of the analysis, waters were added to the model using as criteria a 2σ peak height in the difference maps and hydrogen-bonding distances to protein atoms of 2.0–3.5 Å.
2.4. Model analysis
The quality of the structure was analyzed with PROCHECK (Laskowski et al., 1993 ▶). Hydrogen bonds were calculated with HBPLUS (McDonald & Thornton, 1994 ▶) using donor–acceptor distances of less that 3.5 Å, hydrogen–acceptor distances of less than 2.5 Å and associated angles greater than 90°. Salt bridges between two charged groups corresponded to distances of less than 4.0 Å. A hydrophobic contact was assumed to exist between two apolar residues if the carbon–carbon distance was less than 4.0 Å. DSSP (Kabsch & Sander, 1983 ▶) was used for computing accessible surface areas (ASAs). Structure superpositions were performed with LSQMAN (Kleywegt, 1996 ▶). Figures were prepared with PyMOL (http://pymol.sourceforge.net).
The planar angles between the CP and ASP domains were computed by a modification of the method of Williams et al. (1998 ▶) using the angle between the geometric centers of the two domains and a hinge point. The geometric centers for the CP and ASP domains in M. jannaschii were computed from the Cα atoms of residues 1–131 and 147–280, respectively. The hinge point was taken as the Cα atom of residue 137. The corresponding residues were used in the computation of the angles in the other ATCases.
The global association of two catalytic subunits in a complex is described by the distance between their geometric centers and the torsion angle between the individual chains of the two subunits around the axis defined by this line. The geometric centers were computed from the Cα atoms of residues 1–131 and 147–280.
2.5. Dynamic light-scattering measurements
Dynamic light-scattering measurements were carried out by the W. M. Keck Foundation Biotechnology Laboratory, Yale University, New Haven, Connecticut at 0.66 M ammonium sulfate and a protein concentration of 5.0 mg ml−1 using a DynaPro 99-362.
3. Results and discussion
3.1. Quality of the structure
The final model consists of two crystallographically independent complexes consisting of trimers, four sulfates and 500 waters. Final refinement statistics are presented in Table 1 ▶. All the stereochemical parameters are within the range expected for a model at this resolution. In the Ramachandran plot computed in PROCHECK (Laskowski et al., 1993 ▶), 84.5% of the residues are found in the most favorable regions. Leu263 is in a disallowed region, as is often the case for active-site residues. This residue is also found in disallowed regions in the PALA-liganded and unliganded E. coli catalytic subunit (Endrizzi et al., 2000 ▶; Beernink et al., 1999 ▶) and holoenzyme (Jin et al., 1999 ▶; Stevens et al., 1990a ▶,b ▶) and in the PALA-liganded P. abyssi catalytic trimer (Van Boxstael et al., 2003 ▶). The two complexes have different average temperature factors (complex 1, 54.1 Å2; complex 2, 86.6 Å2), probably because they form different numbers of intermolecular contacts (complex 1, 229; complex 2, 138). The final map, contoured at 1.0σ, shows continuous density for most main-chain and side-chain atoms for both complexes with a few exceptions. Residues 74–82 and 126–129 have fragmented and/or weak density in all monomers. The C-terminus is well defined for chain C4 of complex 1 but poorly defined beyond residue 304 for most other chains. A few residues on the surface of the monomers do not have side-chain electron density. Complex 2 has weaker electron density overall than complex 1.
3.2. Description of the structure
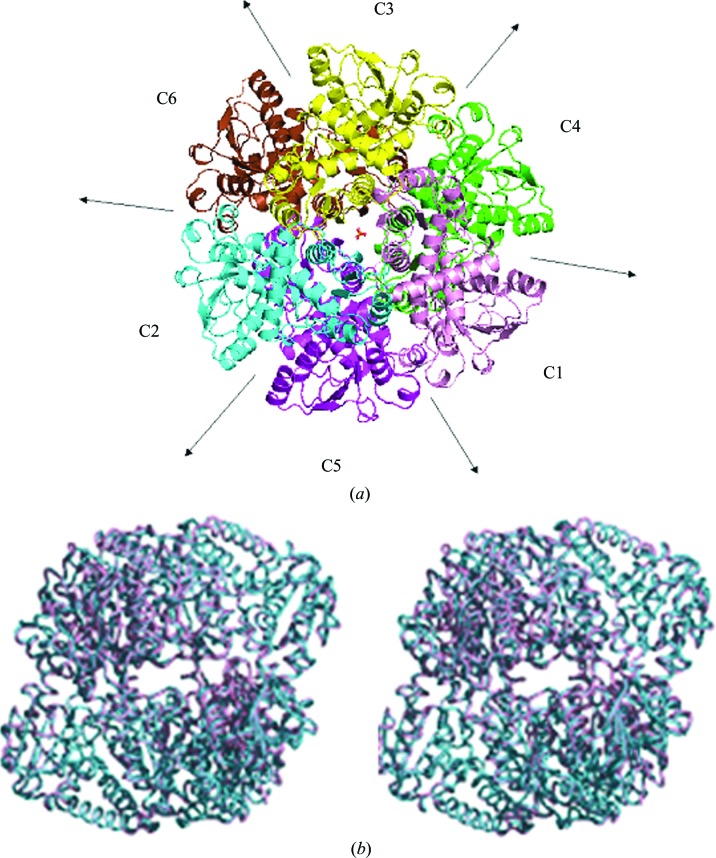
The four crystallographically independent trimers in the asymmetric unit associate in pairs to form two complexes of trimers with 32 point-group symmetry, similar to the complex observed in the monoclinic form (Vitali et al., 2008 ▶; Fig. 1 ▶). The two complexes are related by a rotation of 157.4° around the direction with spherical polar angles (ω, ϕ) = (82.4, −135.2°) and are similar to each other and to the complex in the monoclinic form. The superposition of the two complexes in the present structure is illustrated in Fig. 1 ▶(b). The r.m.s.d. between corresponding Cα atoms is 0.49 Å for the two complexes in the present structure and 1.32 and 1.16 Å for complexes 1 and 2, respectively, compared with the monoclinic complex. The monomers are similar, with an average r.m.s.d. between Cα atoms of all chains of 0.31 Å. The r.m.s.d. between corresponding Cα atoms of the central chains in the present structure and the monoclinic structure is 0.71 Å.
Figure 1.
(a) View of one of the M. jannaschii complexes in the asymmetric unit (complex 1) looking down the noncrystallographic threefold. Each monomer is depicted in a different color. Both sulfates on the noncrystallographic threefold are shown. Arrows denote the noncrystallographic twofold axes. The catalytic chains C1, C2 and C3 comprise the top trimer of the complex, while the C4, C5 and C6 chains comprise the bottom subunit. (b) Side stereoview of the two M. jannaschii complexes in the present structure superimposed on each other. Complex 1 is shown in pink and complex 2 is shown in teal. The view is looking down the noncrystallographic twofold.
The planar angles (Table 2 ▶) are nearly constant in each trimer and are in the ranges 122.9–124.7° in complex 1 and 124.2–125.2° in complex 2. These values are up to 7.4° less than in the monoclinic form and are comparable to those in the E. coli T-form ATCase. Planar angles are sensitive indicators of domain movement and it is unlikely that they are affected by resolution (Stieglitz et al., 2004 ▶; Vitali et al., 2008 ▶). The variation in the planar angles between the crystal forms is large and may suggest high flexibility in solution and high reactivity even though the near-threefold symmetry in each trimer is maintained. The unliganded E. coli catalytic trimer shows variation of this angle within the same trimer and the threefold symmetry is not maintained (Beernink et al., 1999 ▶).
Table 2. Comparison of the planar angles of the M. jannaschii catalytic trimer in the orthorhombic form with the monoclinic form and other systems.
PDB codes used were M. jannaschii monoclinic form, 2rgw (Vitali et al., 2008 ▶); E. coli catalytic trimer, 3csu (Beernink et al., 1999 ▶); P. abyssi catalytic trimer + PALA, 1ml4 (Van Boxstael et al., 2003 ▶); E. coli catalytic trimer + PALA, 1ekx (Endrizzi et al., 2000 ▶); S.acidocaldarius T-form ATCase, 1pg5 (De Vos et al., 2004 ▶); E. coli T-form ATCase, 6at1 (Stevens et al., 1990a ▶,b ▶); E. coli R-form ATCase, 1d09 (Jin et al., 1999 ▶). Values for crystallographically independent chains are separated by commas.
| System | Planar angles () |
|---|---|
| M. jannaschii orthorhombic form | |
| Complex 1, C1C2C3 | 124.7, 124.3, 123.8 |
| Complex 1, C4C5C6 | 124.3, 122.9, 124.1 |
| Complex 2, C1C2C3 | 124.7, 125.2, 124.2 |
| Complex 2, C4C5C6 | 125.0, 124.3, 124.8 |
| M. jannaschii monoclinic form | |
| Complex C1C2C3 | 130.2, 129.6, 128.8 |
| Complex C4C5C6 | 129.0, 130.3, 129.0 |
| E. coli catalytic trimer | 141.1, 135.8, 135.0 |
| P. abyssi catalytic trimer + PALA | 118.2 |
| E. coli catalytic trimer + PALA | 115.7, 116.1, 115.9 |
| S. acidocaldarius T form | 122.4 |
| E. coli T form | 123.1, 124.6 |
| E. coli R form | 115.1, 115.7 |
The vertical association of the catalytic subunits in both complexes of the present structure is similar to that observed in the monoclinic form. Monomers of adjacent trimers have a staggered arrangement around the threefold axis in all three complexes, with torsional angles for C1–C4, C1–C6 and C1–C5 (Fig. 1 ▶) of −40.0, −159.8 and 79.9° (complex 1), −40.6, −160.5 and 79.6° (complex 2) and −44.4, −164.5 and 75.6° (monoclinic form). The inter-trimer distance in all three complexes is short at 33.8 Å. These torsional angles are sensitive indicators of the relative orientations of the catalytic subunits and are not likely to depend on resolution. The twist of −4.0° between the complexes of the orthorhombic and monoclinic forms demonstrates some flexibility in the relative rotation around the threefold axis at a constant inter-trimer distance. The interactions (Table 3 ▶) between the two trimers are across C1–C4-type and C1–C5-type interfaces and several are conserved among the M. jannaschii catalytic complexes. The differences in the relative rotations of the trimers between complexes correlate with small differences in the ASAs buried in these interfaces (Table 3 ▶). These parameters are known not to depend on resolution (Novotny et al., 2007 ▶). The similarity between the association of the two catalytic trimers in the present structure and in the monoclinic form in different crystalline environments indicates that this arrangement is stable. However, size-exclusion chromatography studies have shown that the catalytic subunits exist as isolated trimers in Tris solution (Hack et al., 2000 ▶). It is possible that the association we observe in the crystalline state occurs at high concentrations of the protein and/or in the presence of ammonium sulfate. Estimates of the hydrodynamic radius and molecular mass (58 Å and 205 000) from dynamic light-scattering measurements in 0.66 M ammonium sulfate are consistent with the formation of hexamers, as observed in the crystalline state, but these solutions show high polydispersity and the estimated values could be weighted averages of more than one species. Moreover, such molecular-weight estimates from dynamic light scattering involve assumptions about molecular shape that may not accurately represent the particles in solution. It is important to note that this novel association may also be part of the holoenzyme in vivo in the presence of the regulatory subunits.
Table 3. Comparative analysis of the trimertrimer interfaces of the M. jannaschii complexes in the present structure and the monoclinic form.
| Complex 1 | Complex 2 | Monoclinic form | |
|---|---|---|---|
| Hydrogen bonds, ion pairs and hydrophobic contacts in C1C4-type interfaces | |||
| Hydrogen bonds | 25 | 4 | 2 |
| Salt bridges | 0 | 0 | 0 |
| % in networks | 0 | 0 | 0 |
| Hydrophobic | 918 | 915 | 1214 |
| No. of hydrophobic pairs | 812 | 79 | 810 |
| ASA† buried, total | 1014 | 1073 | 1048 |
| ASA† buried, charged | 266 | 244 | 226 |
| ASA† buried, apolar | 480 | 457 | 555 |
| Hydrogen bonds, ion pairs and hydrophobic contacts in C1C5-type interfaces | |||
| Hydrogen bonds | 1‡ | 1‡ | 1‡ |
| Salt bridges | 12§ | 1§ | 1§ |
| % in networks | 100 | 100 | 100 |
| Hydrophobic | 822 | 1216 | 1012 |
| No. of apolar pairs | 36 | 34 | 2 |
| ASA†, buried total | 610 | 572 | 811 |
| ASA†, buried charged | 247 | 209 | 414 |
The ASA of the C1C4-type interfaces was computed as the sum of the ASAs of all the monomers in the C complex minus the sum of the ASAs of the three C1C4-type dimers divided by three. The same procedure was used for the C1C5-type interfaces except that the C1C5-type dimers were used in the calculation.
Only in one interface.
In two of the interfaces.
Each trimer has a sulfate at the center of its three CP domains on the noncrystallographic threefold as was observed in the structure of the monoclinic form (Vitali et al., 2008 ▶; Fig. 1 ▶ a). Each sulfate is involved in an extended ion-pair network with all three monomers of its trimer through charged residues of the α2 helix that point into the central channel. This network provides additional stability to the association of monomers to form a trimer. The sulfates most likely came from the crystallization medium.
3.3. Thermostability
Several structural features that could potentially contribute to the thermostability of the M. jannaschii catalytic trimer were identified in the monoclinic structure (Vitali et al., 2008 ▶). These include (i) changes in the amino-acid composition such as a decrease in the thermolabile residues Gln and Asn, an increase in the charged residues Lys and Glu, an increase in Tyr residues and a decrease in Ala residues, (ii) shortening of the N-terminus and shortening of three loops and (iii) a larger number of salt bridges, in particular the improvement of ion-pair networks. The present structure confirms these conclusions. The possible association of the catalytic subunits with each other and with the regulatory subunits in the holoenzyme may provide additional thermostabilization in vivo.
4. Conclusions
Crystals of the catalytic subunit of M. jannaschii ATCase in an orthorhombic crystal form contain four crystallographically independent trimers in the asymmetric unit which are similar in structure to the catalytic trimers in the monoclinic form of the enzyme (Vitali et al., 2008 ▶). Each trimer has a sulfate in the central channel. The catalytic subunits show large differences in their planar angles from the C2 form, indicating high flexibility and reactivity in solution. In both crystal forms the catalytic subunits associate in pairs to form similar staggered complexes of trimers with a short inter-trimer distance of 33.8 Å. This association shows flexibility in the rotation of the two trimers relative to each other around their common threefold axis. The formation of these complexes in three different crystalline environments in two different crystal forms indicates that they are stable. It is tempting to speculate that this association may be part of the holoenzyme in vivo in the presence of the regulatory subunits. The earlier study of the C2 form provided insight into some of the factors that potentially contribute to the thermostability of the trimer. The possible association of the catalytic subunits with each other, as observed in these complexes, and with the regulatory subunits in the holoenzyme may provide additional thermostabilization in vivo.
Supplementary Material
PDB reference: aspartate transcarbamoylase, 3e2p, r3e2psf
Acknowledgments
This work was supported in part by startup funds from Cleveland State University (JV) and grants GM071512 (JV) and GM008180 (MJC) from the National Institutes of Health. We thank Dr Evan Kantrowitz for providing the EK1911 strain that was used for this study, Dr Dieter Schneider of Brookhaven National Laboratory for use of his beamline X12B and for instruction and help during data collection and Dr Ewa Folta-Stogniew for the dynamic light-scattering measurements and discussions.
References
- Allewell, N. M. (1989). Annu. Rev. Biophys. Chem. 18, 71–92. [DOI] [PubMed]
- Beernink, P. T., Endrizzi, J. A., Alber, T. & Schachman, H. K. (1999). Proc. Natl Acad. Sci. USA, 96, 5388–5393. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- De Vos, D., Van Petegem, F., Renaut, H., Legrain, C., Glansdorff, N. & Van Beeumen, J. J. (2004). J. Mol. Biol. 339, 887–900. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Endrizzi, J. A., Beernink, P. T., Alber, T. & Schachman, H. K. (2000). Proc. Natl Acad. Sci. USA, 97, 5077–5082. [DOI] [PMC free article] [PubMed]
- England, P., Leconte, C., Tauc, P. & Herve, G. (1994). Eur. J. Biochem. 222, 775–780. [DOI] [PubMed]
- Hack, E. S., Vorobyova, T., Sakash, J. B., West, J. M., Macol, C. P., Herve, G., Williams, M. K. & Kantrowitz, E. R. (2000). J. Biol. Chem. 275, 15820–15827. [DOI] [PubMed]
- Herve, G. (1989). Allosteric Enzymes, edited by G. Herve, pp. 61–79. Boca Raton: CRC Press.
- Jin, L., Stec, B., Lipscomb, W. N. & Kantrowitz, E. R. (1999). Proteins, 37, 729–742. [PubMed]
- Jones, M. E., Spector, L. & Lipmann, F. (1955). J. Am. Chem. Soc. 77, 819–820.
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Ke, H. M., Lipscomb, W. N., Cho, Y. J. & Honzatko, R. B. (1988). J. Mol. Biol. 204, 725–747. [DOI] [PubMed]
- Kim, R., Sandler, S. J., Goldman, S., Yokota, H., Clark, A. J. & Kim, S.-H. (1998). Biotechnol. Lett. 20, 207–210.
- Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999). Acta Cryst. D55, 484–491. [DOI] [PubMed]
- Kleywegt, G. J. (1996). Acta Cryst. D52, 842–857. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lipscomb, W. N. (1992). Proceedings of the Robert A. Welch Foundation Conference on Chemical Research, Vol. XIII, pp. 103–143. Houston: Robert A. Welch Foundation.
- Lipscomb, W. N. (1994). Adv. Enzymol. 68, 67–152. [DOI] [PubMed]
- McDonald, I. & Thornton, J. (1994). J. Mol. Biol. 238, 777–793. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Novotny, M., Seibert, M. & Kleywegt, G. J. (2007). Acta Cryst. D63, 270–274. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Stevens, R. C., Gouaux, J. E. & Lipscomb, W. N. (1990a). Biochemistry, 29, 7691–7701. [DOI] [PubMed]
- Stevens, R. C., Gouaux, J. E. & Lipscomb, W. N. (1990b). Biochemistry, 29, 11146. [DOI] [PubMed]
- Stieglitz, K., Stec, B., Baker, D. P. & Kantrowitz, E. R. (2004). J. Mol. Biol. 341, 853–868. [DOI] [PubMed]
- Van Boxstael, S., Cunin, R., Khan, S. & Maes, D. (2003). J. Mol. Biol. 326, 203–216. [DOI] [PubMed]
- Vitali, J., Colaneri, M. J. & Kantrowitz, E. R. (2008). Proteins, 71, 1324–1334. [DOI] [PubMed]
- Williams, M. K., Stec, B. & Kantrowitz, E. R. (1998). J. Mol. Biol. 281, 121–134. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: aspartate transcarbamoylase, 3e2p, r3e2psf