M. tuberculosis tetrahydrodipicolinate-N-succinyltransferase, the enzyme that catalyses the fifth reaction step of the lysine-biosynthesis pathway, has been cloned, expressed, purified and crystallized.
Keywords: tetrahydrodipicolinate-N-succinyltransferase, Mycobacterium tuberculosis, DapD
Abstract
Tetrahydrodipicolinate-N-succinyltransferase from Mycobacterium tuberculosis (DapD, Rv1201c) has been cloned, heterologously expressed in Escherichia coli, purified using standard chromatographic techniques and crystallized in the cubic space group I23 or I213. Preliminary diffraction data analysis indicates the presence of five molecules per asymmetric unit. Furthermore, the data exhibit icosahedral point-group symmetry. One possible explanation for this is that the enzyme assembles into a 60-mer exhibiting 235 point-group symmetry and crystallizes as such in space group I23. In this case, the combination of crystallographic and noncrystallographic symmetry elements results in an arrangement of the icosahedrons in the cubic crystal with one pentamer in the asymmetric unit. Another explanation is that the packing of the molecules itself mimics icosahedral symmetry. In this case both space groups I23 and I213 would be possible.
1. Introduction
Tuberculosis is an airborne bacterial infection which is predominantly caused by Mycobacterium tuberculosis (Mtb). Every year, around ten million people worldwide are newly infected with this disease and several million patients die from it. As a consequence, tuberculosis was declared a global health emergency by the World Health Organization in 1993. A vaccine against tuberculosis is available and efficient antibiotics such as isoniazid, rifampin, ethambutol and streptomycin were introduced in the 1950s and 1960s. Nonetheless, the need for new drugs is becoming ever more urgent in the light of emerging Mtb strains that are resistant to many and sometimes to all known antibiotics (Hutton et al., 2003 ▶; Parish & Stoker, 2002 ▶; Smith et al., 2000 ▶; Snider et al., 1994 ▶). Promising new antibacterial drug targets may be found in biosynthetic pathways that are present in the pathogenic bacterium but not in humans. The lysine-biosynthetic pathway has been discussed as one such possibility (Hutton et al., 2003 ▶), since both the final product of the pathway l-lysine and the lysine precursor meso-diaminopimelate (meso-DAP) are essential for bacterial growth and survival.
In the lysine-biosynthetic pathway, the amino acid l-lysine is synthesized from the amino acid l-aspartate in nine consecutive steps (Umbarger, 1978 ▶). In the fifth reaction step, the ring of the heterocyclic compound (S)-2,3,4,5-tetrahydropyridine-2,6-dicarboxylate (tetrahydrodipicolinate; THDP) is opened up and a succinyl group is transferred from succinyl-CoA to the now present amino group, yielding the product N-succinyl-l-2-amino-6-oxoheptanedioate. This step is catalysed by the enzyme 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (succinyl-CoA:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase; DapD; EC 2.3.1.117). In Mtb strain H37Rv (Cole et al., 1998a ▶,b ▶), the gene coding for this enzyme is denoted Rv1201c.
The enzyme DapD has been identified in a wide range of different organisms, but biochemical and enzymological data are only available for DapD from Escherichia coli (Simms et al., 1984 ▶; Berges et al., 1986 ▶). Three-dimensional structures have been described of DapD from M. bovis (PDB codes 1tdt, 2tdt, 3tdt, 1kgq and 1kgt; Beaman et al., 1997 ▶, 1998 ▶, 2002 ▶), E. coli (PDB code 3bxy; Nguyen et al., 2008 ▶), Campylobacter jejuni (PDB code 2rij; unpublished results) and Enterococcus faecalis V583 (PDB code 3cj8; unpublished results). In all of these structures, DapD appears as a homotrimer. Each individual chain is divided into three distinct domains: an N-terminal globular domain, a three-stranded left-handed β-helix and a small C-terminal domain. Whereas the β-helix structure is relatively well conserved between DapDs from different organisms, the N-terminal domain varies dramatically in size and architecture.
In this report, we describe the cloning of Mtb-DapD (Rv1201c) from genomic DNA, the heterologous overexpression of the protein in E. coli, its purification to homogeneity and successful crystallization. Following Mtb-Asd (Vyas et al., 2008 ▶), Mtb-DapA (Kefala & Weiss, 2006 ▶, 2008 ▶), Mtb-DapB (Kefala, Janowski et al., 2005 ▶), Mtb-DapC (Weyand et al., 2006 ▶) and Mtb-LysA (Kefala, Perry et al., 2005 ▶), Mtb-DapD represents the sixth enzyme of the lysine-biosynthetic pathway of Mtb for which diffraction-quality crystals have been obtained in our laboratory. This constitutes an important step towards our goal of structurally characterizing all the proteins involved in lysine biosynthesis in M. tuberculosis.
2. Experimental methods
2.1. Cloning
The genomic DNA of the bacterial strain Mtb H37Rv was used as template for the polymerase chain reaction. The following oligonucleotides (Invitrogen) were used as forward and reverse primers, respectively: 5′-GGGGCATATG GCTGTGTCGACCGTGACTGGAGCAGCAGGCATCG-3′ and 5′-GGGGCTCGAG ACCTCGTGGTACACCGTTGGCGTGCAGGTCCTCGTTGAGGGCAATGC-3′. In the forward primer the GCT triplet coding for Ala (shown in italics) was introduced as the second codon to increase the efficiency of expression (Looman et al., 1987 ▶). In the reverse primer a thrombin cleavage site (shown in italics) was introduced. The amplified fragment containing 5′-NdeI and 3′-XhoI restriction sites (in bold) was cloned into the pCR-Blunt II-TOPO vector (Invitrogen). After sequence confirmation, the insert was subcloned into the pET22b expression vector (Novagen), which adds a C-terminal His6 tag to the expressed recombinant protein directly after the thrombin cleavage sequence. The final protein produced therefore contains the amino-acid sequence GVPRGLEHHHHHH at the C-terminus immediately following its coded sequence and exhibits a total molecular weight of 34 213 Da. Both the TOPO and the final pET22b constructs were sequenced to confirm the cloning of the Rv1201c gene sequence in frame.
2.2. Expression and purification
The recombinant plasmid was used to transform E. coli BL21 (DE3) cells modified to co-express several chaperones in order to increase the yield of soluble protein expression (de Marco et al., 2007 ▶). The strain used was the chaperone combination 5 (cc5) strain. Cells from a 5 ml overnight culture were used for the inoculation of 1 l LB broth medium containing ampicillin (50 µg ml−1), spectinomycin (50 µg ml−1) and chloramphenicol (17 µg ml−1). Cultivation was carried out at 310 K and 220 rev min−1 until an OD600nm of 0.6–0.7 was reached. The temperature was subsequently lowered to 293 K and protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 500 µM. Cells were harvested by centrifugation about 20 h after induction and stored at 253 K until further processing.
1 g of cell pellet was dissolved in 10 ml buffer A [20 mM Tris pH 8.0, 1000 mM NaCl, 40 mM imidazole, 3 mM β-mercaptoethanol (β-ME)] containing one Complete Mini EDTA-free Protease Inhibitor Cocktail tablet (Roche) per 20 ml and lysed by sonication three times for 5 min at a time using 0.4 s pulses at 277 K. The cell debris was pelleted by centrifugation for 60 min at 277 K and 20 000 rev min−1. The crude lysate was filtered through a 0.22 µm membrane and was loaded onto a 1 ml NiII HiTrap Chelating HP column (Qiagen) equilibrated with three column volumes of buffer A. In order to remove unbound proteins, the column was first washed with 50 ml buffer A and then with 25 ml buffer B (20 mM Tris pH 8.0, 200 mM NaCl, 40 mM imidazole, 3 mM β-ME). The protein was eluted using a linear imidazole gradient to 500 mM imidazole in buffer A. The peak fractions were pooled and the protein was transferred to buffer C (20 mM Tris pH 8.0, 200 mM NaCl, 3 mM DTT) by a three-step centrifugation buffer exchange using a filter with a 30 kDa molecular-weight cutoff. The protein solution was adjusted to a total volume of 5 ml and filtered through a 0.22 µm membrane. Subsequently, the protein was purified by size-exclusion chromatography (Superdex 200 16/60, GE Healthcare) using buffer C for both equilibration and elution. It eluted with an apparent molecular weight of approximately 100 kDa, which is consistent with the expected molecular weight of a homotrimer. The peak fractions were pooled and concentrated to 15 mg ml−1. The protein concentration was determined based on the UV absorption at 280 nm and a calculated extinction coefficient of 20 065 M −1 cm−1 for the full-length protein including the C-terminal tag (Gasteiger et al., 2005 ▶). The purity of the protein was estimated by SDS–PAGE and its oligomeric state was analysed by static light scattering (SLS) using an ÄKTA Purifier (GE Healthcare) equipped with a size-exclusion chromatography column (Superdex 200 16/60, GE Healthcare) and an SLS detector (Wyatt Technology miniDAWN Tristar).
2.3. Crystallization
Purified protein (with the additional Ala residue in position 2 and the C-terminal extension GVPRGLEHHHHHH) was set up for crystallization at a concentration of 15 mg ml−1. Initial crystallization experiments were carried out at the High Throughput Crystallization Facility at the EMBL Hamburg Outstation (Mueller-Dieckmann, 2006 ▶). Initial crystals appeared within 4 d in Hampton Crystal Screen Lite condition No. 18 [200 mM magnesium acetate tetrahydrate, 100 mM sodium cacodylate pH 6.5, 10%(w/v) PEG 8000] at room temperature. Optimization of the initial hit was carried out using the hanging-drop vapour-diffusion method. 1 µl protein solution and 1 µl reservoir solution were equilibrated against 500 µl reservoir solution in 24-well Linbro plates. Crystals grew in the presence of 190 mM magnesium acetate tetrahydrate, 100 mM sodium cacodylate pH 6.5, 10%(w/v) PEG 8000 at room temperature within 5–7 d to a maximum dimensions of 150 × 150 × 150 µm (Fig. 1 ▶). They diffracted X-rays to about 2.0 Å resolution.
Figure 1.
Cubic crystal of DapD (Rv1201c) from M. tuberculosis.
2.4. Diffraction data collection and processing
Diffraction data collection was carried out using a single crystal on beamline I911-3 at the MAX-lab synchrotron in Lund, Sweden. The beamline is equipped with a MAR Mosaic CCD detector (225 mm) mounted on a MAR desktop beamline (MARdtb). A crystal which had been flash-cooled and stored for transport to the synchrotron was mounted on the beamline in a cold nitrogen stream at 100 K. A total of 300 diffraction images were then collected from this crystal in 0.3° steps. Indexing and integration of the data was performed using the program DENZO (Otwinowski & Minor, 1997 ▶), followed by scaling with SCALEPACK (Otwinowski & Minor, 1997 ▶). The R factors R r.i.m. (redundancy-independent merging R factor) and R p.i.m. (precision-indicating merging R factor) (Weiss, 2001 ▶) were obtained using the program RMERGE (available from http://www.embl-hamburg.de/~msweiss/projects/msw_qual.html or from MSW upon request). In Table 1 ▶, all relevant data-collection and processing parameters are given. Intensities were converted to structure-factor amplitudes using the program TRUNCATE (French & Wilson, 1978 ▶; Collaborative Computational Project, Number 4, 1994 ▶). The optical resolution of the data set was calculated with SFCHECK (Vaguine et al., 1999 ▶) and the self-rotation function was computed using the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶) based on structure-factor amplitudes to a maximum resolution of 6.0 Å.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Wavelength (Å) | 1.0 |
| Crystal-to-detector distance (mm) | 220 |
| Rotation range per image (°) | 0.3 |
| Total rotation range (°) | 90 |
| Exposure time per image (s) | 10 |
| Resolution range (Å) | 99.0–2.15 (2.19–2.15) |
| Space group† | I23 or I213 |
| Unit-cell parameter (Å) | a = 216.99 |
| Mosaicity (°) | 0.58 |
| Total No. of reflections | 950871 |
| Unique reflections | 91783 |
| Redundancy | 10.4 (8.9) |
| I/σ(I) | 19.7 (3.1) |
| Completeness (%) | 100.0 (99.7) |
| Rmerge (%) | 12.5 (74.2) |
| Rr.i.m. (%) | 13.1 (78.7) |
| Rp.i.m. (%) | 4.0 (26.1) |
| Overall B factor from Wilson plot (Å2) | 30.2 |
| Optical resolution (Å) | 1.67 |
The final space-group assignment has to await successful structure determination.
3. Results and discussion
From 1 g of wet cell pellet, approximately 2 mg pure protein was obtained after the two-step chromatographic purification procedure. The purity of the sample was at least 95% as estimated by SDS–PAGE. From SLS experiments, a molecular weight of approximately 92 kDa was deduced (data not shown), which is consistent with the value obtained by gel filtration and which indicates that Mtb-DapD is a homotrimer in solution. This in turn concurs with the observation that DapDs from other organisms also occur as homotrimers (Beaman et al., 1997 ▶, 1998 ▶, 2002 ▶; Nguyen et al., 2008 ▶).
After setting up the initial crystallization experiments, most of the protein precipitated. However, within about one week crystals appeared and grew out of the precipitate. As the crystals grew, the precipitant dissolved partially or sometimes completely. The crystallization conditions were then optimized and produced nice-looking single crystals of cubic shape (Fig. 1 ▶).
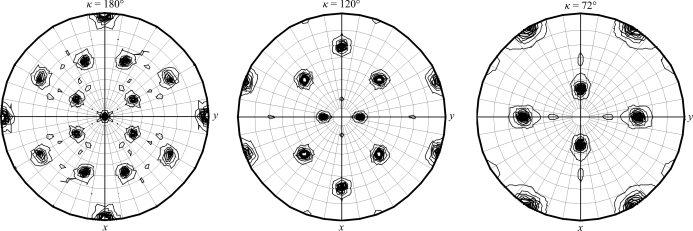
In preparation for diffraction data collection, a single crystal was mounted in a nylon loop, cryoprotected using 20%(v/v) MPD in reservoir solution and stored in liquid nitrogen. Diffraction data were then collected from the crystal to a resolution of 2.15 Å. The relevant data-collection and processing parameters are given in Table 1 ▶. The crystal belonged to the cubic space group I23 or I213. Since the merging R-factor values in the high-resolution shell are higher than expected based on the I/σ(I) value of 3.1, the data were also scaled in space group P1 in order to exclude the possibility of misassigned space-group symmetry (too high symmetry). In P1, the observed I/σ(I) value at high resolution is 1.1 and the R merge is 58%, which is close to the value expected from statistics. As a matter of fact, the values of R r.i.m. are practically identical when scaling the data in P1 and I23, confirming the presence of the the cubic 23 symmetry. Based on the total molecular weight of 34 213 Da and the unit-cell parameter shown in Table 1 ▶, calculation of the Matthews parameter (Matthews, 1968 ▶) suggests the presence of three to six subunits per asymmetric unit, corresponding to one or two complete trimers. With one trimer, the solvent content would be approximately 70% (V M = 4.2 Å3 Da−1), whereas with two trimers it would be approximately 40% (V M = 2.1 Å3 Da−1). However, calculation of the self-rotation function (Fig. 2 ▶) revealed the unexpected and surprising presence of fivefold rotation axes. This observation strongly suggests the presence of five subunits in the asymmetric unit. The combination of the crystallographic (twofold and threefold axes) and noncrystallographic (twofold, threefold and fivefold axes) symmetry elements results in the formation of the icosahedral point-group symmetry 235 (Hahn, 2002 ▶). Assuming space group I23, the appearance of the icosahedral symmetry of the diffraction pattern may be explained by the protein assembling into 60-mers exhibiting 235 point-group symmetry similar to some virus particles or the enzyme lumazine synthase (Ritsert et al., 1995 ▶). Two of these nearly spherical 60-mers of diameter 188 Å would then pack in a body-centred fashion to form the cubic I lattice. Alternatively, the packing itself of the Mtb-DapD monomers or trimers may mimic the icosahedral symmetry suggested by the self-rotation function (Fig. 2 ▶). In such a case, both space groups I23 and I213 would be possible. The final answer to this interesting puzzle will have to await the successful determination of the three-dimensional structure of Mtb-DapD. With sequence identities of Mtb-DapD to the DapDs of known structure from M. bovis, E. coli, C. jejuni and E. faecalis of 28%, 28%, 43% and 14%, respectively, structure determination will be attempted by molecular replacement. This is currently in progress.
Figure 2.
Self-rotation function based on data collected from a cubic crystal of Mtb-DapD. Shown are the κ = 180°, κ = 120° and κ = 72° sections indicating the relative locations of the twofold, threefold and fivefold symmetry axes, respectively. The 15 twofold, ten threefold and six fivefold axes describe an icosahedral body of point group 235 (Hahn, 2002 ▶). This figure was produced using the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶).
Acknowledgments
We would like to thank Dr L. Jeanne Perry (University of California at Los Angeles, USA) for providing genomic Mtb H37Rv DNA, Dr Arie Geerlof (EMBL) for providing the E. coli BL21 Star (DE3) cc5 cells, the TB consortium (http://www.doe-mbi.ucla.edu/TB) for postdoctoral exchange grants to GK and the X-Mtb consortium (http://www.xmtb.org) for funding through BMBF/PTJ grant No. BIO/0312992A. SW was supported by an E-STAR fellowship funded by the EC’s FP6 Marie Curie Host fellowship for Early Stage Research Training under contract No. MEST-CT-2004-504640. We would also like to acknowledge the support by the European Community Research Infrastructure Action under the FP6 ‘Structuring the European Research Area’ Programme through the Integrated Infrastructure Initiative ‘Integrating Activity on Synchrotron and Free Electron Laser Science’ for access to synchrotron beam time at MAX-lab (Lund, Sweden) and the beamline scientist Dr Johan Unge for help.
References
- Beaman, T. W., Binder, D. A., Blanchard, J. S. & Roderick, S. L. (1997). Biochemistry, 36, 489–494. [DOI] [PubMed]
- Beaman, T. W., Blanchard, J. S. & Roderick, S. L. (1998). Biochemistry, 37, 10363–10369. [DOI] [PubMed]
- Beaman, T. W., Vogel, K. W., Drueckhammer, D. G., Blanchard, J. S. & Roderick, S. L. (2002). Protein Sci.11, 974–979. [DOI] [PMC free article] [PubMed]
- Berges, D. A., DeWolf, W. E. Jr, Dunn, G. L., Newmann, D. J. II, Schmidt, S. J., Taggart, J. J. & Gilvar, C. (1986). J. Biol. Chem.261, 6160–6167. [PubMed]
- Cole, S. T. et al. (1998a). Nature (London), 393, 537–544.
- Cole, S. T. et al. (1998b). Nature (London)396, 190.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- De Marco, A., Deuerling, E., Mogk, A., Tomoyasu, T. & Bukau, B. (2007). BMC Biotechnol.7, 32–40. [DOI] [PMC free article] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Hahn, T. (2002). Editor. International Tables for Crystallography, Vol. A, 5th ed. Dordrecht: Kluwer Academic Publishers.
- Hutton, C. A., Southwood, T. J. & Turner, J. J. (2003). Mini Rev. Med. Chem.3, 115–127. [DOI] [PubMed]
- Kefala, G., Janowski, R., Panjikar, S., Mueller-Dieckmann, C. & Weiss, M. S. (2005). Acta Cryst. F61, 718–721. [DOI] [PMC free article] [PubMed]
- Kefala, G., Perry, L. J. & Weiss, M. S. (2005). Acta Cryst. F61, 782–784. [DOI] [PMC free article] [PubMed]
- Kefala, G. & Weiss, M. S. (2006). Acta Cryst. F62, 1116–1119. [DOI] [PMC free article] [PubMed]
- Kefala, G. & Weiss, M. S. (2008). Acta Cryst. F64, 62.
- Looman, A. C., Bodlaender, J., Comstock, L. J., Eaton, D., Jhurani, P., de Boer, H. A. & van Knippenberg, P. H. (1987). EMBO J.6, 2489–2492. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mueller-Dieckmann, J. (2006). Acta Cryst. D62, 1446–1452. [DOI] [PubMed]
- Nguyen, L., Kozlov, G. & Gehring, K. (2008). FEBS Lett.582, 623–626. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Parish, P. & Stoker, N. G. (2002). Microbiology, 148, 3069–3077. [DOI] [PubMed]
- Ritsert, K., Huber, R., Turk, D., Ladenstein, R., Schmidt-Base, K. & Bacher, A. (1995). J. Mol. Biol.253, 151–167. [DOI] [PubMed]
- Simms, S. A., Voige, W. H. & Gilvarg, C. (1984). J. Biol. Chem.259, 2734–2741. [PubMed]
- Smith, D., Wiegeshaus, E. & Balasubramanian, V. (2000). Clin. Infect. Dis.31, S77–S80. [DOI] [PubMed]
- Snider, D. E. Jr, Raviglione, M. & Kochi, A. (1994). Tuberculosis: Pathogenesis, Protection and Control, edited by B. R. Bloom, pp. 2–11. Washington DC: American Society for Microbiology.
- Umbarger, H. E. (1978). Annu. Rev. Biochem.47, 532–606. [DOI] [PubMed]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Vyas, R., Kumar, V., Panjikar, S., Karthikeyan, S., Kishan, K. V. R., Tewari, R. & Weiss, M. S. (2008). Acta Cryst. F64, 167–170. [DOI] [PMC free article] [PubMed]
- Weyand, S., Kefala, G. & Weiss, M. S. (2006). Acta Cryst. F62, 794–797. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.