Abstract
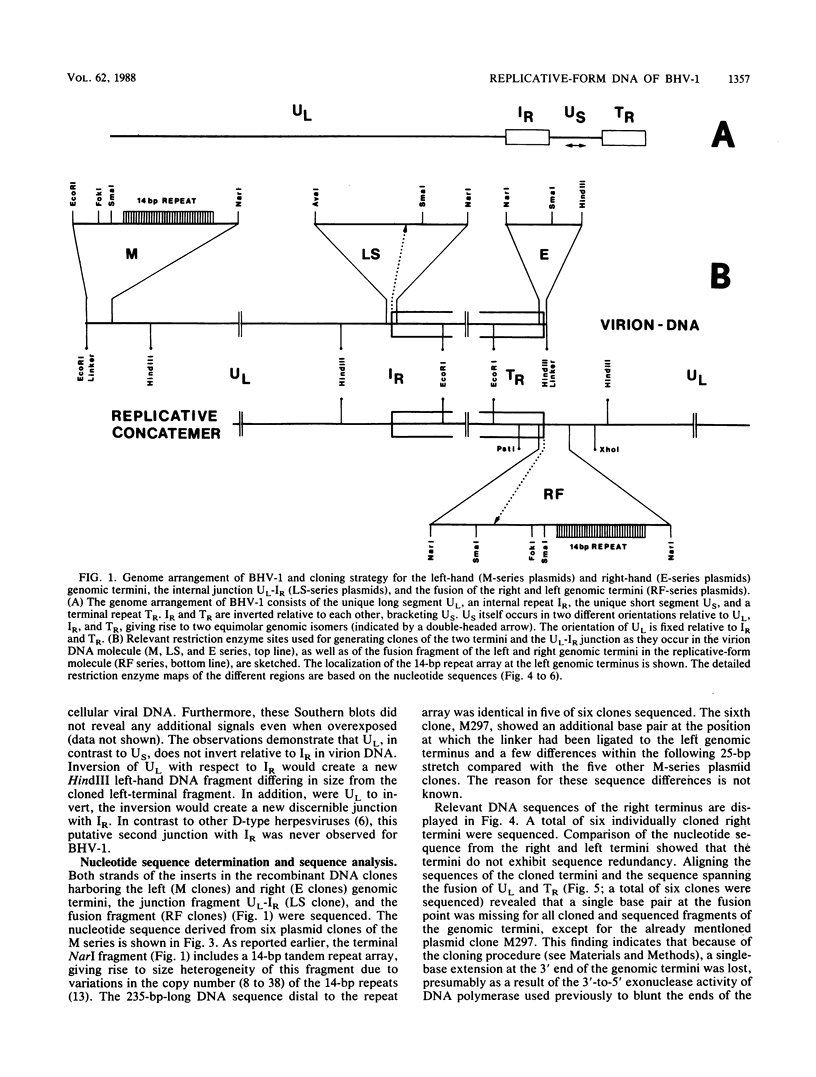
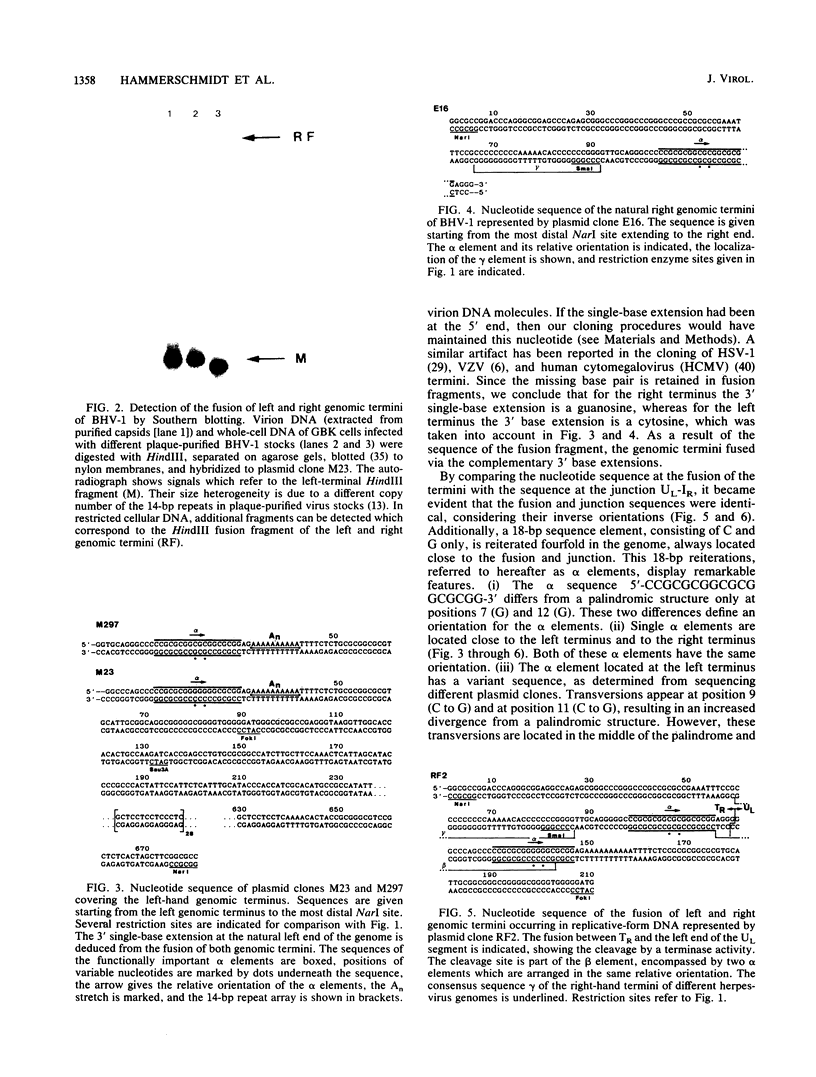
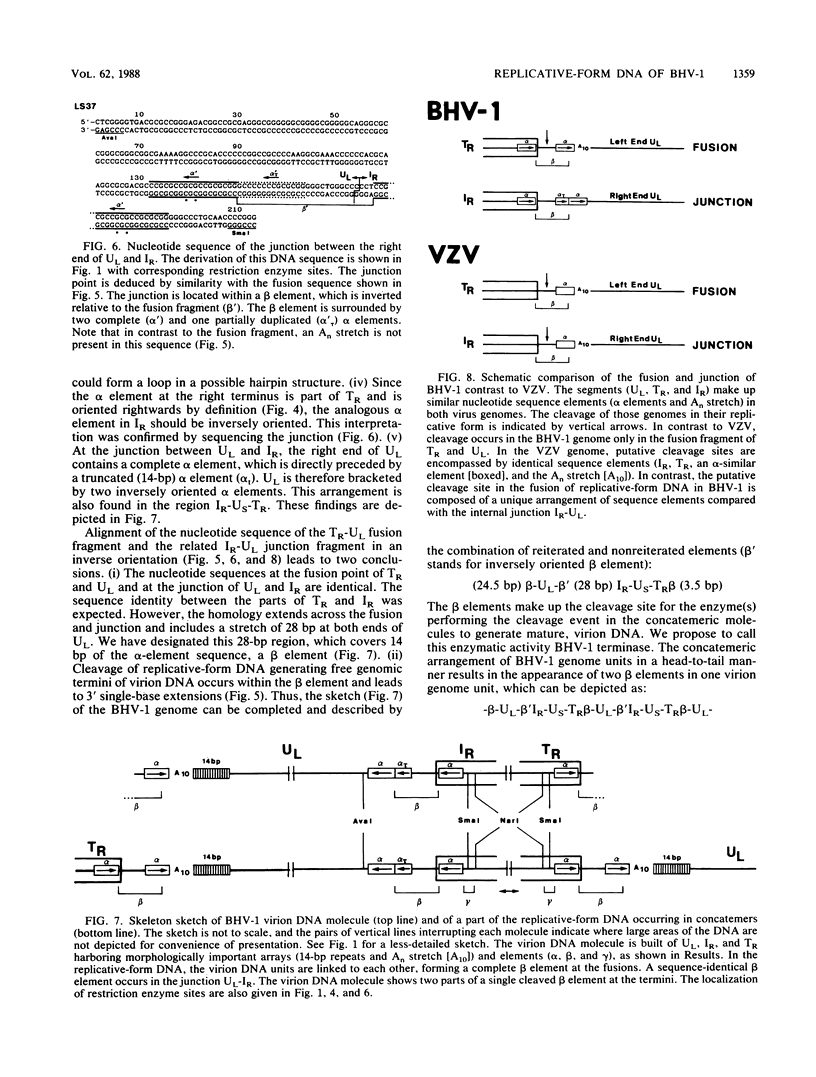
The linear double-stranded DNA genome of herpesvirus as it is present in infectious virions needs to be circularized after infection of host cells and before DNA replication. Replicative-form genomes have to be cleaved into linear unit-length molecules during virion maturation and are most probably the substrate for inversion of the short segment relative to the long segment of the bovine herpesvirus 1 (BHV-1) genome. Those regions of the BHV-1 genome which are functionally involved in these processes have been analyzed at the molecular level by cloning and sequencing the genomic termini, the fusion of both termini from replicative-form molecules, and the junction between the short and the long genome segment. On the basis of the simple genome arrangement of BHV-1, it was inferable that the cleavage of replicative-form genomes by a hypothetical BHV-1 terminase activity may be specified by a sequence at the left end of UL (An element), which is located proximal to a reiterated beta element that makes up the cleavage site itself. The relationship of those elements in BHV-1 and the comparison to similar regions of other herpesviruses indicate consensus sequence elements which are functionally important for cleavage and isomerization of viral DNA during maturation of virions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bankier A. T., Dietrich W., Baer R., Barrell B. G., Colbère-Garapin F., Fleckenstein B., Bodemer W. Terminal repetitive sequences in herpesvirus saimiri virion DNA. J Virol. 1985 Jul;55(1):133–139. doi: 10.1128/jvi.55.1.133-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Kaplan A. S., Stehn B., Rubenstein A. S. Concatemeric forms of intracellular herpesvirus DNA. Virology. 1976 Feb;69(2):547–560. doi: 10.1016/0042-6822(76)90484-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J., Blankenship M. L. Analysis of the structure of the genome of pseudorabies virus. Virology. 1979 Jun;95(2):285–294. doi: 10.1016/0042-6822(79)90484-7. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Chowdhury S. I., Kubin G., Ludwig H. Equine herpesvirus type 1 (EHV-1) induced abortions and paralysis in a Lipizzaner stud: a contribution to the classification of equine herpesviruses. Arch Virol. 1986;90(3-4):273–288. doi: 10.1007/BF01317376. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Davison A. J. Structure of the genome termini of varicella-zoster virus. J Gen Virol. 1984 Nov;65(Pt 11):1969–1977. doi: 10.1099/0022-1317-65-11-1969. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss L. P., Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986 Mar;57(3):933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels M., Giuliani C., Wild P., Beck T. M., Loepfe E., Wyler R. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res. 1986 Oct;6(1):57–73. doi: 10.1016/0168-1702(86)90057-2. [DOI] [PubMed] [Google Scholar]
- Gregersen J. P., Pauli G., Ludwig H. Bovine herpesvirus 1: differentiation of IBR- and IPV-viruses and identification and functional role of their major immunogenic components. Arch Virol. 1985;84(1-2):91–103. doi: 10.1007/BF01310556. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Ludwig H., Buhk H. J. Short repeats cause heterogeneity at genomic terminus of bovine herpesvirus 1. J Virol. 1986 Apr;58(1):43–49. doi: 10.1128/jvi.58.1.43-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Harper L., Demarchi J., Ben-Porat T. Sequence of the genome ends and of the junction between the ends in concatemeric DNA of pseudorabies virus. J Virol. 1986 Dec;60(3):1183–1185. doi: 10.1128/jvi.60.3.1183-1185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Huang E. S., Pagano J. S. Human cytomegalovirus. II. Lack of relatedness to DNA of herpes simples I and II, Epstein-Barr virus, and nonhuman strains of cytomegalovirus. J Virol. 1974 Mar;13(3):642–645. doi: 10.1128/jvi.13.3.642-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob R. J., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979 Feb;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Heller M., Petti L., O'Shiro E., Kieff E. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science. 1984 Dec 14;226(4680):1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mayfield J. E., Good P. J., VanOort H. J., Campbell A. R., Reed D. E. Cloning and cleavage site mapping of DNA from bovine herpesvirus 1 (Cooper strain). J Virol. 1983 Jul;47(1):259–264. doi: 10.1128/jvi.47.1.259-264.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler A. E., Matile H., Gassmann U., Engels M., Wyler R. European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch Virol. 1985;85(1-2):57–69. doi: 10.1007/BF01317006. [DOI] [PubMed] [Google Scholar]
- Misra V., Babiuk L. A., Darcel C. L. Analysis of bovine herpes virus-type 1 isolates by restriction endonuclease fingerprinting. Arch Virol. 1983;76(4):341–354. doi: 10.1007/BF01311201. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Poffenberger K. L., Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985 Feb;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. The alpha sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985 Jun;54(3):817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D. C., Atherton S. S., Staczek J., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 3. Virology. 1984 Jan 30;132(2):352–367. doi: 10.1016/0042-6822(84)90041-2. [DOI] [PubMed] [Google Scholar]
- Tamashiro J. C., Filpula D., Friedmann T., Spector D. H. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J Virol. 1984 Nov;52(2):541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro J. C., Spector D. H. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J Virol. 1986 Sep;59(3):591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]




