Abstract
We have addressed the mechanisms governing the activation and trafficking of G protein-coupled receptors (GPCRs) by analyzing constitutively active mating pheromone receptors (Ste2p and Ste3p) of the yeast Saccharomyces cerevisiae. Substitution of the highly conserved proline residue in transmembrane segment VI of these receptors causes constitutive signaling. This proline residue may facilitate folding of GPCRs into native, inactive conformations, and/or mediate agonist-induced structural changes leading to G protein activation. Constitutive signaling by mutant receptors is suppressed upon coexpression with wild-type, but not G protein coupling-defective, receptors. Wild-type receptors may therefore sequester a limiting pool of G proteins; this apparent “precoupling” of receptors and G proteins could facilitate signal production at sites where cell surface projections form during mating partner discrimination. Finally, rather than being expressed mainly at the cell surface, constitutively active pheromone receptors accumulate in post-endoplasmic reticulum compartments. This is in contrast to other defective membrane proteins, which apparently are targeted by default to the vacuole. We suggest that the quality-control mechanism that retains receptors in post-endoplasmic reticulum compartments may normally allow wild-type receptors to fold into their native, fully inactive conformations before reaching the cell surface. This may ensure that receptors do not trigger a response in the absence of agonist.
INTRODUCTION
G protein-coupled receptors (GPCRs) are integral membrane proteins that are inserted into the membrane of endoplasmic reticulum (ER), folded into their native, inactive conformations, and transported through the secretory pathway to the cell surface where they can be activated by hormones, neurotransmitters or sensory stimuli. Mechanisms that control the activation or biogenesis of GCPRs therefore have critical roles in governing cellular responsiveness to an array of extracellular signals.
GPCR activation has been investigated in many systems, leading to the following model (Baldwin, 1993; Lefkowitz et al., 1993; Coughlin, 1994). In the absence of ligands, GPCRs are thought to exist in equilibrium between inactive and active conformations, usually favoring the inactive state. Agonists bind and stabilize receptors in their active conformations, leading to G protein activation. Inverse agonists bind and stabilize the inactive conformation of GPCRs, precluding receptor activation by agonists. Antagonists bind receptors without significantly affecting the equilibrium distribution between inactive and active conformations, which also blocks agonist-induced signaling.
Recent studies have begun to reveal structural changes that distinguish the active and inactive states of GPCRs. Mutations affecting cytoplasmic loops I, II, or III, or transmembrane segments (TMS) I, II, VI, or VII, constitutively activate GPCRs by destabilizing the inactive state or stabilizing the active state (Kjelsberg et al., 1992; Robinson et al., 1992; Parma et al., 1993; Robbins et al.; 1993, Samama et al., 1993; Shenker et al., 1993; Konopka et al., 1996; Scheer et al., 1996). Indeed, conformational changes accompanying GPCR activation occur in cytoplasmic loops, near the cytoplasmic terminus of TMS III or VII and within TMS VI (Ganter et al., 1992; Farahbakhsh et al., 1993; Bukusoglu and Jenness, 1996; Lin and Sakmar, 1996). Furthermore, the distance between TMS III and VI increases when rhodopsin is activated (Farrens et al., 1996; Yang et al., 1996). However, the specific kinds of secondary or tertiary structural changes that occur in activated GPCRs are poorly understood because high-resolution structural information is unavailable.
Less is understood about the mechanisms governing the biogenesis and trafficking of GCPRs, although insights are emerging from studies of visual opsins. Opsin biogenesis is facilitated by the action of cyclophilin-related proteins, which apparently function as prolyl isomerases and chaperones in the ER (Colley et al., 1991, 1995; Baker et al., 1994; Ferreira et al., 1996). However, the specific steps in the folding, assembly, and transport of opsins that are facilitated by cyclophilin homologs or other components of the quality control apparatus in the secretory pathway have not been clearly established.
Receptors for the oligopeptide mating pheromones, α-factor and a-factor of the yeast Saccharomyces cerevisiae, are useful models with which to study the function and biogenesis of GPCRs (Dohlman et al., 1991; Sprague and Thorner, 1992). Mating pheromones trigger a G protein-linked signal transduction pathway that induces expression of mating-specific genes, arrests cells in the G1 phase of the cell cycle, and alters cell morphology, culminating in cell and nuclear fusion. Mating pheromone receptors use their third cytoplasmic loops to couple with heterotrimeric G proteins (Boone et al., 1993; Weiner et al., 1993; Clark et al., 1994; Stefan and Blumer, 1994); they use their C-terminal cytoplasmic domains to promote receptor endocytosis and desensitization (Konopka et al., 1988; Reneke, et al., 1988; Rohrer et al., 1993), indicating that yeast and mammalian GCPRs function in similar ways.
Here we describe mutations that constitutively activate the receptors for the pheromones α-factor and a-factor. Characterization of these constitutively active receptors suggests that a conserved proline residue in transmembrane segment VI has a critical role in governing the activity and trafficking of GPCRs and provides genetic evidence that pheromone receptors and G proteins are precoupled before agonist stimulation.
MATERIALS AND METHODS
Materials, Media, and Isotopes
Enzymes used for recombinant DNA methods were purchased from commercial sources and used according to the suppliers’ recommendations. Sources of growth media for yeast and bacterial cells have been described previously (Blumer et al., 1988; Reneke et al., 1988). [35S]H2SO4 (carrier free) was obtained from Du Pont-New England Nuclear (Boston, MA). Sources of antibodies were as follows: rabbit polyclonal antibodies specific for Kar2p (Rose et al., 1989) (S. Wente of this department); rabbit polyclonal antisera specific for Gda1p (Berninsone et al., 1995) (C. Hirschberg, (University of Massachusetts, Amherst, MA); mouse monoclonal antibody C56 specific for the plasma membrane ATPase (Pma1p) (Aris and Blobel, 1988; Schandel and Jenness, 1994) (D. Jenness [University of Massachusetts] and J. Aris [University of Florida, Gainesville, FL]); mouse monoclonal antibodies specific for dolichol phosphate mannose transferase (Dpm1p), and the vacuolar ATPase (Vph1p) (Molecular Probes, Eugene, OR); peroxidase-, fluorescein isothiocyanate- and rhodamine-conjugated secondary antibodies (goat anti-rabbit and goat anti-mouse) (Organon Teknika, Durham, NC).
Plasmids and Yeast Strains
A plasmid that was used to create an unmarked chromosomal deletion of the STE2 gene was constructed by inserting a 1.6-kilobase (kb) EcoRI–HindIII fragment from pRS314STE2 (Weiner et al., 1993) into YIp5 that had been cleaved with EcoRI and HindIII, creating YIp5STE2-5′-UTR. Polymerase chain reaction (PCR) was used to generate a 0.7-kb HindIII–SphI fragment containing sequences downstream of the STE2-coding region. This fragment was digested with HindIII and SphI and inserted into YIp5STE2-5′-UTR that had been cleaved with HindIII and SphI to create YIp5ste2Δ. Thus, in plasmid YIp5ste2Δ the entire STE2-coding region was removed from a HindIII site 538 base pairs (bp) upstream of the start codon to a HindIII site 195 bp downstream of the stop codon.
To facilitate construction of plasmids that express various STE2 alleles, we deleted the PstI site in the polylinker of pRS314STE2 to create a plasmid (pRS314ΔP-STE2) with a unique PstI site in the STE2-coding region. For coexpression of various STE2 alleles, plasmid pRS313STE2 was constructed by isolating a 3.6 kb EcoRI–XbaI fragment encompassing the STE2 locus from pRS314STE2 and inserting it into pRS313 that had been cleaved with EcoRI and XbaI. Plasmid pRS313ste2L236R was constructed by isolating a 3.6-kb EcoRI–XbaI fragment containing the ste2L236R allele and inserting it into pRS313 that had been cleaved with EcoRI and XbaI. To overexpress various STE2 alleles, we inserted 4.3-kb ApaI–SacI fragments carrying either the wild-type allele or various codon 258 mutations into the high copy plasmid pRS424 that had been cleaved with ApaI and SacI. To express various STE3 alleles, we constructed plasmid pRS425STE3 by isolating a 2.4-kb HpaI–SacI fragment encompassing the STE3 locus and inserting it into pRS425 that had been cut with EcoRV and SacI.
To remove the C-terminal cytoplasmic domain of the α-factor receptor, we used PCR to create a nonsense mutation at codon 300 in the STE2-coding sequence and to introduce unique BglII and PstI sites immediately upstream of this stop codon. This PCR product was cloned, sequenced, digested with AatII and SacI, and introduced into pRS314ΔP-STE2 that had been cleaved with AatII and SacI to create pRS314ste2-300ter. Plasmid pRS314ste2P258L,300ter was created by inserting a 0.4-kb AatII–PstI fragment containing the ste2P258L allele into pRS314ste2-300ter that had been cleaved with AatII and PstI.
To detect α-factor receptors by immunological methods, we constructed plasmids that express wild-type and constitutively active receptors containing three c-myc epitopes at their extreme C termini. PCR was used to change the BclI site overlapping the natural stop codon of the STE2-coding sequence to a unique BglII site, destroying the translational stop codon. This PCR product was cloned, sequenced, digested with PstI and SacI, and inserted into pRS314ΔP-STE2 that had been cleaved with PstI and SacI to create pRS314STE2Δter. Plasmid pRS314STE2-3xmyc was created by inserting a BamHI fragment containing three c-myc epitopes in the appropriate reading frame into the BglII site of pRS314STE2Δter. A 0.6 kb AatII-PstI fragment carrying various mutations affecting codon 258 of the STE2 gene was inserted into pRS314STE2-3xmyc that had been cleaved with AatII and PstI to generate plasmids that express myc-tagged constitutively active receptors.
The S. cerevisiae strains used in these studies were: KBY16 (MATa ura3–52 trp1–903 his3-Δ200 ade2–101 leu2–3, 112 lys2–801 mfα1::LYS2 mfα2::LEU2 ste2Δ::HIS3 sst1-Δ5) (Stefan and Blumer, 1994), KBY17 (same as KBY16 but sst2Δ), KBY18 (same as KBY16 but far1Δ), KBY20 (same as KBY16 except it contains an unmarked ste2Δ allele), KBY22 (same as KBY16 except ste4::URA3), and SY1985 (MATα ste3Δ::URA3 ste2Δ mfa1Δ mfa2Δ::FUS1-lacZ FUS1::HIS3 ura3–52 leu2–3, 112 ade1 sst2Δ). They were constructed as follows. KBY17 was constructed by using NheI-cut pBC14 (Dohlman et al., 1996) to disrupt the SST2 gene in KBY16 by two-step gene replacement. Plasmid pFC13 (Chang and Herskowitz, 1990) digested with NotI was used to disrupt the FAR1 gene in KBY16; a 5-fluoroorotic acid-resistant derivative of this far1Δ strain was selected to create KBY18. KBY22 was constructed by using pAG3 (Grishin et al., 1994) cut with PstI and XhoI to disrupt the STE4 gene in KBY16. An unmarked deletion of STE2, ste2Δ, was made in KBY16 by two-step gene deletion using ClaI-cut YIp5ste2Δ to create KBY20; this disruption was confirmed by loss of HIS3. Strain SY1985 is a ste3Δ::URA3 sst2Δ derivative of SY1937 (Boone et al., 1993), which was provided by G.F. Sprague Jr. (University of Oregon, Eugene, OR).
Mutagenesis and Genetic Screening
Generation of mutations throughout the STE2-coding region was performed by hydroxylamine treatment (Sikorski and Boeke, 1991) and low fidelity PCR (Kocher et al., 1989) of pRS314STE2. Mutations isolated by genetic screens were identified by using primers to sequence the region encoding the Ste2p polypeptide. Site-directed mutagenesis of codon 258 in STE2 and codon 222 in STE3 was performed by PCR as described previously (Kjelsberg, et al., 1992). PCR products were digested with AatII and PstI to generate 0.6-kb fragments carrying various STE2 mutations and were inserted into pRS314ΔP-STE2 that had been cleaved with AatII and PstI. In experiments involving STE3, PCR products were digested with NheI and NdeI to generate 0.8- kb fragments carrying the various codon 222 mutations, which were inserted into pRS425STE3 that had been cleaved with NheI and NdeI. The resultant plasmids were sequenced across the relevant regions of the STE2- or STE3-coding regions to confirm the presence of codon 258 or 222 mutations and the absence of secondary mutations.
A library of hydroxylamine-treated plasmids (pRS314STE2) carrying mutations in sequences coding for Ste2p was introduced by transformation into a ste2Δ::HIS3 far1Δ mfα1::LYS2 mfα2::LEU2 strain (KBY18) containing FUS1-lacZ on plasmid pSL307 (McCaffrey, et al., 1987). In addition, four pools of fragments carrying random mutations in the STE2 gene that had been generated by low-fidelity PCR were independently introduced into KBY18 (containing pSL307) by gap repair of pRS314STE2 that had been cleaved with NdeI and AatII. Cells were plated on selective media (SD-tryptophan and uracil) lacking pheromone. Transformant colonies were replica plated onto filters impregnated with X-gal and assayed for expression of β-galactosidase as described previously (Fields and Song, 1989). Under the assay conditions employed (1 h incubation at 30°C), cells expressing the wild-type STE2 gene remained white. Plasmids isolated from transformants that were blue (expressed FUS1-lacZ) were transferred to Escherichia coli and introduced again into KBY18 containing the FUS1-lacZ plasmid. These transformants were subjected to quantitative assays to measure the strength of the constitutive signal, as described below.
Pheromone Response Assays and Dominance Tests
The level of pathway activation was determined by measuring the expression of the pheromone-inducible FUS1-lacZ reporter gene in plasmid pSL307. Cells carrying pSL307 and expressing various STE2 alleles were grown in selective media to a density of 107 cells/ml. Cultures were split into aliquots: one was a control, and the other received α-factor (1 μM final concentration). After a 2-h incubation at 30°C, cells were permeabilized and assayed for β-galactosidase activity (McCaffrey et al., 1987). Dominance tests were performed by using centromeric plasmids to coexpress various constitutively active α-factor receptors (pRS314 derivatives) and the wild type STE2 or ste2L236R alleles (pRS313 derivatives) in a ste2Δ mutant (KBY20) that also carried the FUS1-lacZ gene on pSL307; pathway activation in the absence or presence of α-factor was determined as described previously.
Ligand Binding and Receptor Internalization Assays
Methods used to purify [35S]α-factor and perform ligand- binding assays with inviable, intact cells have been described (Blumer et al., 1988). Assays of cells expressing wild-type receptors employed [35S]α-factor (20 Ci/mmol) at concentrations ranging from 0.1 to 10 nM, and those of cells expressing constitutively active receptors used α-factor concentrations from 0.05 to 20 nM. Assays of cells overexpressing various STE2 alleles from high-copy plasmids used [35S]α-factor (15 Ci/mmol) at concentrations ranging from 0.05 to 30 nM. Ligand-binding data were plotted according to the method of Scatchard and fitted by nonlinear least mean square regression. Nonspecific binding was determined in the presence of a 500-fold excess of unlabeled α-factor.
Rates of ligand-independent and ligand-induced loss of α-factor binding sites from the cell surface were measured as previously described (Stefan and Blumer, 1994), with the following modifications. Cultures were grown at 22°C in selective media (SD-tryptophan) to a density of 107 cells/ml and treated with cycloheximide (20 μg/ml) for 5 min. Basal rates of receptor internalization were determined in the absence of α-factor. Pheromone-induced rates of receptor internalization were determined by adding unlabeled α-factor to a final concentration of 50 nM. Aliquots of cells were removed at various times, treated with 10 mM NaN3 and 10 mM KF, and washed in YP (Blumer et al., 1988) containing 100 mM H3PO4, pH 2.5, to remove cell surface-bound α-factor. After cells were washed in 10 mM PIPES (pH 6.0), 1 mM MgCl2, 0.1 mM EDTA, 10 mM NaN3, 10 mM KF in YP media (Blumer et al., 1988), they were incubated with [35S]α-factor (10 nM, 30 Ci/mmol) with or without a 250-fold excess of unlabeled α-factor, which was used to determine levels of nonspecific binding. To determine whether α-factor–binding sites were preserved by these manipulations, we treated control cells with metabolic inhibitors (NaN3 and KF) immediately after treatment with cycloheximide and before addition of unlabeled pheromone and treated them as described above.
Immunoblotting and Indirect Immunofluorescence
Cultures were grown to a density of 2 × 107 cells/ml in synthetic medium (SD-tryptophan) to select for plasmid pRS314ΔPSTE2-3xmyc and its derivatives encoding myc-tagged constitutively active α-factor receptors. Methods used to detect myc-tagged Ste2p in yeast whole-cell extracts by immunoblotting were based on those previously described (Blumer et al., 1988). The protein concentration of yeast whole-cell lysates was determined by the Bradford method and adjusted to 2 mg/ml with Laemmli sample buffer before SDS-PAGE.
Preparation of cells for antibody incubations and immunofluorescence was performed essentially as described (Pringle et al., 1991). Cultures were grown at 30°C in selective medium (SD-tryptophan) to a density of 107 cells/ml. Formaldehyde was added to a final concentration of 3.7%. Cells were incubated 5 min at room temperature, washed, and suspended in 0.1 M potassium phosphate buffer, pH 7.0, containing 1 M sorbitol (buffer A). Spheroplasts were generated by incubating cells with glusulase and zymolyase 20T and washed with buffer A. Spheroplasts were bound to polylysine-coated slides, washed with Tris-buffered saline (TBS) containing 0.02% Tween 20, 0.01% Triton X-100, and 2% nonfat milk. Samples were incubated with antibodies for 16 h (9E10 tissue culture supernatant and/or Kar2p antibodies diluted 1:2 or 1:200, respectively, in TBS containing 0.01% Tween-20 and 2% nonfat milk [dilution buffer]). Slides were washed seven times with dilution buffer and incubated 2 h with rhodamine-conjugated goat anti-rabbit IgG and/or fluorescein isothiocyanate-conjugated goat anti-mouse IgG antibody diluted 1:500 or 1:1000, respectively, in TBS containing 0.01% Tween 20 and 1% bovine serum albumin (BSA). Slides were washed 10 times with TBS containing 0.01% Tween 20 and 1% BSA and incubated 5 min with 2.5 μg/ml 4′,6-diamino-2-phenylindole in TBS containing 0.01% Tween 20 and 1% BSA. Slides were washed once with TBS containing 0.01% Tween 20 and 1% BSA, and cells were observed under an Olympus epifluorescence microscope.
Subcellular Fractionation
Subcellular fractionation was carried out by equilibrium density gradient centrifugation essentially as described previously (Kölling and Hollenberg, 1994). Cells were grown in selective medium (SD-tryptophan) to a density of 107 cells/ml. Cultures were treated with 10 mM sodium azide and 10 mM KF. Cells were collected by centrifugation and washed once with 25 ml of sorbitol buffer (10 mM Tris, pH 7.6, 0.8 M sorbitol, 10 mM NaN3, 10 mM KF, 1 mM EDTA, pH 8.0). Cells were collected by centrifugation and washed once with 1 ml sorbitol buffer, once with 1 ml sucrose buffer (10 mM Tris pH 7.6, 1 mM EDTA, 10% [wt/vol] sucrose), and suspended in 1 ml sucrose buffer containing protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM benzamidine, 20 μM tosyl-phenylalanine chloromethyl ketone, 5 μM pepstatin A, and 5 μM leupeptin). Glass beads were added, and the cells were lysed by mechanical disruption. Unbroken cells were removed from the lysate by centrifugation at 300 × g for 5 min. The supernatant fraction (0.5 ml) was mixed with 0.5 ml of 50% (wt/vol) sucrose in 10 mM Tris, pH 7.6, 1 mM EDTA, and layered on top of a 4 ml, 35–60% linear sucrose gradient prepared in 10 mM Tris, pH 7.6, 1 mM EDTA. Gradients were centrifuged 20 h at 150,000 × g in a SW50.1 rotor at 4°C. Fractions (350 μl) were collected from the top of the gradient and diluted 1:2 with 2× Laemmli sample buffer containing 8 M urea. Samples were heated for 10 min at 37°C before SDS-PAGE. Ste2p-myc, Vph1p, Gda1p, Dpm1p, and Pma1p were detected by immunoblotting.
RESULTS
Identification of Constitutively Active α-Factor Receptors
To identify mutations in the α-factor receptor structural gene (STE2) that activate the response pathway in the absence of pheromone, we used a genetic screen to identify cells that constitutively express a pheromone-inducible reporter. The yeast strain used for this purpose (KBY18) had the following important features: a ste2Δ mutation, which ensures that cells express receptors only from mutagenized plasmids; deletions of the two α-factor structural genes (MFα1, MFα2), which ensure that apparent constitutive signaling is not due to autocrine stimulation of a hypersensitive mutant receptor; a deletion of the gene (FAR1) encoding a cyclin-dependent kinase inhibitor, which prevents growth arrest from occurring in cells expressing strongly constitutively active receptors; and a plasmid-borne pheromone-inducible reporter (FUS1-lacZ), which enables the identification of cells that signal constitutively (stain blue with X-gal in the absence of α-factor). This strain was transformed with pools of a mutagenized (hydroxylamine treatment or error-prone PCR) single-copy plasmid in which the STE2 gene is expressed from its normal promoter. Approximately 60,000 transformants were screened in the absence of pheromone for elevated expression of FUS1-lacZ by staining colony filter lifts with X-gal. Plasmids from 10 transformants that were positive in this assay were recovered in Escherichia coli and rescreened in yeast for the ability to induce FUS1-lacZ in the absence of pheromone. Eight of the 10 plasmids passed this test. The STE2-coding regions of these eight plasmids were sequenced.
Three classes of mutations were obtained. The first class (four plasmids) contained a mutation that resulted in substitution of proline-258 for leucine (P258L) in transmembrane segment six (TMS VI). This ste2P258L allele was chosen for further study because it caused the strongest constitutive signaling phenotype (C. Stefan, unpublished data). Furthermore, the P258L substitution was particularly interesting because proline residues in transmembrane domains have been proposed to control the activity of receptors, ion channels, and transporters (Williams and Deber, 1991), and a proline residue is present in TMS VI in >90% of all GPCRs (Table 1; Baldwin, 1993), suggesting that it may have a conserved function. The second class (one plasmid) carried a single mutation that resulted in a serine-to-proline substitution at position 259 in TMS VI, and a third class (three plasmids) contained several mutations within the STE2 gene; analysis of these two classes will be described elsewhere.
Table 1.
TMS VI sequences of representative GPCRs
| Receptor | TMS VI sequence |
|---|---|
| AChM3 bovine (493-513) | SAILLAFIITWTPYNIMVLV |
| α1B-AR rat (296-319) | LGIVVGMFILCWLPFFIALPLGSL |
| β2AR human (275-298) | LGIIMGTFTLCWLPFFIVNIVHVI |
| BK-1R human (248-272) | TTALILTLVVASLVCWAPYHFFAFL |
| FMLF-R human (243-266) | LSFVAAAFFLCWSPYQVVALIATV |
| FSH-R human (574-597) | MAMLIFTDFLCMAPISFFAISASL |
| δOpioid R-1 human (262-284) | MVLVVVGAFVVCWAPIHIFVIVW |
| Rhodopsin bovine (253-276) | MVIIMVIAFLICWLPYAGVAFYIF |
| PGE2-R2 human (263-286) | LILLAIMTITFAVCSLPFTIFAYM |
| NK1R rat (249-270) | MMIVVVCTFAICWLPFHVFFLL |
| ETR1 bovine (307-328) | TVFCLVVIFALCWFPLHLSRIL |
| THR-R rat (319-341) | LFLSAAVFCIFIVCFGPTNVLLI |
| MGR1 rat (768-808) | YIAFTMYTTCIIWLAFVPIYFGS |
| CASR rat (806-828) | ICFFFAFKSRKLPENFNEAKFIT |
| GUSB bovine (242-265) | LFTVVIVFIVTQLPYNIVKFCQAI |
| STE2 S. cerevisiae (244-266) | FHILLIMSCQSLLVPSIIFILAY |
| STE3 S. cerevisiae (206-228) | FARLLIFCFIIILVMFPFSVYTF |
| CALR human (360-377) | ATMILVPLLGIQFVVFPW |
| GIP-R human (342-362) | STLTLVPLLGVHEVVFAPVTE |
| PTR-R human (410-428) | TLVLMPLFGVHYIVFMATP |
| OLF1 rat (237-260) | IFSTCGSHLSVVSLFYGTIIGLYL |
| VN2 rat (241-259) | ILMLRSLFGLMSIFDSIAS |
| srb-1 C. elegans (238-260) | FTLIVSFTHILFIGWYLGVTIFI |
| CAR1 D. discoidium. (206-224) | FKLINYIIVFLVCWVFAVV |
| VN1 rat (241-259) | ILMLMSLFVLMSVFDSIVC |
| srg-5 C. elegans (236-260) | LCFASFYMSAAFFSAALFQSYFAFF |
TMS VI sequences from the indicated receptors are shown; members of the first group (which constitutes ∼90% of GPRCs) contain proline residues in TMS VI, those of the second contain glycine and/or proline residues, while those of the third lack proline and glycine residues. The N- and C-terminal boundaries of TMS VI of each receptor (indicated in parentheses) were assigned according to data obtained from the GPCR database (http://receptor.mgh.harvard.edu/GCRDBHOME.html). The single letter amino acid code is used.
Other investigators have shown previously that the P258L substitution in TMS VI constitutively activates the α-factor receptor (Konopka et al., 1996). This finding led Konopka and colleagues to propose that a conserved proline residue at this position is required to stabilize the inactive conformation of the α-factor receptor, and perhaps other GCPRs as well, possibly by inducing a kink in TMS VI. However, there were several reasons why we believed it was important to test this model further. First, these investigators did not determine whether a proline residue is specifically required at position 258 in the α-factor receptor or whether other amino acids at this site can preserve wild-type receptor function. Second, the analogous proline residue has been mutated in other GCPRs, but constitutive activity was not reported (Wess et al., 1993; Kaushal and Khorana, 1994; Kolakowski et al., 1995). Third, a significant number of GPCRs (∼10%) lack a proline at this position (Table 1 lists some examples), indicating that a proline is not always required. Accordingly, to address these points we have determined whether changing proline-258 to any other amino acid constitutively activates the α-factor receptor, investigated cellular regulatory mechanisms that influence detection of a constitutive signal, and determined whether changing the equivalent proline residue in another GPCR results in constitutive signaling.
Role of Proline-258 in TMS VI of the α-Factor Receptor
We constructed a set of mutations that change proline-258 in the α-factor receptor to all other amino acids. Each allele was expressed from the normal STE2 promoter on single-copy plasmids in a ste2Δ mutant that contained a pheromone-inducible reporter (FUS1-lacZ) on a high-copy plasmid (pSL307). We found that several substitutions of proline-258 increased agonist-independent reporter gene expression (Table 2, column 2). Substitution of proline-258 with methionine caused the strongest constitutive signaling phenotype (basal expression of FUS1-lacZ was increased approximately 50-fold above wild-type basal levels).
Table 2.
Substitutions of proline-258 in TMS VI of the α-factor receptor: effects on receptor signal transduction, agonist binding affinity, and cell surface expression
| STE2 allele |
FUS1-lacZ expression (% wild type
+ α-factor)
|
α-Factor binding sites | ||||
|---|---|---|---|---|---|---|
|
SST2
|
sst2Δ
|
SST2
|
||||
| −α-factor | +α-factor | −α-factor | +α-factor | Kd (nM) | Bmax (sites/cell) | |
| ste2Δ | 0.2 | 0.2 | 1.6 | 1.4 | − | − |
| wild type | 0.2 | 100 | 1.3 | 100 | 3.3 | 15,000 |
| P258A | 2.7 | 78 | 14 | 100 | 0.37 | 1,000 |
| P258I | 4.2 | 110 | 18 | 120 | 0.44 | 1,100 |
| P258L | 5.3 | 140 | 13 | 99 | 0.42 | 1,700 |
| P258M | 14 | 120 | 53 | 110 | 1.0 | 480 |
| P258V | 2.8 | 110 | 21 | 70 | 0.34 | 1,500 |
| P258D | 1.0 | 23 | 7.1 | 58 | 0.58 | 260 |
| P258E | 0.2 | 18 | 6.0 | 51 | 0.56 | 70 |
| P258H | 0.2 | 0.5 | 6.1 | 6.9 | n.d. | n.d. |
| P258K | 0.2 | 7.2 | 5.3 | 28 | 1.2 | 40 |
| P258R | 0.4 | 16 | 3.6 | 17 | 0.68 | 60 |
| P258C | 0.5 | 23 | 13 | 56 | 0.45 | 210 |
| P258G | 0.4 | 20 | 5.6 | 110 | 1.0 | 170 |
| P258N | 1.7 | 3.2 | 11 | 12 | n.d. | n.d. |
| P258Q | 0.6 | 10 | 3.5 | 9.4 | 0.58 | 40 |
| P258S | 0.3 | 0.6 | 4.6 | 7.3 | n.d. | n.d. |
| P258T | 0.3 | 0.7 | 6.8 | 7.8 | n.d. | n.d. |
| P258Y | 13 | 47 | 25 | 63 | n.d. | n.d. |
| P258F | 3.7 | 83 | 27 | 91 | 2.5 | 170 |
| P258W | 1.6 | 3.2 | 5.3 | 6.9 | n.d. | n.d. |
| 300ter | 1.0 | 79 | ||||
| P258L,300ter | 45 | 92 | ||||
The indicated STE2 alleles were expressed from centromeric plasmids (pRS314 derivatives) in isogenic MATa ste2Δ mfα1Δ mfα2Δ strains that expressed (KBY16) or lacked (KBY17) the SST2 gene. Cells also contained the pheromone-inducible FUS1-lacZ gene on plasmid pSL307. Cells were treated as indicated with synthetic α-factor (1 μM, 2 h at 30°), and activation of the pheromone response pathway was quantified by performing β-galactosidase assays. Data are expressed as the percent of the activity detected in α-factor–treated cells that expressed the wild-type STE2 gene. Data shown for each STE2 allele are the average obtained from assays of at least four independent transformants, each of which was assayed in duplicate; standard errors were 10-30% of the values shown. Radioligand binding assays were performed using [35S]α-factor and KBY16 cells expressing the indicated STE2 alleles from centromeric plasmids (pRS314 derivatives). The Kd and Bmax values shown for cells expressing each STE2 allele were calculated by nonlinear regression of data obtained from two to three independent transformants assayed in duplicate; standard errors for these determinations were 5-15% of the values shown. n.d., Specific binding was not detected.
However, not all substitutions of proline-258 resulted in a detectable constitutive signal (Table 2, column 2). Although there were several possible explanations, one obvious possibility was that these mutant receptors transduce a weak constitutive signal that is attenuated by mechanisms that normally promote desensitization to pheromone.
To determine whether desensitization mechanisms reduce the apparent strength of the constitutive signal, we expressed mutant receptors in two types of desensitization-defective mutants. First, we blocked phosphorylation-dependent receptor desensitization and endocytosis by removing the C-terminal cytoplasmic domain of receptors bearing the P258L substitution (ste2P258L, 300ter). Combining this truncation mutation with the P258L substitution increased agonist-independent reporter gene expression eightfold over that observed when the P258L substitution was present in the full-length receptor (Table 2, column 2), consistent with the expectation that the apparent strength of the constitutive signal is negatively regulated at the receptor level. Second, we expressed each of the 19 mutant receptors in an sst2Δ mutant, which lacks a regulator of G-protein signaling homolog that apparently promotes desensitization by stimulating the guanosine triphosphatase activity of the yeast G protein α subunit (Gpa1p) (Dohlman et al., 1996; Dohlman and Thorner, 1997; Dohlman, personal communication). In an sst2Δ mutant, substitution of proline-258 in the α-factor receptor with any other amino acid resulted in a detectable constitutive signal (2- to 40-fold above wild-type receptor controls) (Table 2, column 4). This allowed us to compare the phenotypes conferred by various mutations, leading to the following observations. Substitution of proline-258 with aliphatic amino acids generally gave the strongest constitutive signal. Substitution of proline-258 with a charged residue resulted in intermediate constitutive activity and somewhat impaired responses to pheromone. Substitution of proline-258 with uncharged hydrophilic residues resulted in weak constitutive activity and strongly impaired responses to pheromone. Thus, the conserved proline residue at position 258 is essential for normal function of the α-factor receptor, consistent with the suggestion that it is required for the receptor to adopt or maintain a native, fully inactive conformation and to be activated normally by agonist.
Mutations Affecting the Conserved Proline Residue in TMS VI of the a-Factor Receptor
To determine whether the conserved proline residue of TMS VI may generally control the activity of GPCRs, we generated substitutions of the equivalent proline residue (proline-222) in TMS VI of the a-factor receptor of S. cerevisiae (STE3 gene product), which is unrelated in sequence to the α-factor receptor. These experiments employed a strain of a different genetic background with the following key features: a ste3Δ mutation, which ensures that a-factor receptors are expressed only from mutated plasmids; deletions of both a-factor structural genes (MFa1, MFa2), which preclude autocrine stimulation of mutant receptors; an sst2Δ mutation, which eliminates RGS-stimulated guanosine triphosphatase activity of Gα subunits that might otherwise attenuate a weak constitutive signal; and a chromosomally integrated pheromone-inducible reporter gene (FUS1-lacZ), which allows constitutive signals to be detected by performing β-galactosidase assays. Using this strain we examined the effects of substituting proline-222 of the a-factor receptor with leucine (ste3P222L) or tyrosine (ste3P222Y); the analogous substitutions affecting the α-factor receptor gave readily detectable constitutive signals. Although expression of these ste3 alleles from their normal promoters on single-copy plasmids did not result in a detectable constitutive signal (Stefan, unpublished data), expression from high-copy plasmids did result in a twofold increase in constitutive expression of the reporter, relative to wild- type receptor controls (Table 3, column 2). This constitutive signal was significant because it is 50% of the maximal pheromone-stimulated signal in cells expressing wild-type receptors (Table 3, column 3). We also noted that a-factor receptors bearing either substitution of proline-222 did not respond to a-factor (Table 3, column 3), possibly because these substitutions interfere with ligand binding, receptor folding, or cell surface expression (see below). Despite these complex effects on receptor function, the results support the hypothesis that a conserved proline residue in TMS VI helps establish or maintain the inactive conformation of GPCRs.
Table 3.
Effects of substitutions of proline-222 in TMS VI of the a-factor receptor
| STE3 allele |
FUS1-lacZ expression (% wild type
+a-factor)
|
|
|---|---|---|
| −a-factor | +a-factor | |
| ste3Δ | 40 ± 6 | 40 ± 6 |
| wild-type | 32 ± 7 | 100 ± 9 |
| P222L | 60 ± 10 | 72 ± 8 |
| P222F | 60 ± 11 | 76 ± 3 |
The indicated STE3 alleles were overexpressed from their normal promoters on high copy plasmids (pRS425 derivatives) in a ste3Δ mfa1Δ mfa2Δ sst2Δ strain (SY1985) that contained an integrated FUS1-lacZ reporter. Where indicated, cells were diluted 1:1 with culture fluid from MATa cells (source of a-factor), incubated 2 h at 30° and assayed for β-galactosidase activity. Data are expressed as a percent of the activity detected using a-factor-treated cells that expressed the wild-type STE3 gene. At least four independent transformants of each type were assayed in duplicate; standard errors are indicated.
Other Functions of the α-Factor Receptor Influenced by Proline-258
Because various substitutions of proline-258 in the α-factor receptor constitutively activated the response pathway to different degrees, and because some of these substitutions impaired further activation of the pathway by pheromone, it was likely that proline-258 has complex roles in governing receptor function. Therefore, we analyzed other properties of mutant receptors to investigate the mechanisms that may underlie these phenotypic differences.
Level and Affinity of Cell-Surface α-Factor–Binding Sites.
Radioligand binding experiments employing intact, inviable cells revealed differences among the 19 mutant receptors (Table 2, columns 6 and 7). One striking difference was the level of α-factor–binding sites expressed at the cell surface. Six mutants displayed undetectable levels of agonist-binding activity. Relative to wild-type cells, the remaining mutants expressed 10- to 400-fold fewer ligand-binding sites per cell. A second difference was that mutant receptors displayed increased affinity for α-factor, ranging from a 50% increase (P258F) to nearly 10-fold (P258A, P258I, P258L, P258V, P258C, P258Q), similar to the properties of constitutively active GPCRs in mammalian cells (e.g., Kjelsberg et al., 1992). These differences in α-factor–binding affinity could reflect the extent that various amino acid substitutions destabilize the inactive conformation of the receptor, affecting agonist-binding affinity indirectly; alternatively, they could be due to alterations of the ligand-binding site, directly affecting pheromone-binding affinity. Further experiments will be needed to address these questions.
Receptor Protein Expression and Trafficking.
Substitutions of proline-258 could reduce the expression of cell-surface α-factor–binding sites by affecting receptor endocytosis, degradation, retention within the cell, or folding to form an active ligand-binding site. To address these possibilities we performed several experiments with a subset of the mutant receptors.
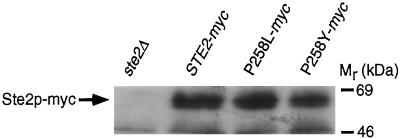
Initially, immunoblotting was used to examine the expression of wild-type and constitutively active receptor polypeptides. These experiments employed receptors that were tagged at their C termini with three tandem copies of the c-myc epitope, a modification that did not alter the signaling, ligand-binding, or internalization properties of the receptors (Stefan, unpublished data). The results indicated that although substitutions of proline-258 caused severe reductions in the level of cell-surface ligand-binding sites (P258L, eightfold reduction; P258Y, undetectable ligand-binding activity), they had relatively little effect on receptor protein expression levels (Figure 1).
Figure 1.
Expression of wild-type and constitutively active α-factor receptors. Various myc-tagged α-factor receptors were expressed from their normal promoters on centromere-containing plasmids (pRS314 derivatives) in a ste2Δ mutant (KBY16). Equivalent amounts of protein extract (50 μg) prepared from cells carrying a control plasmid (lane 1), or plasmids expressing wild-type STE2-myc (lane 2) or constitutively active α-factor receptors (ste2P258L-myc, lane 3; ste2P258Y-myc, lane 4) were resolved by SDS-PAGE. Immunoblotting was performed by using 9E10 antibodies specific for the c-myc epitope and a chemiluminescence detection system.
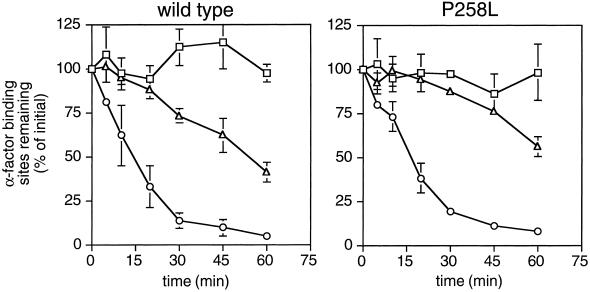
Subsequently, we examined whether reduced cell surface expression of receptors was due to increased rates of receptor internalization. This was studied in two ways. First, we examined rates of receptor internalization from the cell surface. These experiments were done by following the loss of cell-surface α-factor–binding sites over time under conditions in which new receptor synthesis is blocked. The results indicated that the P258L substitution did not increase the basal or α-factor–stimulated rates of receptor internalization (Figure 2). However, this does not necessarily rule out that mutant receptors have increased basal internalization rates. During the course of the internalization experiment there could be two balancing, competing processes occurring simultaneously: increased basal internalization of mutant receptors from the cell surface, and the delivery of mutant receptors from intracellular pools to the cell surface. If so, removal of the C-terminal domain of mutant receptors, which is required for endocytosis (Reneke et al., 1988), should restore cell surface expression of mutant receptors to wild-type levels. Accordingly, we truncated wild-type and mutant receptors bearing the P258L substitution immediately after TMS VII by changing codon 300 to a nonsense codon, creating the ste2-300ter and ste2P258L,300ter alleles, respectively. We found that the level of ligand-binding sites on the surface of cells expressing truncated constitutively active receptors (encoded by the ste2P258L, 300ter allele) was 10-fold lower than that on the surface of cells expressing truncated wild-type receptors (ste2-300ter) (5,000 sites/cell and 49,000 sites/cell, respectively). Thus, there were no indications that increased basal rates of receptor internalization are responsible for reducing the levels of mutant receptors at the cell surface.
Figure 2.
Internalization of wild-type and constitutively active α-factor receptors. The wild-type STE2 or ste2P258L alleles were expressed from their normal promoters on centromere-containing plasmids (pRS314 derivatives) in a ste2Δ mutant (KBY16). Assays of basal and agonist-induced internalization of wild-type receptors (left panel) and constitutively active α-factor receptors (expressed from the ste2P258L allele; right panel) were performed by determining the number of cell surface α-factor–binding sites remaining as a function of time. Rates of receptor internalization were determined in the absence of α-factor (triangles) and in response to unlabeled α-factor (circles) that was stripped from cells before radioligand binding assays were performed. As a control, the stability of cell surface receptors (squares) was determined by inhibiting internalization (with NaN3 and KF) and determining the number of ligand-binding sites remaining over time. Data shown are the average of duplicate assays of two independent transformants expressing wild-type or constitutively active receptors; standard deviations are indicated.
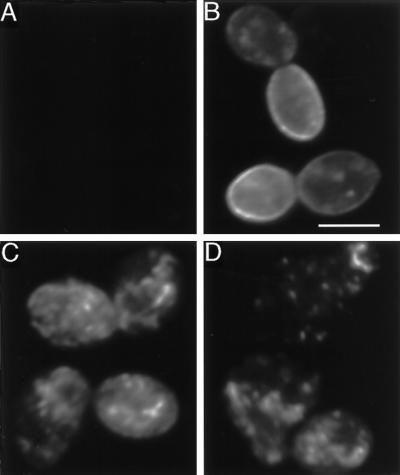
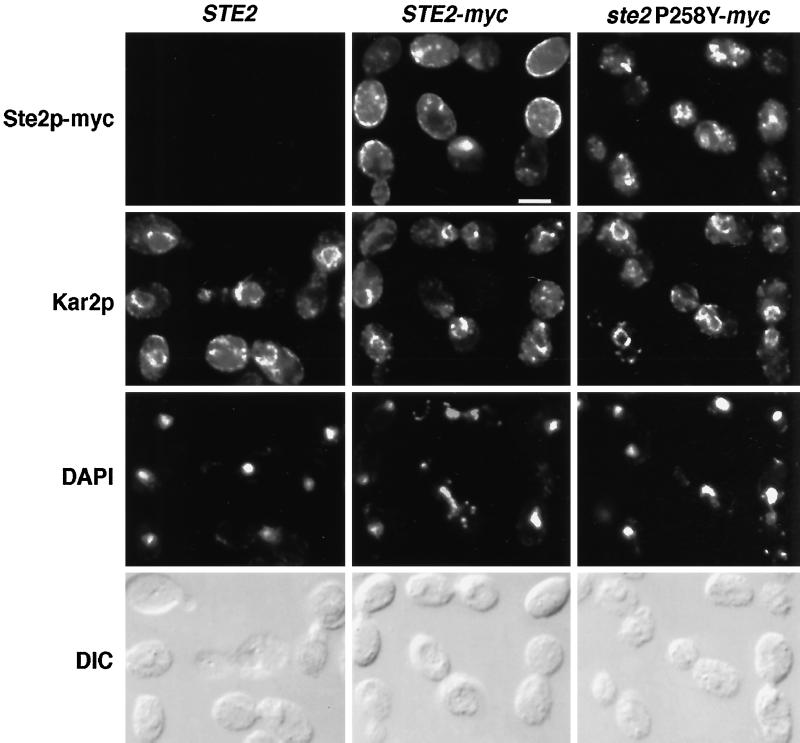
Because decreased protein expression or increased endocytosis rates appeared insufficient to account for the low level of cell-surface ligand-binding sites in cells expressing constitutively active receptors, we examined the subcellular localization of wild-type and constitutively active receptors by performing indirect immunofluorescence experiments. These experiments employed the full-length myc-tagged wild-type and constitutively active receptors described previously. Cells expressing wild-type myc-tagged receptors displayed intense cell-surface staining (Figure 3B), and less extensive staining of intracellular compartments, consistent with previous studies using untagged wild-type receptors (Jackson et al., 1991). In contrast, cells expressing myc-tagged receptors harboring either the P258L or P258Y substitution displayed weak or undetectable cell-surface staining (Figure 3, C and D); however, staining of intracellular compartments was observed. Therefore, the low level of α-factor–binding sites detected in cells expressing constitutively active receptors is correlated with the retention of receptor polypeptides in intracellular organelles.
Figure 3.
Immunofluorescence localization of myc-tagged wild-type and constitutively active α-factor receptors. Various myc-tagged α-factor receptors were expressed from their normal promoters on centromere-containing plasmids (pRS314 derivatives) in a ste2Δ mutant (KBY16). Cells carrying a control plasmid (expressing untagged α-factor receptors; panel A), or plasmids expressing myc-tagged wild-type (panel B) or constitutively active α-factor receptors (ste2P258L-myc, panel C; ste2P258Y-myc, panel D) were prepared for indirect immunofluorescence using 9E10 monoclonal antibodies specific for the c-myc epitope. Cells expressing the ste2P258Y-myc allele were misshapen, resembling the morphology of pheromone-treated wild-type cells. Bar, 5 μm.
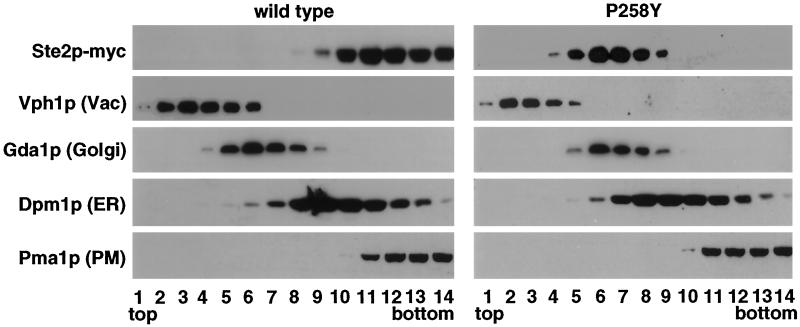
To characterize the intracellular compartment(s) where mutant α-factor receptors accumulate, we performed subcellular fractionation and double-label immunofluorescence experiments. The former experiments used sucrose density gradients to fractionate lysates prepared from cells expressing myc-tagged wild-type or mutant (P258Y) receptors. Immunoblotting was used to detect myc-tagged receptors and various marker proteins in gradient fractions (Figure 4). As expected, the fractionation of wild-type receptors most closely resembled that of the plasma membrane ATPase (Pma1p). In contrast, the fractionation of mutant receptors most closely resembled that of the Golgi-localized guanosine diphosphatase (Gda1p). This conclusion was further supported by the results of double-label immunofluorescence experiments using anti-Kar2p antibodies and anti-myc monoclonal antibodies (Figure 5). Kar2p immunofluorescence was restricted mainly to perinuclear rings characteristic of the ER. In contrast, staining of myc-tagged mutant receptors was more widely distributed in a punctate pattern that did not overlap considerably with that of Kar2p. These results therefore suggested that mutant α-factor receptors bearing substitutions of proline-258 accumulate in post-ER compartments.
Figure 4.
Subcellular fractionation of wild-type and constitutively active α-factor receptors. Myc-tagged forms of wild-type and constitutively active (P258Y) α-factor receptors were expressed from their normal promoters on centromere-containing plasmids (pRS314 derivatives) in a ste2Δ mutant (KBY16). Cell extracts were fractionated by sucrose gradient centrifugation. Gradient fractions (1 = top; 14 = bottom) were analyzed by immunoblotting using anti-myc antibodies and antibodies specific for the following marker proteins: Vph1p (vacuole), Gda1p (Golgi), Dpm1p (ER), and Pma1p (plasma membrane).
Figure 5.
Double-label immunofluorescence localization of α-factor receptors and Kar2p. Untagged wild-type receptors (STE2; first column) and myc-tagged forms of wild type (STE2-myc ; second column) and constitutively active (ste2P258Y-myc; third column) α-factor receptors were expressed from their normal promoters on centromere-containing plasmids (pRS314 derivatives) in a ste2Δ mutant (KBY16). Cells were costained with anti-myc monoclonal antibodies (first row), a rabbit antisera specific for the ER marker protein Kar2p (second row), and 4′,6-diamino-2-phenylindole (third row). Bar, 5 μm.
The intracellular accumulation of constitutively active receptors could occur for various reasons. One possibility is that receptor retention is caused directly or indirectly by activation of the pheromone-response pathway. To test this possibility we inactivated the signal transduction pathway by deleting the STE4 gene, which encodes the G protein β subunit required for receptor-G protein coupling and signal propagation (Whiteway et al., 1989; Grishin et al, 1994), and examined the expression and localization of wild-type and constitutively active receptors by performing radioligand binding and immunofluorescence experiments. In the ste4 mutant, wild-type and constitutively active (P258L) receptors were expressed at 2500 sites/cell and 350 sites/cell, respectively. Similarly, immunofluorescence experiments indicated that wild-type myc-tagged receptors were present primarily at the cell surface in ste4 mutants, whereas myc-tagged constitutively active receptors (bearing the P258L substitution) were localized in intracellular compartments (Stefan, unpublished data). Therefore, activation of the signaling pathway was not required for intracellular localization of constitutively active receptors.
A second possibility is that mutant receptors are retained in intracellular compartments because they have folding defects. However, the folding status of the intracellular pool of mutant receptors cannot be determined by performing ligand-binding assays because the ligand-binding site should be lumenally disposed, and because receptors are inactivated upon detergent treatment of membranes (Blumer, unpublished data). Nevertheless, a folding defect seems likely because similar defects appear to occur when conserved proline residues in other GPCRs are substituted with other amino acids (Wess et al., 1993; Kolakowski et al., 1995).
To determine whether defects in receptor expression at the cell surface limit the strength of the constitutive signal, we examined the effects of overexpressing various STE2 alleles. Overexpression of STE2 alleles (wild type, P258D, P258L, P258Y) from high-copy plasmids increased the levels of cell-surface α-factor binding sites 3- to more than 10-fold (compare Table 4, column 4, with Table 2, column 7). Whereas overexpression of wild type receptors did not increase the basal signal, overexpression of mutant receptors caused a threefold increase (up to 50% of the fully induced level) in the constitutive signal (compare Table 4, column 2, with Table 2, column 2). Furthermore, receptor overexpression completely corrected the defects in agonist-induced signaling of some of these proline-258 substitutions (P258D and P258Y; compare Table 4, column 3 with Table 2, column 3). Thus, defects in receptor expression at the cell surface probably account for the relatively weak constitutive signal and impaired agonist-induced signaling caused by certain substitutions of proline-258.
Table 4.
Effects of overexpressing STE2 alleles on constitutive activation of the pheromome response pathway and expression of cell-surface α-factor binding sites
| STE2 allele expressed |
FUS1-lacZ expression (% wild
type + α-factor)
|
Receptor expression (α-factor binding sites/cell) | |
|---|---|---|---|
| −α-factor | +α-factor | ||
| ste2Δ | 0.7 | 1.0 | − |
| Wild type | 0.4 | 100 | 34,000 |
| P258L | 18 | 96 | 18,000 |
| P258D | 3.9 | 97 | 700 |
| P258Y | 54 | 103 | 250 |
The indicated STE2 alleles were expressed from their normal promoters on high-copy plasmids (pRS424 derivatives) in a ste2Δ mfα1Δ mfα2Δ mutant (KBY16) that contained the FUS1-lacZ gene on a plasmid (pSL307). Where indicated, cells were treated 2 h at 30°C with 1 μM synthetic α-factor (+α-factor); β-galactosidase assays were performed to monitor activation of the pheromone response pathway. Data are expressed as a percent of the activity detected with α-factor–treated cells overexpressing the wild-type STE2 gene. Values shown are the average of a least four independent transformants, each assayed in duplicate; standard errors were 5-30% of the values shown. Radioligand binding assays using [35S]α-factor were performed to determine the number of cell-surface ligand-binding sites (Bmax) in cells (KBY16) overexpressing various STE2 alleles from high-copy plasmids (pRS424 derivatives). The values shown for each STE2 allele were calculated from nonlinear regressions of data obtained from two independent transformants, each of which was assayed two to four times. Standard errors for these determinations were 5-15%
Expression of Wild-Type Receptors Suppresses Constitutive Signaling by Mutant Receptors
Because α-factor receptors bearing substitutions of proline-258 are constitutively active, we anticipated that they would signal constitutively when they are coexpressed with wild-type receptors. Contrary to this expectation, when constitutively active (P258L or P258Y) and wild-type α-factor receptors were coexpressed from their normal promoters on single-copy plasmids, a significant constitutive signal was not detected (Table 5), indicating that mutations resulting in constitutively active receptors are nearly completely recessive. Similarly, Konopka and colleagues showed that the presence of wild-type receptors reduces ability of constitutively active receptors to transduce a signal in the absence of α-factor (Konopka et al., 1996); however, the magnitude of this inhibitory effect was less than we observed, which led these investigators to conclude that the ste2P258L allele is partially dominant. Differences in strain background might account for these quantitative differences (for example, our strains were deleted for the α-factor structural genes, whereas those used by others were not), but this has not been examined directly. Nevertheless, our results agree qualitatively with those published previously (Konopka et al., 1996).
Table 5.
Effects of coexpressing STE2 alleles on constitutive activation of the pheromone response pathway
| STE2 alleles coexpressed |
FUS1-lacZ
expression (% wild type +
α-factor)
|
||
|---|---|---|---|
| −α-factor | +α-factor | ||
| Wild type | Wild type | 0.4 | 100 |
| Wild type | P258L | 0.9 | 128 |
| Wild type | P258F | 0.5 | 103 |
| L236R | Wild type | 0.7 | 123 |
| L236R | P258L | 4.6 | 114 |
| L236R | P258F | 7.0 | 104 |
The wild-type STE2 gene or the ste2L236R allele, which causes a specific defect in receptor coupling with G proteins, was coexpressed with the wild-type STE2 gene, or the ste2P258L or ste2P258F alleles. These genes were coexpressed from their normal promoters on centromeric plasmids (pRS313 and pRS314 derivatives) in a ste2Δ mfα1Δ mfα2Δ mutant (KBY20) that contained the FUS1-lacZ gene on a plasmid (pSL307). Where indicated, cells were treated 2 h at 30°C with 1 μM synthetic α-factor (+α-factor); β-galactosidase assays were performed to monitor activation of the pheromone response pathway. Values are expressed as a percent of the activity detected with α-factor–treated cells expressing the wild-type STE2 gene. Values shown are the average of at least four independent transformants, each assayed in duplicate; standard errors were 5-30% of the values shown.
Wild-type receptors could interfere with the ability of constitutively active receptors to signal by various mechanisms. For example, wild-type and mutant receptors could interact to form oligomers having low agonist-independent activity similar to that of wild-type receptors alone; however, evidence that α-factor receptors form oligomers in the membrane has not been reported. Alternatively, in the absence of agonist, wild-type receptors could associate with and sequester G protein heterotrimers that are present in limiting amounts (i.e., receptors and G proteins are “precoupled”), thereby preventing constitutively active receptors from transmitting a signal. Overexpressing the three G protein subunits could overcome this effect, but this would be difficult to accomplish experimentally because the subunits must be overproduced stoichiometrically. As an alternative, we determined whether receptors that interact inefficiently with G proteins are unable to interfere with the ability of constitutively active receptors to signal. Accordingly, we coexpressed constitutively active receptors with receptors that bear a substitution affecting the third cytoplasmic loop (ste2L236R, which reduces coupling efficiency 10-fold without affecting ligand-binding affinity, receptor cell surface expression, or endocytosis; Weiner et al., 1993). In this situation, constitutively active receptors were able to transduce a constitutive signal (Table 5). Assuming mutations that uncouple receptors do not affect receptor oligomerization or other aspects of receptor function, these results suggest that α-factor receptors and G proteins are precoupled in the absence of pheromone stimulation.
DISCUSSION
GPCR Activation Mechanisms
Our results and those of others (Konopka et al., 1996) indicate that a conserved proline residue in TMS VI controls the equilibrium between the inactive and active states of the α-factor and a-factor receptors. We suggest that the same mechanism is likely to control the activity of other GPCRs, even though substitutions affecting the equivalent proline residues in rhodopsin, m3-muscarinic acetylcholine receptors, and C5a receptors reportedly do not cause a constitutive signal (Wess et al., 1993; Kaushal and Khorana, 1994; Kolakowski et al., 1995). These negative results may be due to the type of amino acid used to replace the proline residue in these receptors, reductions in receptor expression at the cell surface, or the action of RGS proteins, all of which strongly influence the apparent strength of the signal transduced by constitutively active receptors in yeast.
There are several ways, which are not mutually exclusive, whereby the conserved proline residue in TMS VI could control the activity of the α-factor receptor or other GPCRs. It could facilitate the initial folding of the receptor into its native, inactive conformation, stabilize the inactive conformation once it forms, and/or participate directly in the process of agonist-induced activation. It could control these processes by affecting the secondary and/or tertiary structure of the receptor. For example, the conserved proline residue may allow TMS VI to switch between kinked and more helical secondary structures, consistent with studies of artificial proline-containing transmembrane segments that suggest secondary structural changes of this kind could involve relatively modest changes in free energy (Polinsky et al., 1992). Alternatively, the conserved proline residue could allow TMS VI to adopt a relatively fixed secondary structure that favors formation of a native, inactive tertiary structure. Changing the secondary structure of TMS VI by substituting the proline with another amino acid could therefore destabilize the inactive tertiary structure of the receptor, leading to G protein activation.
These hypotheses are consistent with recent biochemical and biophysical studies of rhodopsin. In rhodopsin the region of helix F (TMS VI) containing the conserved proline residue (proline-267) is located near the β-ionone ring of retinal when the chromophore exists in the cis isomer (Nakanishi et al., 1995), which maintains the inactive conformation of rhodopsin. When rhodopsin is activated by light, the environment of tryptophan-265 in helix F changes (Lin and Sakmar, 1996) and a rigid-body motion of helix F relative to helix C (TMS III) appears to occur (Farrens et al., 1996).
The conserved proline residue of TMS VI is probably not the sole determinant governing GPCR activation. We have found that none of the substitutions of proline-258 appear to result in full constitutive activation. Instead, pheromone stimulation was needed to elicit a maximal signal. Mutations affecting domains other than TMS VI of various mammalian GPCRs also result in constitutive activation (Robinson et al., 1992; Parma et al., 1993; Robbins et al., 1993; Samama et al., 1993; Shenker et al., 1993). Thus, the activation process probably involve various subdomains of GPCRs.
GPCR Trafficking
Our results indicate that the conserved proline residue in TMS VI is required for efficient expression of α-factor receptors at the cell surface. Receptors bearing substitutions of proline-258 accumulate in intracellular compartments, achieving steady state levels similar to those of wild-type receptors expressed at the cell surface; however, it is possible that a small proportion of the mutant receptor population is targetted to the vacuole and degraded. Mutant receptors may accumulate in intracellular compartments because they are folded incompletely, although this remains to be established experimentally. Intracellular accumulation of mutant receptors apparently occurs by a mechanism that does not involve receptor internalization from the cell surface. Instead, mutant receptors accumulate mainly in post-ER compartments. Consistent with post-ER accumulation of receptors, loss of Cne1p, a calnexin homolog that is a component of the ER quality control machinery (Parlati et al., 1995), does not suppress the cell surface expression defects of constitutively active α-factor receptors (Stefan, unpublished results).
Various mutant forms of the α-factor receptor appear to have distinct targetting defects. Whereas constitutively active receptors accumulate in post-ER compartments without undergoing extensive degradation, temperature-sensitive receptors are targetted relatively efficiently to the vacuole and degraded (Jenness et al., 1997). Although the mechanisms responsible for achieving these different fates are unknown, there are several possibilities. For example, cells may possess two types of trafficking receptors, one that recognizes more grossly misfolded membrane proteins, such as temperature sensitive α-factor receptors, targetting them to the vacuole, and a second type of trafficking receptor that binds more completely folded membrane proteins, such as constitutively active α-factor receptors, preventing them from reaching the cell surface until folding is complete. In a second model, a single type of trafficking receptor recognizes relatively grossly misfolded membrane proteins and targets them to the vacuole, whereas more completely folded membrane proteins accumulate in the Golgi because they are not packaged or concentrated efficiently into secretory vesicles destined for the plasma membrane. In a third model, a single type of trafficking receptor or chaperone binds membrane proteins that are folded nearly normally, allowing them to be retained in post-ER compartments until folding is complete, whereas grossly defective membrane proteins are not bound and are targetted by default to the vacuole. Of these models, the latter is somewhat more consistent with the general view that protein targetting to the vacuole is the default pathway for defective membrane proteins (e.g., Chang and Fink, 1995; Jenness et al., 1997) or proteins that fail to be retained normally in the ER or Golgi (e.g., Roberts et al., 1992; Wilcox et al., 1992; an alternate interpretation is expressed by Rayner and Pelham, 1997). Regardless of the specific mechanisms involved, these quality control processes may ensure that wild-type pheromone receptors are retained intracellularly until they fold into their native, fully inactive conformations. This may prevent partially folded wild-type receptors, which may have some degree of constitutive activity, from reaching the cell surface and inappropriately triggering a signal in the absence of pheromone.
Similar quality control mechanisms may govern the trafficking and biogenesis of GPCRs in mammalian cells because normal biogenesis of certain mammalian GPCRs appears to require the conserved proline residue in TMS VI. For example, a leucine substitution of the conserved proline residue in TMS VI of human rhodopsin causes autosomal dominant retinitis pigmentosa (Fishman, et al., 1992), which can be caused by defects in rhodopsin biogenesis (Sung et al., 1993, 1994; Kaushal and Khorana, 1994; Colley et al., 1995). Similarly, substitutions affecting the equivalent residues in m3-muscarinic acetylcholine and C5a receptors cause defects in receptor expression at the cell surface (Wess et al., 1993, Kolakowski et al., 1995), although the effects of these mutations on the stability, endocytosis, or transit of these receptors through the secretory pathway have not been established. However, GPCR-targetting defects do not always result from substitutions affecting the conserved proline residue in TMS VI (Hong et al., 1997), suggesting that targetting defects can be receptor- and/or cell type-specific.
Precoupling of Pheromone Receptors and G Proteins?
We have found that coexpression of wild-type, but not G protein coupling-defective receptors, effectively suppresses the ability of constitutively active α-factor receptors to signal in the absence of agonist. Based on this finding, our current working hypothesis is that wild-type pheromone receptors associate with and sequester a limiting pool of G proteins. This “precoupling” model is consistent with pharmacological and biochemical evidence in mammalian systems (Neubig et al., 1988; Siciliano et al., 1990; Tian and Deth, 1993; Shi and Deth, 1994), whereas other potential mechanisms, such as receptor oligomerization, are less well substantiated biochemically. Precoupling of receptors and G proteins may enable cells to respond efficiently and rapidly to low levels of signal and/or facilitate signal propagation at specific sites on the cell surface. In yeast, precoupling could be important for sensing and responding chemotropically to pheromone gradients (Segall, 1993), as is thought to occur during mating partner discrimination (Jackson et al., 1991).
ACKNOWLEDGMENTS
We thank G.F. Sprague, Jr., for providing strains and plasmids, R. Kopan and M. Linder for 9E10 tissue culture supernatant, and D. Dutta for assistance with plasmid constructions. We thank J. Cooper, I. Herskowitz, M. Linder, and A. Muslin for comments on the manuscript. This work was supported by NIH grant GM-44592 (K.J.B.). K.J.B. is an Established Investigator of the American Heart Association.
REFERENCES
- Aris JP, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EK, Colley NJ, Zuker CS. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone P, Lin Z, Kempner E, Hirschberg CB. Regulation of yeast Golgi glycosylation. J Biol Chem. 1995;270:14564–14567. doi: 10.1074/jbc.270.24.14564. [DOI] [PubMed] [Google Scholar]
- Blumer KJ, Reneke JE, Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J Biol Chem. 1988;263:10836–10842. [PubMed] [Google Scholar]
- Boone C, Davis NG, Sprague GF., Jr Mutations that alter the third cytoplasmic loop of the a-factor receptor lead to a constitutive and hypersensitive phenotype. Proc Natl Acad Sci USA. 1993;90:9921–9925. doi: 10.1073/pnas.90.21.9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukusoglu G, Jenness DD. Agonist-specific conformational changes in the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1996;16:4818–4823. doi: 10.1128/mcb.16.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Fink GR. Targeting of the yeast plasma membrane [+H]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Clark CD, Palzkill T, Botstein D. Systematic mutagenesis of the yeast mating pheromone receptor third intracellular loop. J Biol Chem. 1994;269:8831–8841. [PubMed] [Google Scholar]
- Colley NJ, Baker EK, Stamnes MA, Zuker CS. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- Colley NJ, Cassill JA, Baker EK, Zuker CS. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc Natl Acad Sci USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin SR. Expanding horizons for receptors coupled to G proteins: diversity and disease. Curr Opin Cell Biol. 1994;6:191–197. doi: 10.1016/0955-0674(94)90135-x. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, genetic interaction and physical association with Gpa1 (G protein alpha subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- Farahbakhsh ZT, Hideg K, Hubbell WL. Photoactivated conformational changes in rhodopsin: a time-resolved spin label study. Science. 1993;262:1416–1419. doi: 10.1126/science.8248781. [DOI] [PubMed] [Google Scholar]
- Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- Ferreira PA, Nakayama TA, Pak WL, Travis GH. Cyclophilin-related protein ranbp2 acts as chaperone for red/green opsin. Nature. 1996;383:637–640. doi: 10.1038/383637a0. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fishman GA, Vandenburgh K, Stone EM, Gilbert LD, Alexander KR, Sheffield VC. Ocular findings associated with rhodopsin gene codon 267 and codon 190 mutations in dominant retinitis pigmentosa. Arch Ophthalmol. 1992;110:1582–1588. doi: 10.1001/archopht.1992.01080230082026. [DOI] [PubMed] [Google Scholar]
- Ganter UM, Charitopoulos T, Virmaux N, Siebert F. Conformational changes of cytosolic loops of bovine rhodopsin during the transition to metarhodopsin-II: an investigation by Fourier transform infrared difference spectroscopy. J Pharmacol Exp Ther. 1992;56:57–62. doi: 10.1111/j.1751-1097.1992.tb09602.x. [DOI] [PubMed] [Google Scholar]
- Grishin AV, Weiner JL, Blumer KJ. Control of adaptation to mating pheromone by G protein beta subunits of Saccharomyces cerevisiae. Genetics. 1994;138:1081–1092. doi: 10.1093/genetics/138.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Ryu K-S, Oh M-S, Ji I, Ji TH. Roles of transmembrane prolines and proline-induced kinks in the lutropin/choriogonadotropin receptor. J Biol Chem. 1997;272:4166–4171. doi: 10.1074/jbc.272.7.4166. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Konopka JB, Hartwell LH. S. cerevisiae alpha pheromone receptors activate a novel signal transduction pathway for mating partner discrimination. Cell. 1991;67:389–402. doi: 10.1016/0092-8674(91)90190-a. [DOI] [PubMed] [Google Scholar]
- Jenness DD, Li Y, Tipper C, Spatrick P. Elimination of of defective α-factor pheromone receptors. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- Kjelsberg MA, Cotecchia S, Ostrowski J, Caron MG, Lefkowitz RJ. Constitutive activation of the alpha 1B-adrenergic receptor by all amino acid substitutions at a single site. Evidence for a region which constrains receptor activation. J Biol Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakowski LF, Lu B, Gerard C, Gerard NP. Probing the message-address sites for chemoattractant binding to the C5a receptor - mutagenesis of hydrophilic and proline residues within the transmembrane segments. J Biol Chem. 1995;270:18077–18082. doi: 10.1074/jbc.270.30.18077. [DOI] [PubMed] [Google Scholar]
- Kölling R, Hollenberg CP. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka JB, Jenness DD, Hartwell LH. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- Konopka JB, Margarit SM, Dube P. Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc Natl Acad Sci USA. 1996;93:6764–6769. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Cotecchia S, Samama P, Costa T. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol Sci. 1993;14:303–307. doi: 10.1016/0165-6147(93)90048-O. [DOI] [PubMed] [Google Scholar]
- Lin SW, Sakmar TP. Specific tryptophan uv-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996;35:11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Zhang H, Lerro KA, Takekuma S, Yamamoto T, Lien TH, Sastry L, Baek DJ, Moquin PC, Boehm MF, Derguini F, Gawinowicz MA. Photoaffinity labeling of rhodopsin and bacteriorhodopsin. Biophys Chem. 1995;56:13–22. doi: 10.1016/0301-4622(95)00010-u. [DOI] [PubMed] [Google Scholar]
- Neubig RR, Gantzos RD, Thomsen WJ. Mechanism of agonist and antagonist binding to alpha 2-adrenergic receptors: evidence for a precoupled receptor-guanine nucleotide protein complex. Biochemistry. 1988;27:2374–2784. doi: 10.1021/bi00407a019. [DOI] [PubMed] [Google Scholar]
- Parlati F, Dominguez M, Bergeron JJM, Thomas DY. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functionas as a constituent of the ER quality control apparatus. J Biol Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- Parma J, Duprez L, Van SJ, Cochaux P, Gervy C, Mockel J, Dumont J, Vassart G. Somatic mutations in the thyrotropin receptor gene cause hyperfunctioning thyroid adenomas. Nature. 1993;365:649–651. doi: 10.1038/365649a0. [DOI] [PubMed] [Google Scholar]
- Polinsky A, Goodman M, Williams KA, Deber CM. Minimum energy conformations of proline-containing helices. Biopolymers. 1992;32:399–406. doi: 10.1002/bip.360320416. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Pelham HR. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneke JE, Blumer KJ, Courchesne WE, Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Roselli RL, Baack E, Mountjoy KG, Cone RD. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell. 1993;72:827–834. doi: 10.1016/0092-8674(93)90572-8. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PR, Cohen GB, Zhukovsky EA, Oprian DD. Constitutively active mutants of rhodopsin. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Benedetti H, Zanolari B, Riezman H. Identification of a novel sequence mediating regulated endocytosis of the G protein-coupled alpha-pheromone receptor in yeast. Mol Biol Cell. 1993;4:511–521. doi: 10.1091/mbc.4.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JR. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Samama P, Cotecchia S, Costa T, Lefkowitz RJ. A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J Biol Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- Schandel KA, Jennesss DD. Direct evidence for ligand-induced internalization of the yeast α-factor receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer A, Fanelli F, Costa T, De BP, Cotecchia S. Constitutively active mutants of the alpha 1B-adrenergic receptor: role of highly conserved polar amino acids in receptor activation. EMBO J. 1996;15:3566–3578. [PMC free article] [PubMed] [Google Scholar]
- Segall JE. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc Natl Acad Sci USA. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker A, Laue L, Kosugi S, Merendino JJ, Minegishi T, Cutler GJ. A constitutively activating mutation of the luteinizing hormone receptor in familial male precocious puberty. Nature. 1993;365:652–654. doi: 10.1038/365652a0. [DOI] [PubMed] [Google Scholar]
- Shi AG, Deth RC. Precoupling of alpha-2B adrenergic receptors and G-proteins in transfected PC-12 cell membranes: influence of pertussis toxin and a lysine-directed cross-linker. J Pharmacol Exp Ther. 1994;271:1520–1527. [PubMed] [Google Scholar]
- Siciliano SJ, Rollins TE, Springer MS. Interaction between the C5a receptor and Gi in both the membrane-bound and detergent-solubilized states. J Biol Chem. 1990;265:19568–19574. [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sprague GF, Jr, Thorner JW. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones EW, Pringle JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. J.R. Broach. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- Stefan CJ, Blumer KJ. The third cytoplasmic loop of a yeast G-protein-coupled receptor controls pathway activation, ligand discrimination, and receptor internalization. Mol Cell Biol. 1994;14:3339–3349. doi: 10.1128/mcb.14.5.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Davenport CM, Nathans J. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J Biol Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- Sung CH, Makino C, Baylor D, Nathans J. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian WN, Deth RC. Precoupling of Gi/G(o)-linked receptors and its allosteric regulation by monovalent cations. Life Sci. 1993;52:1899–1907. doi: 10.1016/0024-3205(93)90630-l. [DOI] [PubMed] [Google Scholar]
- Weiner JL, Guttierez SC, Blumer KJ. Disruption of receptor-G protein coupling in yeast promotes the function of an SST2-dependent adaptation pathway. J Biol Chem. 1993;268:8070–8077. [PubMed] [Google Scholar]
- Wess J, Nanavati S, Vogel Z, Maggio R. Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. EMBO J. 1993;12:331–338. doi: 10.1002/j.1460-2075.1993.tb05661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Hougan L, Dignard D, Thomas DY, Bell L, Saari GC, Grant FJ, O’Hara P, MacKay VL. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KA, Deber CM. Proline residues in transmembrane helices: structural or dynamic role? Biochemistry. 1991;30:8919–8923. doi: 10.1021/bi00101a001. [DOI] [PubMed] [Google Scholar]
- Yang K, Farrens DL, Altenbach C, Farahbakhsh ZT, Hubbell WL, Khorana HG. Structure and function in rhodopsin - cysteines 65 and 316 are in proximity in a rhodopsin mutant as indicated by disulfide formation and interactions between attached spin labels. Biochemistry. 1996;35:14040–14046. doi: 10.1021/bi962113u. [DOI] [PubMed] [Google Scholar]