Abstract
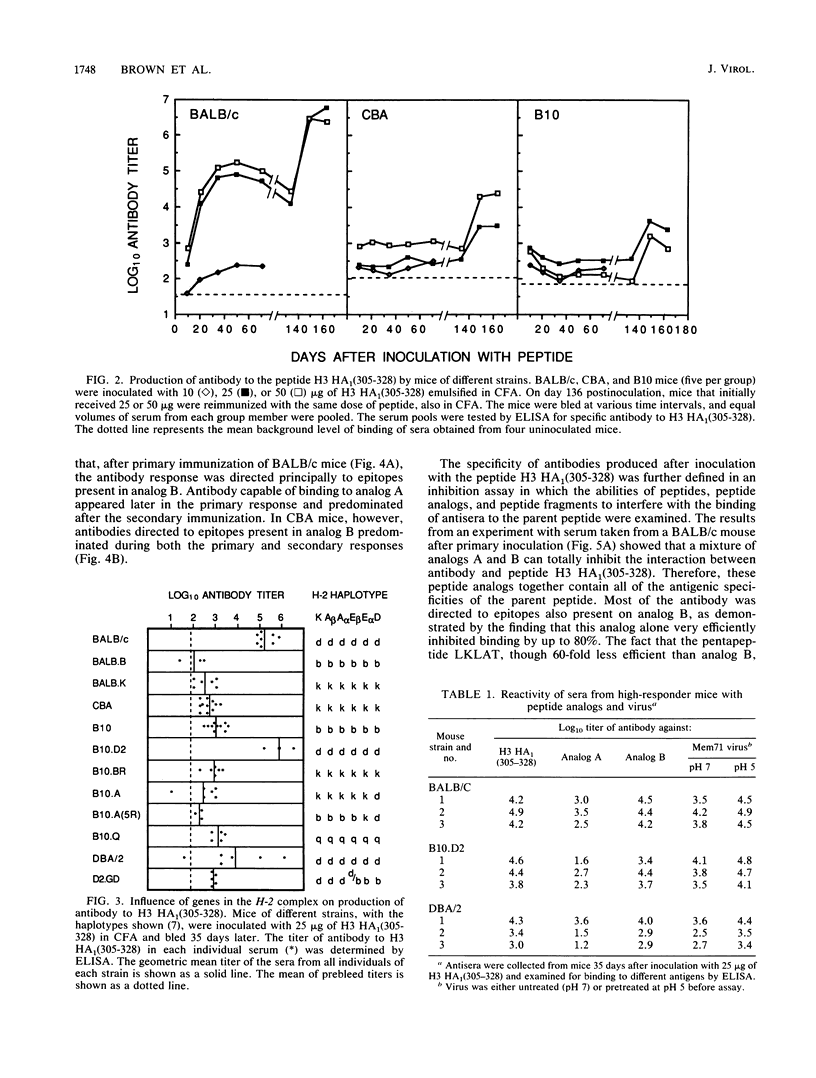
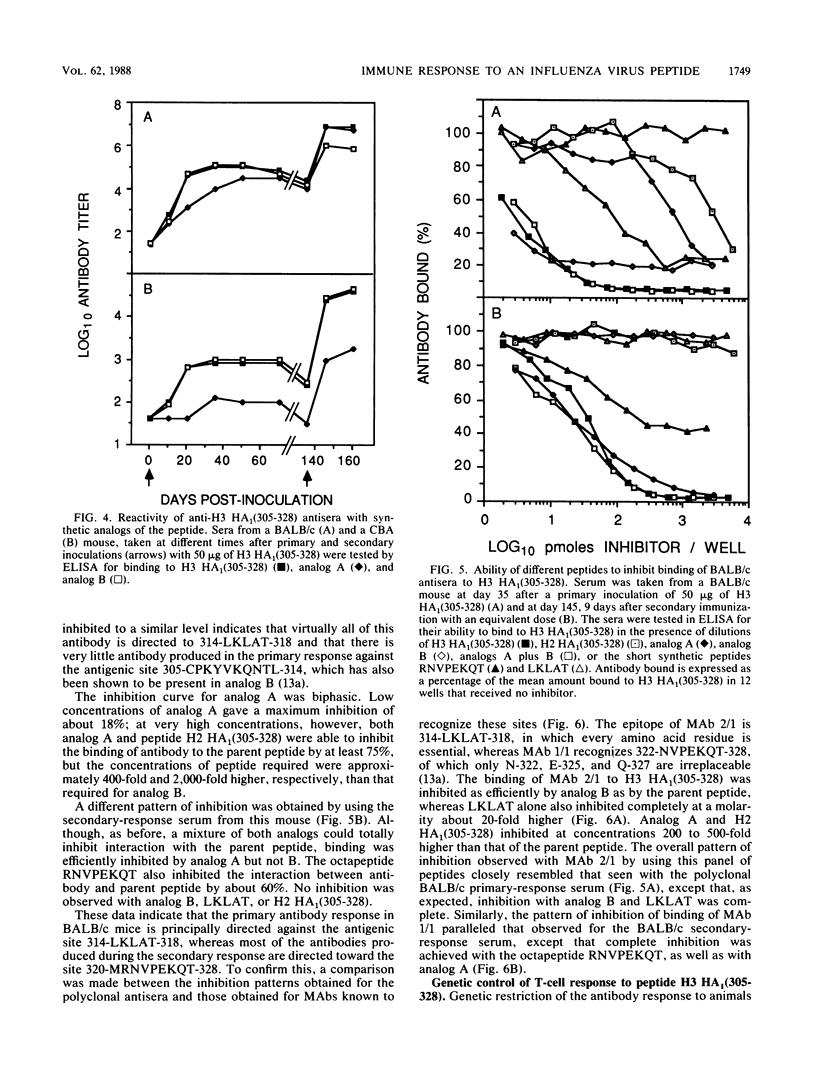
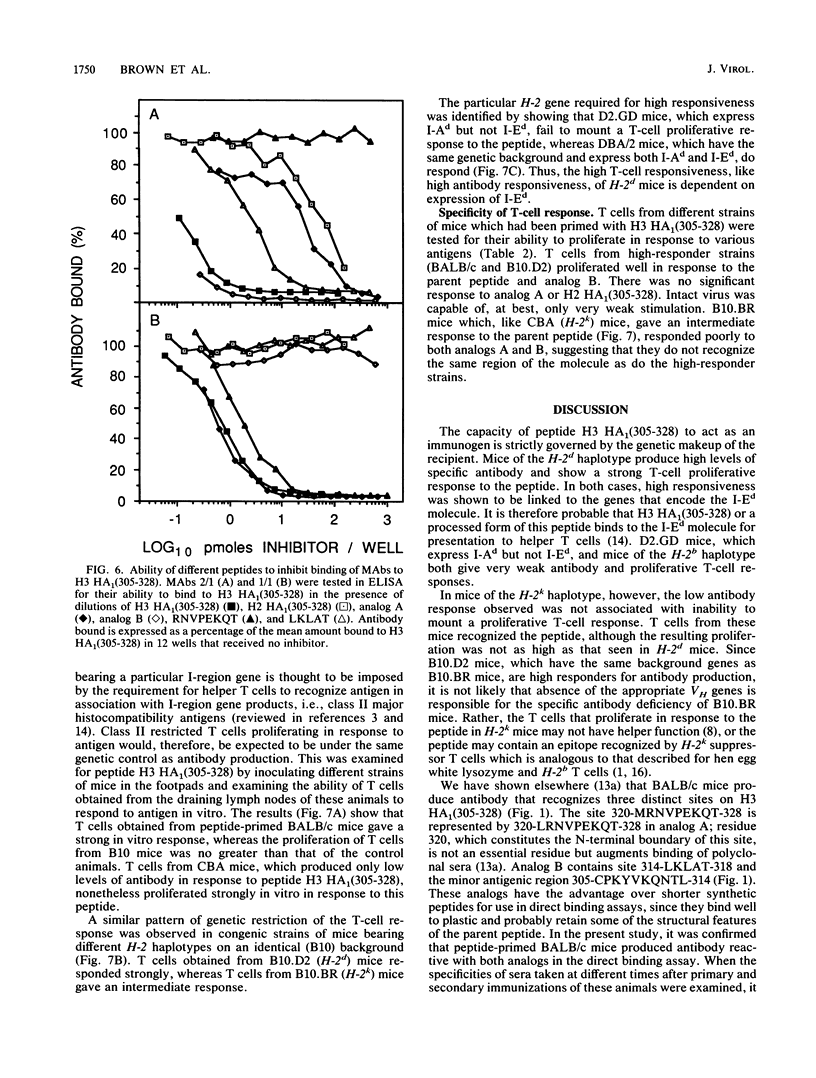
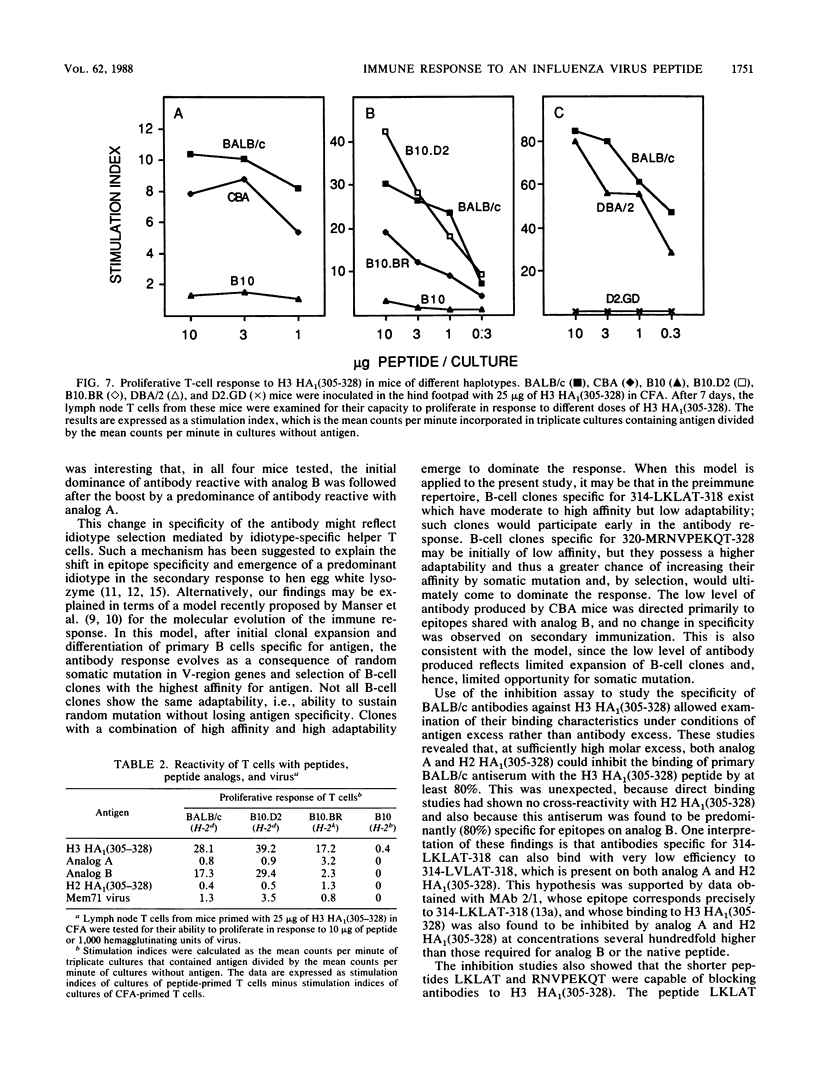
The immune response to a synthetic peptide, H3 HA1(305-328), representing the C'-terminal 24 amino acid residues of the HA1 chain of the hemagglutinin of the H3 subtype of influenza virus is controlled by genes in the I region of the major histocompatibility complex. Mice of the H-2d haplotype are high responders and produce antibody for several months after a single injection of peptide without carrier. Mice of the H-2b, H-2k, and H-2q haplotypes are low antibody responders. Investigation of recombinant and congenic mouse strains revealed that high responsiveness requires the genes that encode the I-Ed molecule. Immunoassays, involving direct binding to analogs of this peptide and inhibition by both these analogs and synthetic epitopes, were used to analyze the specificity of the polyclonal response. In BALB/c mice, the primary antibody response is directed principally against the antigenic site 314-LKLAT-318, whereas the secondary response after a boost is predominantly directed to a distinct site, 320-MRNVPEKQT-328. The T-cell response to the peptide H3 HA1(305-328), as measured by antigen-induced proliferation of primed T cells in vitro, is also I-Ed restricted in high-responder H-2d mice and is directed against an antigenic site that does not require the four C-terminal residues unique to the H3 influenza subtype. A different epitope appears to be recognized by T cells from CBA (H-2k) mice, which proliferate to a moderate extent on exposure to the peptide but, nevertheless, do not provide help for an antibody response.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adorini L., Harvey M. A., Miller A., Sercarz E. E. Fine specificity of regulatory T cells. II. Suppressor and helper T cells are induced by different regions of hen egg-white lysozyme in a genetically nonresponder mouse strain. J Exp Med. 1979 Aug 1;150(2):293–306. doi: 10.1084/jem.150.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. C., Tang X. L., Brown L. E., Murray J. M., White D. O., Tregear G. W. Antigenic determinants of influenza virus hemagglutinin. XII. the epitopes of a synthetic peptide representing the C-terminus of HA1. Virology. 1986 Dec;155(2):625–632. doi: 10.1016/0042-6822(86)90222-9. [DOI] [PubMed] [Google Scholar]
- Julius M. H., Simpson E., Herzenberg L. A. A rapid method for the isolation of functional thymus-derived murine lymphocytes. Eur J Immunol. 1973 Oct;3(10):645–649. doi: 10.1002/eji.1830031011. [DOI] [PubMed] [Google Scholar]
- Kida H., Brown L. E., Webster R. G. Biological activity of monoclonal antibodies to operationally defined antigenic regions on the hemagglutinin molecule of A/Seal/Massachusetts/1/80 (H7N7) influenza virus. Virology. 1982 Oct 15;122(1):38–47. doi: 10.1016/0042-6822(82)90375-0. [DOI] [PubMed] [Google Scholar]
- Klein J., Figueroa F., David C. S. H-2 haplotypes, genes and antigens: second listing. II. The H-2 complex. Immunogenetics. 1983;17(6):553–596. doi: 10.1007/BF00366126. [DOI] [PubMed] [Google Scholar]
- Krzych U., Fowler A. V., Miller A., Sercarz E. E. Repertoires of T cells directed against a large protein antigen, beta-galactosidase. I. Helper cells have a more restricted specificity repertoire than proliferative cells. J Immunol. 1982 Apr;128(4):1529–1534. [PubMed] [Google Scholar]
- Manser T., Wysocki L. J., Margolies M. N., Gefter M. L. Evolution of antibody variable region structure during the immune response. Immunol Rev. 1987 Apr;96:141–162. doi: 10.1111/j.1600-065x.1987.tb00513.x. [DOI] [PubMed] [Google Scholar]
- Metzger D. W., Furman A., Miller A., Sercarz E. E. Idiotypic repertoire of anti-hen eggwhite lysozyme antibodies probed with hybridomas. Selection after immunization of an IdX marker common to antibodies of distinct epitope specificity. J Exp Med. 1981 Sep 1;154(3):701–712. doi: 10.1084/jem.154.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D. W., Miller A., Sercarz E. E. Sharing of an idiotypic marker by monoclonal antibodies specific for distinct regions of hen lysozyme. Nature. 1980 Oct 9;287(5782):540–542. doi: 10.1038/287540a0. [DOI] [PubMed] [Google Scholar]
- Nestorowicz A., Tregear G. W., Southwell C. N., Martyn J., Murray J. M., White D. O., Jackson D. C. Antibodies elicited by influenza virus hemagglutinin fail to bind to synthetic peptides representing putative antigenic sites. Mol Immunol. 1985 Feb;22(2):145–154. doi: 10.1016/s0161-5890(85)80008-0. [DOI] [PubMed] [Google Scholar]
- Schoofs P. G., Geysen H. M., Jackson D. C., Brown L. E., Tang X. L., White D. O. Epitopes of an influenza viral peptide recognized by antibody at single amino acid resolution. J Immunol. 1988 Jan 15;140(2):611–616. [PubMed] [Google Scholar]
- Schwartz R. H. T-lymphocyte recognition of antigen in association with gene products of the major histocompatibility complex. Annu Rev Immunol. 1985;3:237–261. doi: 10.1146/annurev.iy.03.040185.001321. [DOI] [PubMed] [Google Scholar]
- Wicker L. S., Benjamin C. D., Miller A., Sercarz E. E. Immunodominant protein epitopes. II. The primary antibody response to hen egg white lysozyme requires and focuses upon a unique N-terminal epitope. Eur J Immunol. 1984 May;14(5):447–453. doi: 10.1002/eji.1830140512. [DOI] [PubMed] [Google Scholar]
- Wicker L. S., Katz M., Sercarz E. E., Miller A. Immunodominant protein epitopes. I. Induction of suppression to hen egg white lysozyme is obliterated by removal of the first three N-terminal amino acids. Eur J Immunol. 1984 May;14(5):442–447. doi: 10.1002/eji.1830140511. [DOI] [PubMed] [Google Scholar]


