Abstract
Cnm67p, a novel yeast protein, localizes to the microtubule organizing center, the spindle pole body (SPB). Deletion of CNM67 (YNL225c) frequently results in spindle misorientation and impaired nuclear migration, leading to the generation of bi- and multinucleated cells (40%). Electron microscopy indicated that CNM67 is required for proper formation of the SPB outer plaque, a structure that nucleates cytoplasmic (astral) microtubules. Interestingly, cytoplasmic microtubules that are essential for spindle orientation and nuclear migration are still present in cnm67Δ1 cells that lack a detectable outer plaque. These microtubules are attached to the SPB half- bridge throughout the cell cycle. This interaction presumably allows for low-efficiency nuclear migration and thus provides a rescue mechanism in the absence of a functional outer plaque. Although CNM67 is not strictly required for mitosis, it is essential for sporulation. Time-lapse microscopy of cnm67Δ1 cells with green fluorescent protein (GFP)-labeled nuclei indicated that CNM67 is dispensable for nuclear migration (congression) and nuclear fusion during conjugation. This is in agreement with previous data, indicating that cytoplasmic microtubules are organized by the half-bridge during mating.
INTRODUCTION
Faithful segregation of duplicated nuclei is crucial for the precise propagation of genetic material in all eukaryotic cells. In the budding yeast Saccharomyces cerevisiae, nuclear migration comprises two major steps during the mitotic cycle. During the first step, the nucleus moves from a random position in the mother cell to a site close to the bud neck. Insertion of the elongating and separating nucleus into the daughter cell during anaphase marks the second step (Yeh et al., 1995; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Nuclear migration and positioning of the mitotic spindle relative to the mother–daughter axis are strictly dependent on the dynamic action of cytoplasmic (astral) microtubules (Carminati and Stearns, 1997; Shaw et al., 1997). Tubulin mutants affecting predominantly cytoplasmic microtubules thus drastically impair orientation of the spindle and nuclear migration, leading to abnormal nuclear division within mother cells (Huffaker et al., 1988; Sullivan and Huffaker, 1992). As a result, these mother cells accumulate two or more nuclei, whereas daughter cells lack chromosomal DNA. Other proteins required for correct segregation of nuclei include the microtubule-based motor dynein as well as putative components of the dynactin complex and the actin cytoskeleton (Magdolen et al., 1988; Haarer et al., 1990; Palmer et al., 1992; Eshel et al., 1993; Li et al., 1993; Clark and Meyer, 1994; McMillan and Tatchell, 1994; Muhua et al., 1994; Yeh et al., 1995). Recent evidence suggests that several microtubule-based motors act in concert to achieve proper spindle positioning and nuclear migration (Cottingham and Hoyt, 1997; DeZwaan et al., 1997).
Polymerization of microtubules is generally nucleated by microtubule- organizing centers (MTOCs). Although they are morphologically diverse, MTOCs of different organisms serve similar functions in controlling the number, direction, and polarity of attached microtubules (Brinkley, 1985; Kellogg et al., 1994; Kalt and Schliwa, 1996; Pereira and Schiebel, 1997). The MTOC of S. cerevisiae is the spindle pole body (SPB), a multilayered organelle embedded in the nuclear envelope (Byers, 1981; Rose et al., 1993; Winey and Byers, 1993; Kilmartin, 1994; Snyder, 1994; Bullitt et al., 1997). A central electron-dense layer, the central plaque, lies in the plane of the nuclear membrane. On one side of the central plaque is the half-bridge, which early in G1 has a spherical structure called the satellite attached to its cytoplasmic side. This is probably the precursor of the nascent SPB. After duplication the two SPBs are connected by fused half-bridges (now forming the bridge). Three SPB substructures are directly involved in the binding of different sets of microtubules. Intranuclear microtubules are organized by the inner plaque throughout the cell cycle. Cytoplasmic microtubules are initiated by the half-bridge or bridge during the early part of the cell cycle before SPB duplication and continuing until SPB separation. After that and for the rest of the cell cycle, they attach to the outer plaque (Byers and Goetsch, 1975). The mechanism and cellular function of the change in attachment sites for cytoplasmic microtubules are currently not understood.
Nucleation of microtubules at the outer and inner plaque is probably mediated by a protein complex containing the 90-kDa SPB component Spc98p in addition to Tub4p and Spc97p (Rout and Kilmartin, 1990; Sobel and Snyder, 1995; Geissler et al., 1996; Marschall et al., 1996; Spang et al., 1996; Knop et al., 1997). Despite the presence of this complex at both the inner and outer plaque, it seems likely that there are differences in the protein composition of these plaques since electron microscopy reveals structural differences between them. The proteins responsible for this variance remain to be elucidated.
In addition to its essential mitotic functions in spindle formation, chromosome movement, and nuclear migration, the SPB also has key roles in conjugation and sporulation. During mating, the SPBs of fusing haploid cells are connected by cytoplasmic microtubules organized by half-bridges, and nuclear fusion appears to be initiated by fusion of the two SPBs (Byers and Goetsch, 1975). In sporulating yeast cells, spore wall formation is started by an enlargement of the outer plaque, which serves as a nucleation site for spore wall assembly (Moens and Rapport, 1971; Byers, 1981).
In this paper we describe the characterization of a novel gene that is important for spindle orientation and nuclear migration. We propose the name CNM67 (chaotic nuclear migration) since deletion of the gene results in a severe nuclear migration defect and because the gene product was also identified as a 67-kDa protein by mass spectrometric analysis of isolated yeast spindle poles (Jensen, unpublished data). Tagging of Cnm67p demonstrates its localization to the SPB region. We find by electron microscopy that the SPB outer plaque is considerably reduced in cnm67Δ1 cells, indicating a role for Cnm67p in outer plaque formation. Cytoplasmic microtubules are apparently nucleated by the half-bridge throughout the cell cycle, thus providing a rescue pathway for spindle orientation and nuclear migration that does not depend on outer plaque-bound microtubules.
MATERIALS AND METHODS
Strains, Media, and Yeast Transformation
Yeast strains that were used in this study are summarized in Table 2. Yeast media were prepared as described by Guthrie and Fink (1991). The yeast transformation procedure was based on the protocol by Schiestl and Gietz (1989). After the heat shock step, cells were pelleted and resuspended in 5 ml of YPD and incubated for 2 h at 30°C. Cells were again pelleted, resuspended in 1 ml dH2O, and plated on selective YPD-G418 medium (200 mg G418/l). The Escherichia coli strain XL1-blue (Bullock et al., 1987) was used to propagate plasmids.
Table 2.
Yeast strains
| Name | Genotype | Source or reference |
|---|---|---|
| FY1679 | MATα/a ura3-52/ura3-52 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | B. Dujon, diploid from cross FY23 × FY73 (F. Winston) |
| CEN.PK2 | MATα/a ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | K.D. Entian |
| ABY99 | MATα cnm67Δ1::kanMX4 ura3-52 trp1-289 leu2-3,112 his3Δ1 | Brachat et al.a |
| ABY101 | MATα/a cnm67Δ1::kanMX4/CNM67 ura3-52/ura3-52 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | Brachat et al.a |
| ABY102 | MATα cnm67Δ1::kanMX4 ura3-52 TRP1 leu2Δ1 his3Δ200 | Brachat et al.a |
| ABY103 | MATa cnm67Δ1::kanMX4 ura3-52 trp1Δ63 leu2Δ1 HIS3 | Brachat et al.a |
| ABY104 | MATa cnm67Δ1::kanMX4 ura3-52 trp1Δ63 leu2Δ1 his3Δ200 | Brachat et al.a |
| ABY105 | MATα/a cnm67Δ1::kanMX4/CNM67 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3, 112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| ABY106 | MATa cnm67Δ1::kanMX4 ura3-52 trp1Δ63 leu2Δ1 his3Δ200 {pYCGNL225c}b | This study |
| ABY107 | MATα/a cnm67Δ1::kanMX4/cnm67Δ1::kanMX4 ura3-52/ura3-52 trp1Δ63/TRP1 leu2Δ1/leu2Δ1 his3Δ200/HIS3 | This study |
| ABY108 | MATα/a cnm67Δ1::kanMX4/cnm67Δ1::HIS3MX6 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| ABY109 | MATα/a CNM67::GFP-kanMX6/CNM67 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| ABY110 | MATα CNM67::GFP-kanMX6 ura3-52 trp1-289 leu2-3,112 his3Δ1 | This study |
| ABY111 | MATa CNM67::GFP-kanMX6 ura3-52 trp1-289 leu2-3,112 his3Δ1 | This study |
| ABY112 | MATα/a CNM67::GFP-kanMX6/CNM67::GFP-kanMX6 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| ABY132 | MATα/a CNM67::HA-kanMX6/CNM67::HA-kanMX6 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| DHY2 | MATα cnm67Δ1::kanMX4 HHF2::GFP-kanMX6 ura3-52 TRP1 leu2Δ1 his3Δ200 | This study |
| DHY3 | MATa cnm67Δ1::kanMX4 HHF2::GFP-kanMX6 ura3-52 trp1Δ63 LEU2 HIS3 | This study |
| ABY134 | MATα HHF2::GFP-kanMX6 ura3-52 trp1Δ63 HIS3 LEU2 | This study |
| ABY135 | MATa HHF2::GFP-kanMX6 ura3-52 TRP1 leu2Δ1 his3Δ200 | This study |
| 5105 | MATα/a act1-4/act1-4 ura3/ura3 LEU2/leu2 HIS3/his3 | Palmer et al., 1992 |
| DBY2023 | MATa tub2-401 ura3-52 his4-539 lys2-801 | Huffaker et al., 1988 |
| DBY1384 | MATa tub2-104 ura3-52 his4-539 | Thomas et al., 1985 |
| YJC1372 | MATα/a act5Δ1::HIS3/act5Δ1::HIS3 ura3-52/ura3-52 trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 lys2-801/lys2-801 ade2-101/ade2-101 | J. Cooper (Washington University) |
Manuscript in preparation.
pYCGNL225c is a pRS416 (Sikorski and Hieter, 1989) derivative containing CNM67 with promoter and terminator sequences.
DNA Manipulations and Strain Constructions
Standard DNA manipulations were performed as described by Sambrook et al. (1989). We applied a PCR-based method to construct gene deletion cassettes that were used in yeast transformations (McElver and Weber, 1992; Baudin et al., 1993, Wach et al., 1994). DNA of E. coli plasmids pFA6-kanMX4 (Wach et al., 1994) or pFA6-HIS3 MX6 (Wach et al., 1997) served as template for preparative PCR reactions. Oligonucleotides 5′-GGCACTAGTATGCTTGATCCGTAAATTTCTTTAGATTCATTCATCGATGAATTCGAGCTC-3′ and 5′-GCGCAGCTGATTTCGATTTAATGAATTTTCCATTTCATGAGCCGTACGCTGCAGGTCGAC-3′ were used as primers to produce CNM67 gene deletion cassettes that replaced codons 14–537 of the gene upon integration into the genome. During a systematic analysis of novel gene functions, we constructed several GFP fusions to the 3′-ends of open-reading frames (ORFs) of unknown function (Brachat, Duesterhoeft, Moestl, Rebischung, Wach, and Philippsen, unpublished data). For amplification of GFP gene fusion constructs, the template DNA was pFA6-GFP-kanMX6 (Wach et al., 1997). We utilized oligonucleotide primers that introduced short flanking regions of homology to the gene’s 3′-end. The PCR product was then integrated into the genome at the 3′-end of the gene by homologous recombination, creating a fusion gene that excluded the gene’s stop codon. The fusion gene was thus expressed from the original promoter, minimizing the risk of non–wild-type expression levels. Primers 5′-CTGGACCATCTGTATGATCATATCCTGGAGAAGATGGTGAAGGGTCGACGGATCCCCGGG-3′ and 5′-TATACATACTTCCTAGAATATAATTTAATCTTATACCTTAACATCGATGAATTCGAGCTCG-3′ were used for generation of GFP-kanMX6 with flanking homology to the 3′-end of CNM67. Correct genomic integration of the corresponding construct was verified by analytical PCR (Huxley et al., 1990; Wach et al., 1994). To facilitate the production of 3HA epitope fusions to protein C termini, a pFA-3HA-kanMX6 plasmid was constructed. The 3HA antigen was amplified by PCR with primers 5′-GCGTTAATTAACTACCCATACGATGTTCCT-3′ and 5′-CCGGGCGCGCCGCACTGAGCAGCGTAATC-3′ and plasmid pSKIIHA1 (provided by B. Futcher) as template. The PCR product was cleaved with AscI and PacI and cloned into pFA6-GFP-kanMX6 from which the GFP sequence had been released by AscI and PacI digestion. The resulting plasmid was then used as template for generation of a 3HA-kanMX6 gene fusion cassette to tag CNM67. Primers for this reaction were the same as for production of the GFP fusion cassette.
Heterozygous diploid strains expressing the CNM67-GFP or CNM67–3HA fusion genes were sporulated and analyzed by tetrad dissection. Cells carrying only the gene fusion allele and no wild-type copy of CNM67 grew at the same rate as the corresponding wild-type cells. Microscopic observation of DAPI-stained cells also did not reveal any morphological or nuclear migration defects caused by the Cnm67-GFPp or Cnm67-HAp fusion protein. We used strain ABY112 to characterize the Cnm67-GFPp distribution pattern and strain ABY132 for the Cnm67-HAp pattern.
C-terminal fusion of GFPp to Hhf2p was performed as described by Wach et al. (1997) and caused a slight reduction in growth rates of haploid strains compared with the wild-type, but this was considered as acceptable since both strains that were to be compared in mating experiments carried the same Hhf2-GFPp label. Thus, defects visible in labeled cnm67Δ1 but not in labeled CNM67 cells should be specifically caused by the cnm67Δ1 mutation. Hhf2-GFPp fluorescence produced a bright nuclear staining that proved to be very suitable for in vivo studies. The major advantage of this label was the fact that very short excitation times (0.12 s) could be used to obtain strong GFP fluorescence. This, in turn, drastically reduced light-induced damage to the cells and allowed for observation of cells over many generations.
Yeast strains were grown on YPD-geneticin (200 μg geneticin/ml) to select for transformants that had integrated kanMX4-, GFP-kanMX6-, or 3HA-kanMX6-derived constructs. Growth on SD plates lacking histidine selected for HIS3 MX6 integration.
We first produced a diploid homozygous cnm67Δ1 strain by crossing haploid cnm67Δ1 strains ABY102 and ABY103 with selection on SD medium lacking tryptophan and histidine. To obtain independent evidence for the sporulation deficiency of cnm67Δ1 mutants in a second strain background, we constructed a diploid CEN.PK2 derivative with two deleted CNM67 alleles by two successive PCR-based gene deletions. The first allele was deleted by integration of a kanMX4 marker cassette at the CNM67 locus. The resulting heterozygous strain was then used for transformation with a second PCR-amplified HIS3 MX6 marker cassette that carried the respective CNM67 homology regions and rendered transformed cells His+. Colonies that were His+ and also still G418 (geneticin) resistant were subjected to analytical PCR for verification of both deletion alleles (ABY108). Positive clones were then incubated under sporulation conditions.
For complementation analysis of a cnm67Δ1 strain, we subcloned a 2.4 kb HindIII restriction fragment from a chromosome XIV cosmid clone into the HindIII site of pRS 416 (Sikorski and Hieter, 1989), a single-copy vector with the URA3 gene as selectable marker. The subclone plasmid (pYCGNL225c) included the complete CNM67 coding sequence plus 218 base pairs (bp) upstream of the CNM67 start codon and 361 bp downstream of the stop codon. The CNM67 3′-region included the first 23 N-terminal codons of ORF YNL227c, whereas the 5′-region did not include any predicted coding sequences. After transformation of haploid strain (ABY104) with the plasmid, we compared growth of the resulting Ura+ strain (ABY106) and the corresponding wild-type strain on YPD at 30°C (Figure 1D).
Figure 1.
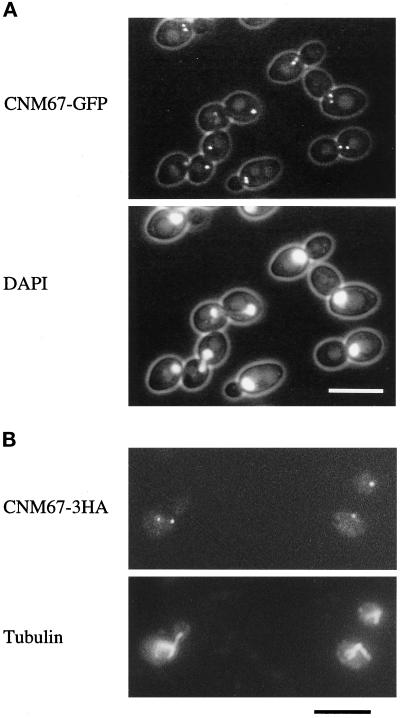
Subcellular localization of functional Cnm67p fusion proteins visualized by fluorescence microscopy. (A) Cnm67-GFPp fluorescence is visible at one or two spots at the nuclear periphery, consistent with a localization to the SPB. Nuclear DNA was stained with DAPI. (B) Immunofluorescence microscopy of Cnm67–3HAp–labeled cells. Dot-shaped staining with anti-HA antibody coincides with the spindle poles visualized by tubulin staining with anti-tubulin antibody.
Test for Benomyl Sensitivity and Synthetic Lethality
Sensitivity to benomyl on solid YPD was assayed as described by Interthal et al. (1995). Cells from exponentially growing CEN.PK2 and ABY108 cultures were spotted as serial dilutions onto plates containing increasing concentrations of benomyl (5–50 μg/ml). Plates were incubated at 30°C.
Double-mutant strains were constructed by CNM67 replacement in the corresponding mutant strain background by a PCR-generated kanMX4 cassette as described above. Nuclear migration mutant strains were DBY2023 (tub2–401), DBY1384 (tub2–104), YJC1371 (act5Δ/act5 Δ), and 5105 (act1–4/act1–4). If the original nuclear migration mutant was temperature sensitive, we incubated the double mutant at a temperature very close to the restrictive temperature to test for a synthetic aggravation of defects by the cnm67Δ,mdul1 deletion. To test for synthetic lethality, we incubated the tub2–401, cnm67Δ1 strain at 20°C, a temperature at which the cold- sensitive tub2–401 strain was barely able to grow (Sullivan and Huffaker, 1992). tub2–104 (Thomas et al., 1985; Huffaker et al., 1988) was also tested for synthetic lethality with cnm67Δ1 at the permissive temperature of 16°C. We also introduced the cnm67Δ1 deletion in a homozygous act1–4 (Palmer et al., 1992) background and sporulated the resulting heterozygous cnm67Δ1/CNM67, act1–4/act1–4 strain. Incubation of derived haploid double mutants was on YPD medium at 30°C or 33°C. A cnm67Δ1/act5Δ double-mutant strain was also constructed by CNM67 deletion in a homozygous diploid act5Δ strain (Muhua et al., 1994) and subsequent sporulation followed by tetrad analysis and incubation on YPD at 30°C.
Fluorescence Microscopy and Electron Microscopy (EM)
For DAPI staining, yeast cells were grown in YPD to early log phase. Approximately 5 × 107 cells were pelleted, resuspended in 50 μl dH2O, and fixed for 5 min by addition of 1 ml 70% ethanol before DAPI staining. One microliter of a 1 mg/ml DAPI stock solution was added, and the suspension was incubated for 5 min at room temperature. Cells were subsequently washed twice in dH2O and mounted on a poly-L-lysine-treated slide for microscopy. If DAPI staining was performed for quantification of nuclear distribution, cells were first digested with Zymolyase 100T (250 μg/ml in 40 mM KH2PO4, pH 6.5, 1.2 M sorbitol) for 1 h at 37°C. Four microliters of the 1 mg/ml DAPI stock solution were directly added per 1 ml of the growth medium if DAPI and GFP fluorescence were to be observed in the same cells.
of strains ABY108, ABY132, and CEN.PK2 was performed essentially as described by Kilmartin et al. (1993). Fixation was for 20 min for tubulin staining and 1.5 min for hemagglutinin antigen (HA)-tubulin double staining. Cells were incubated with either rabbit anti-yeast tubulin immunoglobulin G and/or with mouse monoclonal 12CA5 anti-HA antibody overnight at 4°C. Anti-tubulin primary antibody was detected with Texas-Red–labeled goat anti-rabbit immunoglobulin G. Anti-HA–treated cells were further incubated with fluorescein-labeled anti-mouse antibody.
Fixation and embedding of mutant and wild-type cells for serial thin section electron microscopy was performed as described by Byers and Goetsch (1991) with slight modifications (Goh and Kilmartin, 1993). CEN.PK2 was used as the wild-type control and ABY108 as cnm67Δ1 mutant strain for EM analysis.
In Vivo Time-Lapse Microscopy
Mutant or wild-type Hhf2-GFPp labeled a and α strains (DHY2, DHY3, ABY134, ABY135) were grown overnight in YPD medium to early logarithmic phase. Approximately 1 × 105 cells of the corresponding a and α strains were mixed in a glass reaction tube and incubated without shaking for 1.5 h at 30°C. One milliliter of that culture was then transferred to a 1.5-ml reaction tube and centrifuged for 5 s to sediment the cells. Most of the supernatant was discarded, leaving approximately 20 μl of cell suspension in the tube. Three microliters of this suspension were then placed on top of the agarose surface of a ground well microscopy slide (Huber & Co., Reinach, Switzerland). This had been prepared by placing a drop of growth medium (SD complete medium plus 1.7% agarose) onto the slide and covering it with a coverslip to give a planar agarose surface. The coverslip was then removed, and cells were placed on top and covered by another coverslip that was sealed with nail hardener to allow for long investigation times without liquid loss. GFP fluorescence could then be followed for more than 10 h without significant loss of intensity when very short excitation times were used and excitation was repeated only every 3 or every 5 min. Fluorescence pictures were captured and stored electronically. Fluorescence excitation was controlled by a shutter controller in combination with a MAC2000 shutter system (Ludl controller MAC 2000, Ludl Electronics, Hawthorne, NY). We used a VI-470 video camera and controller (Optronics Engineering, Goleta, CA) for picture taking.
Computer Programs and Hardware
We used the following computer programs for sequence analysis: FASTA (Pearson and Lipman, 1988) (word size = 2), PERCOIL (Berger et al., 1995) with default parameters (probability cutoff = 0.5), ISOELECTRIC (Genetics Computer Group program package [1991]). Image acquisition and processing for fluorescence microscopy was performed on a Power Macintosh 7600/120 computer using the public domain NIH image 1.60 program (developed by Wayne Rasband at the US National Institutes of Health and available from the Internet by anonymous FTP from zippy.nimh.nih.gov or on floppy disk from the National Technical Information Service, Springfield, VA, part number PB95–500195GEI). Picture files were eventually contrast enhanced using Photoshop 4.0 (Adobe Systems Europe, Edinburgh, Scotland). Custom macros were used to control the microscope, the video camera, the MAC2000, and a LG-3 framegrabber (Scion, Frederick, MD).
RESULTS
CNM67 Encodes a Novel Putative Coiled-Coil Protein That Localizes to the SPB
Originally, CNM67 (YNL225c) was identified as one of the novel ORFs on chromosome XIV of S. cerevisiae during systematic sequencing (Philippsen et al., 1997). To analyze the subcellular localization of gene products of such novel ORFs, we used PCR targeting (Wach et al., 1997) to construct genomic GFP fusions that were expressed from the original ORF promoters.
In vivo fluorescence microscopy detected Cnm67-GFPp staining as either one or two intense spots at the nuclear periphery (Figure 1A), suggesting localization to the SPB region. Unbudded cells usually contained one spot, whereas cells with intermediate sized buds showed two spots close to the DAPI- stained region of the nucleus. The maximal distance between them was never greater than the diameter of a nucleus. Large budded cells with elongating nuclei contained one spot at either pole of the DAPI-stained region, consistent with a localization to the poles of an anaphase B spindle. This result was confirmed by an independent experiment in which we constructed a CNM67-3HA fusion gene. Cnm67–3HAp cells were double stained with anti-HA and anti-tubulin antibodies (Figure 1B). HA staining was very similar to the Cnm67-GFPp fluorescence pattern and was always found at the poles of mitotic spindles. Cnm67-GFPp and Cnm67–3HAp fusion proteins were functional because corresponding cells carrying the fusion allele and lacking the wild-type allele showed none of the phenotypes associated with CNM67 deletion (see below). Thus, the observed GFP- or immunofluorescence pattern of these constructs should reflect the wild-type localization of Cnm67p, and we conclude that Cnm67p is either a central component of the yeast SPB or closely associated to the SPB.
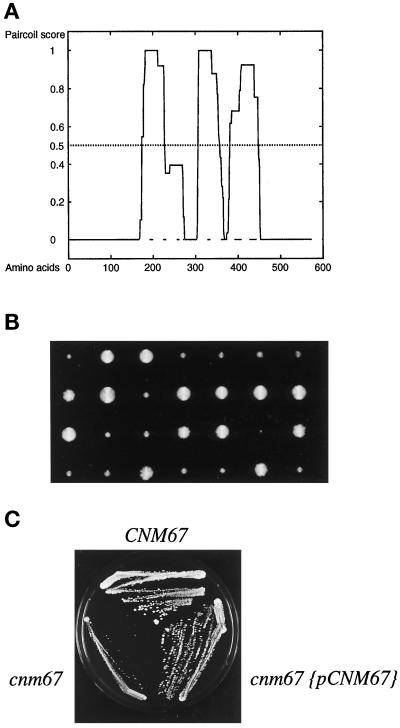
Analysis of the presumptive 581-amino acid sequence of Cnm67p with the Paircoil program (Berger et al., 1995) suggests the presence of three separate coiled-coil–forming regions (see Figure 2A). Cnm67p may therefore stabilize a homo-oligomer or a complex with other proteins via a central coiled-coil region.
Figure 2.
Cnm67p coiled-coil prediction, tetrad analysis, and complementation test. (A) Probability plot for coiled-coil formation of the 581 amino-acid sequence of Cnm67p. The central part of the protein was predicted to contain three α helical coiled-coil regions (residues 174–226, p = 1; residues 306–357, p = 1; residues 382–448, p = 0.924) by the Paircoil program (Berger et al., 1995). The CNM67 sequence is available from EMBL/GenBank/DDBJ under accession number Z71501. (B) Tetrad dissection of a heterozygous CNM67-deleted strain (ABY101). Tetrad colonies were transferred on G418 containing medium to test for segregation of the kanMX4 gene deletion marker. Slow growing colonies always carried the cnm67::kanMX4 allele (cnm67Δ1). (C) Complementation of the cnm67Δ1 growth defect by transformation with a plasmid carrying the CNM67 wild-type allele.
Comparison of the predicted Cnm67p amino acid sequence with sequences in the EMBL and PIR databases did not detect proteins of high homology. The highest FASTA (Pearson and Lipman, 1988) scores were found for other coiled-coil proteins and probably only reflected a structural similarity of the respective coiled-coil regions.
The Role of CNM67 in Nuclear Migration and Sporulation
A complete deletion of CNM67 was found to be viable but resulted in a slow-growth phenotype (Figure 2B). When CNM67-deleted cells were stained with DAPI, a severe nuclear migration defect was observed (Figure 3). Table 1 summarizes the frequency of abnormal distribution of nuclei in ABY108, a homozygous diploid cnm67Δ1 deletion strain. Many mother cells (26%) contained two separated chromosome masses, indicating a completion of nuclear division without migrating the daughter nucleus to the bud; 14% of the cells even showed three to eight nuclei and about 5% carried no nucleus. Some of the bi- or multinucleated cells also displayed morphological defects. Although a substantial portion of cells showed pronounced defects, we also observed many cells (56%) with a single nucleus, suggesting the presence of a protein with an overlapping function to that of Cnm67p or correct alignment of the nuclei occurring by chance. The slow growth behavior, nuclear migration defect, and impaired morphogenesis were found for haploid and homozygous diploid cnm67Δ1 cells derived from two different genetic backgrounds (ABY99, ABY102, ABY103, ABY104, ABY107, ABY108 in Table 2). Time-lapse analysis of cnm67Δ1 cells with GFP-labeled nuclei by fluorescence microscopy suggested that misorientation of the mitotic spindle was the primary defect whereas morphological abnormalities were later consequences of CNM67 loss (Hoepfner, personal communication).
Figure 3.
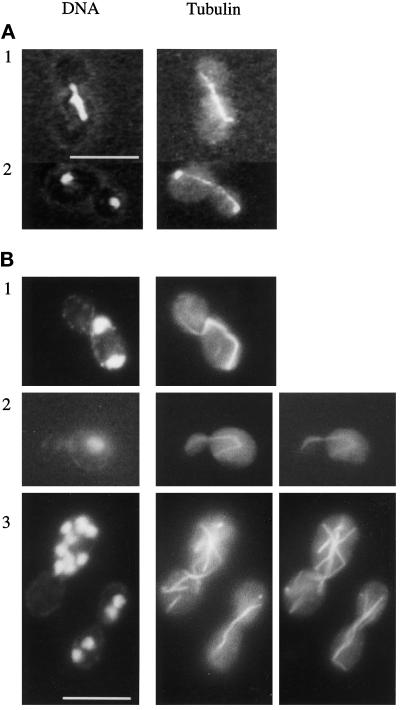
Nuclear distribution and microtubule structure in wild-type and cnm67Δ1. Cells were double stained for nuclear DNA with DAPI and for tubulin with anti-tubulin antibody. (A) Wild-type. (B) cnm67Δ1 cells. Panels B2 and B3 show different focal planes of the respective cells to visualize the complete microtubular structures. B1 and B2 show cytoplasmic microtubules that direct from one spindle pole into the bud. The spindle in B1 is highly elongated although it is restricted to the mother cell. Panel B3 depicts multinucleated cells. Bars, 10 μm.
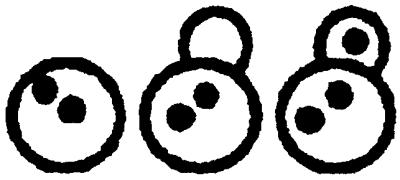
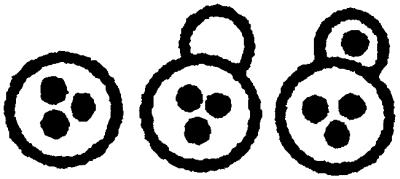
Table 1.
Nuclear migration in diploid cnm67Δ1 (ABY108)- and wild-type (CEN.PK2) strains
| Nuclear migration phenotype
|
||||||
|---|---|---|---|---|---|---|
 |
 |
 |
 |
 |
||
| Yeast strain | 0 Nucleus per cell | 1 Nucleus per cell | 2 Nuclei per cell | 3 Nuclei per cell | >3 Nuclei per cell | |
| 16°C | CNM67 | 0 % | 99.6% | 0.4% | 0 % | 0 % |
| cnm67Δ1 | 4.4% | 57.5% | 25.3% | 7 % | 5.8% | |
| 30°C | CNM67 | 0 % | 98 % | 2 % | 0 % | 0 % |
| cnm67Δ1 | 4.9% | 55.6% | 25.7% | 6.6% | 7.2% | |
| 37°C | CNM67 | 0.1% | 99.5% | 0.4% | 0 % | 0 % |
| cnm67Δ1 | 8.3% | 52.3% | 32.2% | 5.7% | 1.5% | |
The number of nuclei per cell in diploid yeast strains was determined by DAPI staining of asynchronous cultures in exponential growth phase. Greater than 500 cells were counted for each strain. Cells of different morphologies were grouped according to their nuclear distribution as indicated at the top.
To test for complementation of the observed defects, we transformed cnm67Δ1 cells with a plasmid containing the CNM67 wild-type allele. The resulting transformants grew at the same rate as the corresponding wild-type strain (Figure 2C), and fluorescence microscopy of DAPI-stained cells showed wild-type distribution of nuclei. We conclude that the CNM67 clone fully complemented the cnm67Δ1 defects, and thus the observed defects in cnm67Δ1 strains were due to deletion of the CNM67 gene.
We also investigated the sporulation efficiency of a homozygous diploid cnm67Δ1 strain. The wild-type control strain (CEN.PK2) sporulated efficiently (70% four spored asci after 5 d on sporulation medium) whereas we did not observe asci with spores among more than 400 cnm67Δ1 cells (ABY108). Even after several weeks on sporulation plates, no asci were visible, indicating that the CNM67 gene is essential for sporulation. This result was further confirmed with another homozygous cnm67Δ1 deletion strain of different genetic background (ABY107).
Characterization of the Microtubule Cytoskeleton of cnm67Δ1 Cells
Immunofluorescence staining of tubulin in cnm67Δ1 cells revealed several defects in microtubule structure (Figure 3). In large budded cells, spindle microtubules that connected two chromosome masses were frequently restricted to the mother cell without penetrating the bud neck (Figure 3B1). Among these cells some had aligned the spindle along the mother–bud axis, whereas in others the spindles were oriented randomly relative to this axis. Many cells clearly showed cytoplasmic microtubules directing into the bud (Figure 3B, 1 and 2). Multinucleated cells frequently contained spindles that crossed over one another (Figure 3B3), indicating independent orientation of spindles relative to each other within one cell. Elongation of spindles usually appeared to be synchronized in cells that contained more than one nucleus, suggesting that the coupling of spindle dynamics to the general control of cell cycle progression was still functional (Adams and Pringle, 1984; Kilmartin and Adams, 1984). The length of some spindles drastically exceeded the diameter of the mother cell, leading to bent spindles (Figure 3B1), which was probably due to a continued spindle elongation without accompanying migration of the spindle toward the daughter cell. This could also be seen in mother cells containing a spindle that was oriented along the mother–bud axis, suggesting that pushing forces of an elongating spindle were not sufficient to drive effective penetration of the spindle through the bud neck. In other cnm67Δ1 cells, several DAPI- stained regions appeared to be connected by microtubules. Here, we could not definitely distinguish between a cytoplasmic or nuclear origin of these microtubular structures. A clear quantitative description of frequencies of different microtubular structures in cnm67Δ1 cells was made impossible by the extreme heterogeneity of spindle morphologies and numbers. We suppose that the observed defect in nuclear migration is a consequence of spindle misorientation in combination with inefficient pulling of the spindle through the bud neck.
Test for Benomyl Sensitivity and Synthetic Lethality with Nuclear Migration Mutants
Mutations in tubulin-interacting proteins or in tubulin itself often cause an altered sensitivity to the microtubule-destabilizing drug benomyl (Thomas et al., 1985; Stearns et al., 1990; Solomon, 1991; Interthal et al., 1995). Since Cnm67p is involved in the microtubule-dependent process of nuclear migration, we tested the benomyl tolerance of a cnm67Δ1 strain by spotting serial dilutions of cells onto plates with increasing concentrations of benomyl. We did not observe any significant difference between the sensitivity of the cnm67Δ1 strain and the isogenic wild-type strain. Growth of both strains on solid YPD medium at 30°C was completely inhibited at benomyl concentrations >20 μg/ml.
A number of mutants that influence the fidelity of spindle orientation and nuclear movement during mitosis are known. To search for genetic interactions of CNM67, we tested synthetic lethality of the CNM67 deletion with the nuclear migration mutants tub2–401, tub2–104, act5, or act1–4 (Huffaker et al., 1988; Sullivan and Huffaker, 1992; Clark and Meyer, 1994; Muhua et al., 1994; Palmer et al., 1992). None of the tested double mutants resulted in a synthetic lethal phenotype.
SPB Ultrastructure of cnm67Δ1 Cells
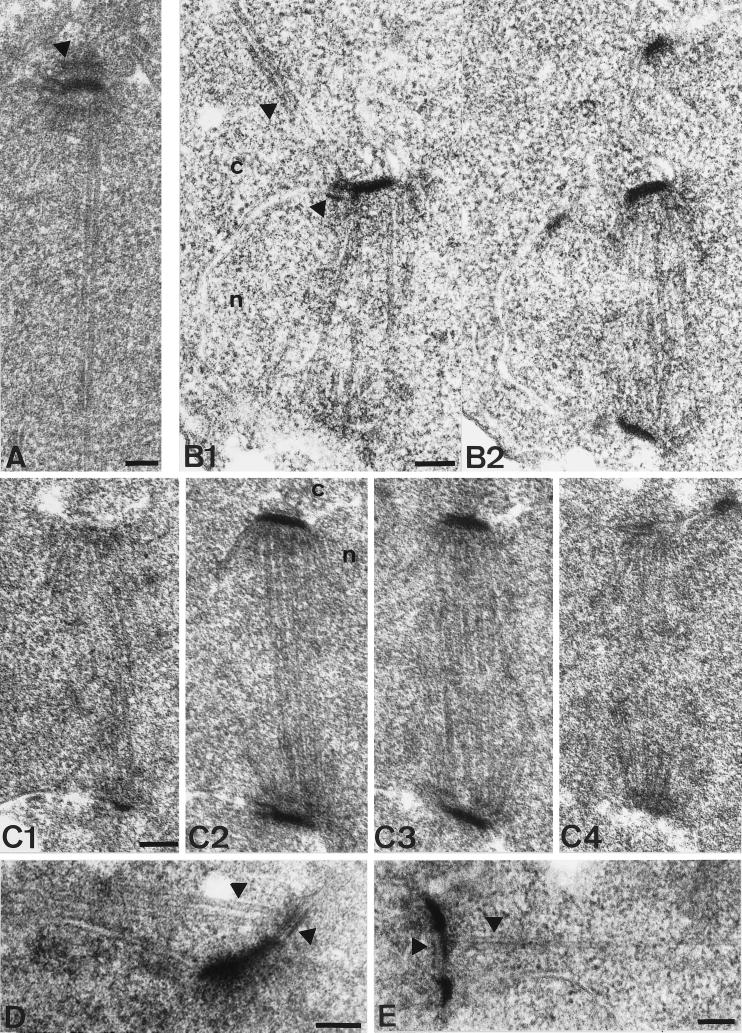
Impaired nuclear migration of cnm67Δ1 cells in combination with the localization of Cnm67p to the SPB pointed to a role of Cnm67p at the SPB substructures that organize the attachment of cytoplasmic microtubules. We investigated more than 300 SPBs of cnm67Δ1 cells by serial thin section EM to determine the spindle and SPB ultrastructure (Figure 4). Wild-type SPBs clearly showed a central plaque embedded in the nuclear envelope in addition to an inner and outer plaque on the nuclear or cytoplasmic side of the nuclear membrane, respectively (Figure 4A). In contrast, the outer plaque of cnm67Δ1 cells could not be clearly visualized in any of the cells. The central and inner plaques were visible and appeared to be structurally normal (Figure 4B). Nuclear microtubules and SPB half-bridges also showed no obvious defect. These observations suggested a specific role of Cnm67p in outer plaque formation or stabilization. The prediction of Cnm67p being a coiled-coil–forming protein favors the idea of a structural role in the establishment of outer plaque integrity.
Figure 4.
Thin section electron microscopy of wild-type and cnm67Δ1 spindles. The nucleus is marked by ’n’ and the cytoplasm by ’c’. (A) SPB of a wild-type cell with elongated spindle. The SPB half-bridge as well as the central, inner, and outer plaque of the SPB are visible. The arrowhead shows the outer plaque. Cytoplasmic microtubules do not lie within the plane of the section. (B1–2) Serial thin sections of a complete cnm67Δ1 spindle after SPB separation. Unlike in wild-type cells, cytoplasmic microtubules are attached to the half-bridge of the upper SPB. Arrowheads show cytoplasmic microtubules and the half-bridge. (C1–2) Serial thin sections of a cnm67Δ1 spindle. The SPB outer plaque is not detectable in any of the sections, although the half-bridge, central, and inner plaques appear normal. The same was found for all cnm67Δ1 SPBs that we investigated (n>300). (D) Cytoplasmic microtubules are attached to the half-bridge of a cnm67Δ1 cell with a single SPB, presumably a G1 cell. Arrowheads show the half-bridge and cytoplasmic microtubules. (E) Side- by-side SPBs are connected by a bridge that nucleates cytoplasmic microtubules directing into the bud. The bridge and cytoplasmic microtubules are indicated. Bars, 0.1 μm.
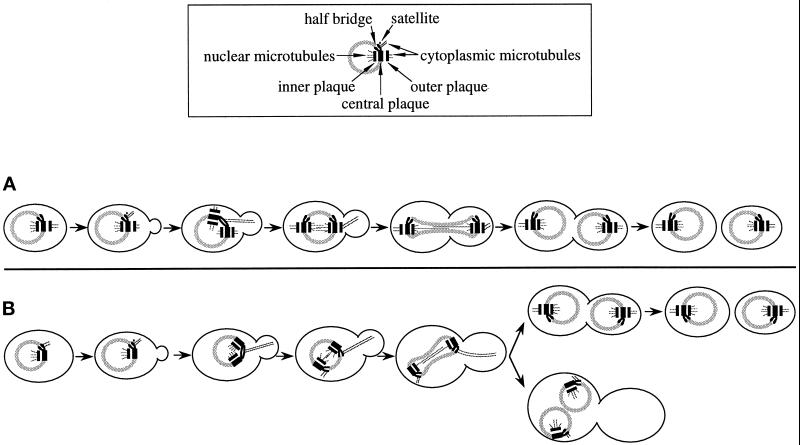
Where do cytoplasmic microtubules attach themselves in cells with a nonfunctional outer plaque? Cytoplasmic microtubules are difficult to detect by serial thin section EM of S. cerevisiae since there are only a few per SPB and, unlike nuclear microtubules, they emerge from the SPB at many different angles and thus rarely lie in the same plane of the section as the nuclear microtubules. Despite the apparent reduction of the SPB outer plaque, which normally initiates cytoplasmic microtubules after SPB separation, cytoplasmic microtubules were still detectable in 36 of the cnm67Δ1 cells examined by EM. In 22 of these cells the cytoplasmic microtubules were clearly connected to the half-bridge (Figure 4, B and D), the bridge (Figure 4E), or the region between the half-bridge and the central plaque. In the remainder the microtubules ended before the SPB, and thus it was not clear what substructure they were connected to. In 10 of the 22 cells cytoplasmic microtubules originated from the bridge of side-by-side SPBs as seen in wild-type cells. However, interestingly, the connection to the half-bridge was still visible after SPB separation in 8 cells (Figure 4B). The remaining 4 cells had cytoplasmic microtubules connected to a half-bridge of a single SPB, but in three cases it was not possible to determine whether a second SPB was present in that cell. One cell (Figure 4D) presumably was in G1 since no second SPB and no bud were observed. In wild-type cells all cytoplasmic microtubules were attached to the outer plaque after SPB separation, as originally described by Byers and Goetsch (1975). It is probable that in cnm67Δ1 cells, unlike in wild-type, cytoplasmic microtubules stay bound to the SPB half-bridge after SPB separation. In this way the mutant may partially compensate for the inability to grow cytoplasmic microtubules from the outer plaque and thus allow cells to maintain a limited nuclear migration capability. In Figure 5 our observations on spindle orientation and microtubule binding in cnm67Δ1 cells are combined to a model for a nuclear migration-rescue mechanism that relies on half-bridge–attached microtubules.
Figure 5.
Model for a salvage mechanism for spindle orientation and nuclear migration in cnm67Δ1 cells. (A) Wild-type: SPB half-bridges and bridges bind cytoplasmic microtubules only when the satellite forms and SPB duplication takes place. After SPB separation, cytoplasmic microtubules are no longer found at the half-bridge (Byers and Goetsch, 1975). During all cell cycle stages, the outer plaque acts as an attachment site. The first phase of nuclear migration begins in S when cytoplasmic microtubules originate from the bridge of side-by-side SPBs. The second phase of nuclear migration in G2/M coincides with cytoplasmic microtubule attachment at the outer plaque. (B) In cnm67Δ1 cells cytoplasmic microtubules originate from the half-bridge throughout the cell cycle while the outer plaque is much reduced or absent and thus incapable of microtubule binding. Consequently, the second phase of nuclear migration is impaired, frequently leading to nuclear division within the mother cell. However, half-bridge–organized cytoplasmic microtubules still have a limited capability to mediate spindle positioning and nuclear migration forces that allow many cells to finally migrate the daughter nucleus into the bud. Cells in which nuclear migration failed double in ploidy and often undergo another round of nuclear division.
Nuclear Fusion in cnm67Δ1 Crosses
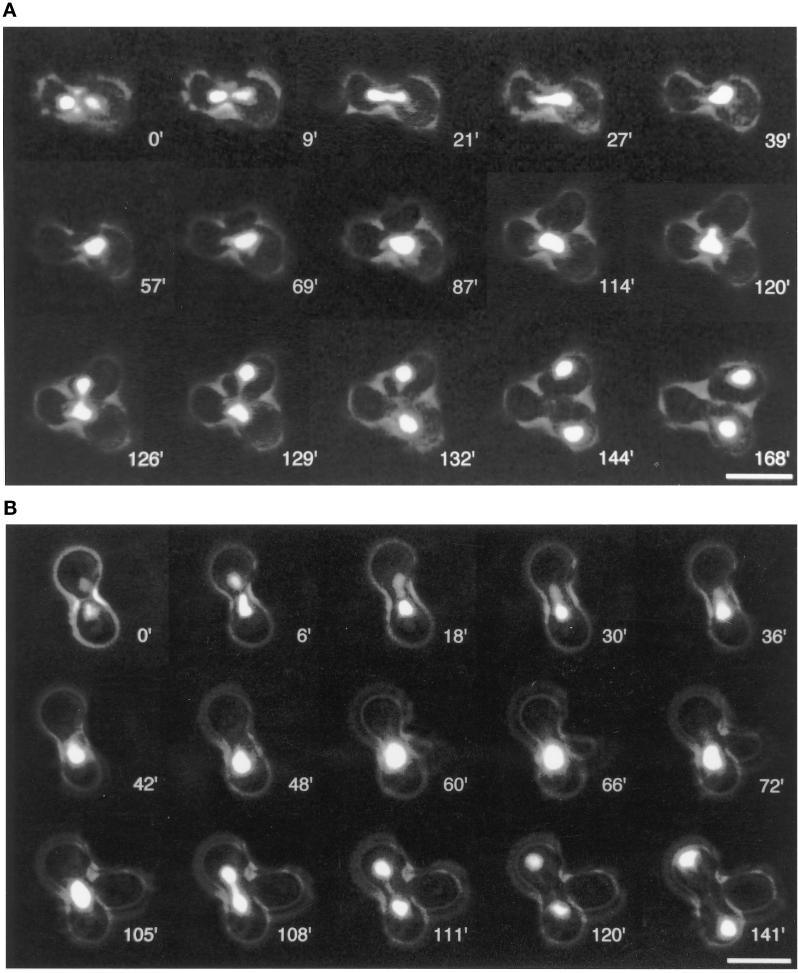
During karyogamy, haploid nuclei migrate toward each other before they fuse their SPBs, nuclear envelopes, and nucleoplasms to produce a diploid zygote nucleus. This step was termed nuclear congression (Kurihara et al., 1994; Rose, 1996) and strictly depends on intact cytoplasmic microtubules that are attached to the SPB half-bridges of haploid nuclei (Byers and Goetsch, 1975; Delgado and Conde, 1984; Hasek et al., 1987; Rose and Fink, 1987). Our EM data indicated that CNM67 loss considerably impairs SPB outer-plaque formation but not half-bridge structure. Hence, nuclear congression should be less impaired than mitotic nuclear migration since it relies on half-bridge–organized microtubules rather than on those that are nucleated by the outer plaque. We tested this hypothesis in two independent experiments. Initially, we examined the ability of haploid cnm67Δ1 strains to mate by crossing an α cnm67Δ1, his3Δ200, TRP1 and an a cnm67Δ1, HIS3, trp1Δ63 strain. Incubation on synthetic minimal medium, lacking histidine and tryptophan, selected for diploid cells. Resulting prototroph colonies were additionally checked for diploidy by analytical PCR as described by Huxley et al. (1990). To confirm that prototroph colonies did not arise from heterokaryotic cells in which only cytoplasmic fusion had occurred without nuclear fusion (cytoductants), we stained the corresponding cultures with DAPI and determined the amount of mono- and multinucleated cells. We found that 60% of cells were mono- and 34% were multinucleated. Prototroph, mononucleated cells are presumably derivatives of haploid cells that had fused their nuclei to produce true diploids. These preliminary data suggested that CNM67 is not required for nuclear fusion during mating.
We attempted to obtain direct evidence for the nuclear fusion capability of cnm67Δ1 cells and to gain insight into the dynamics and efficiency of nuclear migration during conjugation and the following mitosis. For this purpose, we constructed cells with a strongly fluorescent nuclear GFP label and observed conjugating cells in vivo with a time-lapse video microscopy setup. Our recently described Hhf2GFPp (histone H4-GFP) fusion construct (Wach et al., 1997) proved to be suitable for following nuclear dynamics during the mating process and subsequent mitotic cell cycles. Figure 6A shows pictures of selected time points during mating and the first mitotic division of Hhf2-GFPp-labeled wild-type cells, and Figure 6B gives an example for a typical mating event of cnm67Δ1 cells. Eight individual wild-type conjugation events were followed. In all cases, nuclear fusion took place as described by Cross et al. (1988) and Rose (1996). Nuclei moved toward each other in a directed manner and fused in the central region of the newly formed zygote. The first zygote bud developed either in the proximity of the previous cell fusion boundary or at one of the distal tips of the zygote. In any case, the nucleus reliably elongated into the zygote bud during the first mitotic division. Cytokinesis took place about 30 min after the two chromosome masses had completely separated. Nuclear migration during following mitotic divisions also occurred reliably. We recorded 32 mating events of cnm67Δ1 cells. Most importantly, almost all cases clearly indicated that nuclear fusion also occurs in cnm67Δ1 zygotes. In 28 of 32 cases, both mating partners contained one nucleus before cell fusion. In 27 of these 28 cases nuclear fusion occurred with about the same dynamics as seen for the wild-type. As in wild-type cells, nuclei initially adopted a characteristic, roughly triangular, shape before they started to fuse. Presumably, this change of nuclear morphology reflected microtubule-mediated forces that acted on SPBs to pull both SPBs together, before nuclear envelope fusion. Thus, these results confirmed that CNM67 is dispensable for the congression and nuclear envelope fusion steps of karyogamy. In one of the 28 cases mentioned above, nuclei migrated toward each other but did not fuse or show any further activity, suggesting a severe cellular malfunction that may or may not have been due to CNM67 loss. A major difference of cnm67Δ1 cells compared with wild-type cells became visible during the first mitotic division of newly formed diploid nuclei. In 19 of 27 cases, nuclear elongation was restricted to the zygote, without migrating the daughter nucleus into the zygote bud. The direction of nuclear elongation was usually different from the zygote–bud axis. Thus, the first mitotic division of cnm67Δ1 zygotes was strikingly similar to divisions of vegetativly growing cnm67Δ1 cells. Although nuclear migration before nuclear fusion was obviously unaffected, nuclear migration after nuclear fusion was heavily impaired. In 3 of 4 observable cases in which the zygote bud obtained a nucleus, nuclear division was followed by cytokinesis. Importantly, offspring of the first daughter cell clearly budded in a bipolar pattern that is typical for diploid cells (Freifelder, 1960). Those zygotes that did not succeed in migrating the nucleus into the first zygote bud often formed one or more additional buds at the distal ends of the cell. In 3 of 12 cases that could be followed, this second zygote bud obtained a nucleus. Zygotes that did not succeed in migrating daughter nuclei in buds often lysed after further nuclear divisions. Four mating events were observed in which one mating partner carried two nuclei. Nuclear fusion occurred in 3 of these 4 cases, but cells then died without dividing their nucleus.
Figure 6.
Time-lapse fluorescence video microscopy of wild-type and cnm67Δ1 conjugations. Nuclei of haploid cells were labeled with a strongly fluorescent histone H4-GFPp (Hhf2-GFPp). (A) A nuclear fusion event of wild-type cells is followed by the first mitotic division of the zygote nucleus. Both daughter nuclei are correctly segregated between the zygote and the bud. Nuclear division is then followed by cytokinesis. (B) Conjugation of cnm67Δ1 cells. Nuclear migration (congression) before nuclear fusion and nuclear fusion occur as in wild-type cells in the central region of the zygote and also with about the same dynamics. The subsequent mitotic division, however, fails to segregate one of the daughter nuclei into the zygote bud. Presumably, congression is not affected because it is mediated by half-bridge–organized microtubules as in wild-type, whereas the first mitotic nuclear migration is impaired by the outer plaque defect of cnm67Δ1 cells. Cells that surrounded mating cells in panels A and B were removed electronically for clarity. Bars, 10 μm.
Our data indicate that nuclear congression and envelope fusion are not significantly impaired in cnm67Δ1 mutants. Nevertheless, the overall efficiency of mating is reduced due to defects in the segregation of diploid zygote nuclei that often lead to cell death. Fusion events between bi- or multinucleated cells also have frequently fatal consequences.
DISCUSSION
We identified and characterized Cnm67p, a novel yeast protein that plays a crucial role in spindle orientation and mitotic nuclear migration. DNA and tubulin staining of CNM67 null mutants reveals the frequent misorientation of mitotic spindles, indicating that cytoplasmic forces acting on spindle poles are impaired. This deficiency then often leads to a restriction of anaphase spindle elongation to the mother cell and hence to an accumulation of nuclei in mother cells.
Interestingly, the spindle assembly checkpoint (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996) does not stop cell cycle progression in cnm67Δ1 strains, although outer plaque structure is impaired. The most likely explanation for the failure of cell cycle arrest in cnm67Δ1 cells is that this checkpoint does not monitor the structural integrity of the outer plaque, thus allowing the cell cycle to progress even with imperfect SPBs.
Fluorescence microscopy of cells with two independent Cnm67p labels showed its localization to the SPB region. This is consistent with data from mass spectrometric analysis of isolated SPBs identifying Cnm67p (Jensen, unpublished data). Two lines of evidence suggest a role of Cnm67p at the cytoplasmic side of the SPB. First, nuclear migration as a cytoplasmic microtubule-dependent process is heavily impaired in cnm67Δ1 mutants, whereas nuclear microtubule-dependent processes such as SPB separation and spindle elongation are apparently normal. Second, serial thin section electron microscopy of cnm67Δ1 cells indicates a loss of SPB outer plaque integrity while central plaque, inner plaque, half-bridge, and nuclear microtubules are indistinguishable from those of wild-type cells. A CDC37 mutant was recently described to be defective in outer plaque formation during SPB duplication (Schutz et al., 1997). Genetic data on that mutant suggest a general role of CDC37 function in G1 control rather than a structural role as in the case of CNM67. We assume that Cnm67p is a component of the outer plaque itself or a region between outer and central plaque that anchors the outer plaque to the central plaque (Bullitt et al., 1997). A model that summarizes phenotypes of cnm67Δ1 cells is shown in Figure 5.
A further defect that might be expected in a mutant with an impaired outer plaque structure is in spore wall formation, since the outer plaque serves as initiation site for spore wall assembly (Moens and Rapport, 1971; Horesh et al., 1979). Our finding that cnm67Δ1 cells are unable to sporulate is consistent with this defect, although we did not further investigate the meiotic defect of cnm67Δ1 strains, and it is probable that Cnm67p is also critical for meiotic spindle orientation. The sporulation deficiency cannot be due to the multinucleate phenotype per se since about 60% of cnm67Δ1 cells contain only one nucleus. We might also expect that cells with a reduced outer plaque would have less pronounced defects in nuclear congression during mating as compared with nuclear migration in mitosis, since cytoplasmic microtubules are organized by the half-bridge during mating (Byers and Goetsch, 1975). This is exactly what we observed for cnm67Δ1 strains as nuclear congression is not significantly impaired, although orientation of mitotic spindles is clearly inefficient. In vivo observation of nuclei during conjugation allowed us to conclude that kinetics of nuclear congression and fusion are normal in cnm67Δ1 cells. The observed reduced frequency of diploid formation is due to a substantial number of mating cells carrying more than one nucleus and to frequent mitotic missegregation of diploid nuclei.
The apparent reduction of the outer plaque in cnm67Δ1 mutants raises the question of how cells manage to keep a partial nuclear migration capability during mitosis as orientation of the nucleus within the cell and movement through the bud neck are strictly dependent on the function of cytoplasmic microtubules (Huffaker et al., 1988; Palmer et al., 1992; Sullivan and Huffaker, 1992), and yet immunofluorescence of cnm67Δ1 cells clearly shows the presence of cytoplasmic microtubules. In addition, these microtubules in cells with misoriented spindles are often directed into the bud, suggesting that factors controlling overall microtubule directionality are at least partially functional. Examination of cnm67Δ1 SPBs by serial thin section electron microscopy indicated that cytoplasmic microtubules fail to transfer to the outer plaque after SPB separation as in wild-type cells (Byers and Goetsch, 1975), but remain attached to the half-bridge throughout the cell cycle. Presumably, this interaction allows motor proteins to generate pulling or pushing forces that, to a certain extent, position the nucleus and thus allow cells to propagate, although force transfer to the SPB is less efficient than in cells with cytoplasmic microtubules attached to the outer plaque.
Since cnm67Δ1 cells have cytoplasmic microtubules, why then is nuclear migration less efficient in the mutant? It seems possible that different motor proteins act on microtubules initiated from the SPB outer plaque compared with those from the half-bridge. Thus, a motor protein might function less efficiently if the microtubules were attached to an inappropriate structure for that particular stage of the cell cycle, which might change the dynamic behavior of the microtubules (Carminati and Stearns, 1997; Shaw et al., 1997). Interestingly, DeZwaan et al. (1997) recently found that the kinesin-related motor Kip3p predominantly acts in the first phase of nuclear migration whereas dynein mainly contributes to the second phase. During the first, Kip3p mediated- phase microtubules are presumably attached at the half-bridge or bridge whereas they are organized by the outer plaque during the second, dynein-mediated phase. Cottingham and Hoyt (1997) demonstrated the involvement of yet another kinesin-related protein, Kip2p, in proper spindle positioning. Future studies may reveal how different motor protein functions relate to differential attachment of cytoplasmic microtubules to different SPB substructures.
ACKNOWLEDGMENTS
We are grateful to Douglas Kershaw for cutting the serial thin sections for EM and to Dominic Hoepfner and Florian Schärer for help with construction of the Hhf2-GFPp-labeled strains and software development. We also thank M.N. Hall, W.D. Heyer, I. Adams, and S. Souès for discussion. A. Duesterhoeft is gratefully acknowledged for providing sequence information and subclones of cosmid XIV-6. Thanks are also due to D. Botstein, J. Cooper, and D. Koshland for strains. This work was supported by grants to P.P. and A.W. from the Swiss Federal office for Education and Science (grants 95.0191–1 and 95.0191–12).
REFERENCES
- Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier KO, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR. Microtubule organizing centers. Annu Rev Cell Biol. 1985;1:145–172. doi: 10.1146/annurev.cb.01.110185.001045. [DOI] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Bullock WO, Fernandez JM, Short JM. XLl-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern, J.N. JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces-Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin section electron microscopy. Methods Enzymol. 1991;194:602–607. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiae is required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F, Hartwell LH, Jackson C, Konopka JB. Conjugation in Saccharomyces cerevisiae. Annu Rev Cell Biol, 1988;44:29–57. doi: 10.1146/annurev.cb.04.110188.002241. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Conde J. Benomyl prevents nuclear fusion in Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:188–189. doi: 10.1007/BF00327435. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin related KIP3 of Saccharomyces cerevisiae is required for a distinct step of nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Bud formation in Saccharomyces cerevisiae. J Bacteriol, 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin JV, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Goh P-Y, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol, 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:14–15. [PubMed] [Google Scholar]
- Haarer BK, Lillie SH, Adams AE, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasek J, Rupes I, Svobodova J, Streiblova E. Tubulin and actin topology during zygote formation of Saccharomyces cerevisiae. J Gen Microbiol. 1987;133:3355–63. doi: 10.1099/00221287-133-12-3355. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horesh O, Simchen G, Friedmann A. Morphogenesis of the synapton during yeast meiosis. Chromosoma. 1979;75:101–115. doi: 10.1007/BF00330628. [DOI] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Interthal H, Bellocq C, Bahler J, Bashkirov VI, Edelstein S, Heyer WD. A role of Sep1 (= Kem1, Xrn1) as a microtubule-associated protein in Saccharomyces cerevisiae. EMBO J. 1995;14:1057–1066. doi: 10.1002/j.1460-2075.1995.tb07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalt A, Schliwa M. A novel structural component of the Dictyostelium centrosome. J Cell Sci. 1996;109:3103–3112. doi: 10.1242/jcs.109.13.3103. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiae spindle poly body whose transcript is cell cycle-regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV. Genetic and biochemical approaches to spindle function and chromosome segregation in eukaryotic microorganisms. Curr Opin Cell Biol. 1994;6:50–54. doi: 10.1016/0955-0674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen V, Oechsner U, Muller G, Bandlow W. The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol Cell Biol. 1988;8:5108–5115. doi: 10.1128/mcb.8.12.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Weber S. Flag N-terminal epitope overexpression of bacterial alkaline phosphatase and Flag C-terminal epitope tagging by PCR one-step targeted integration. Yeast. 1992;8(special issue):627. [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen) J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Centrosome-microtubule nucleation. J Cell Sci. 1997;110:295–300. doi: 10.1242/jcs.110.3.295. [DOI] [PubMed] [Google Scholar]
- Philippsen, P. et al. (1997). The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature 387 (suppl), 93–98. [PubMed]
- Rose MD. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1996;12:663–695. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- Rose MD, Biggins S, Satterwhite LL. Unravelling the tangled web at the microtubule-organizing center. Curr Opin Cell Biol. 1993;5:105–115. doi: 10.1016/s0955-0674(05)80015-8. [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schutz AR, Giddings TH, Steiner E, Winey M. The yeast CDC37 gene interacts with MPS1 and is required for proper execution of spindle pole body duplication. J Cell Biol. 1997;136:969–982. doi: 10.1083/jcb.136.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The spindle pole body of yeast. Chromosoma. 1994;103:369–380. doi: 10.1007/BF00362281. [DOI] [PubMed] [Google Scholar]
- Sobel SG, Synder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, F. (1991). Analyses of the cytoskeleton in Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 633–662. [DOI] [PubMed]
- Spang A, Geissler S, Grein K, Schiebel E. gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Neff NF, Botstein D. Isolation and characterization of mutations in the beta-tubulin gene of Saccharomyces cerevisiae. Genetics. 1985;111:715–734. doi: 10.1093/genetics/111.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for Saccharomyces cerevisiae transformation with SFH-PCR products. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]