Abstract
The catalytic promiscuity of E. coli alkaline phosphatase (AP) and many other enzymes provides a unique opportunity to dissect the origin of enzymatic rate enhancements via a comparative approach. Here we use kinetic isotope effects (KIEs) to explore the origin of the 109-fold greater catalytic proficiency by AP for phosphate monoester hydrolysis relative to sulfate monoester hydrolysis. The primary 18O KIEs for the leaving group oxygen atoms in the AP-catalyzed hydrolysis of p-nitrophenyl phosphate (pNPP) and p-nitrophenylsulfate (pNPS) decrease relative to the values observed for nonenzymatic hydrolysis reactions. Prior linear free energy relationship results suggest that the transition states for AP-catalyzed reactions of phosphate and sulfate esters are ‘loose’ and indistinguishable from that in solution, suggesting that the decreased primary KIEs do not reflect a change in the nature of the transition state but rather a strong interaction of the leaving group oxygen atom with an active site Zn2+ ion. Furthermore, the KIEs for the two reactions are identical within error, suggesting that the differential catalysis of these reactions cannot be attributed to differential stabilization of the leaving group. In contrast, AP perturbs the KIE for the nonbridging oxygen atoms in the reaction of pNPP but not pNPS, suggesting a differential interaction with the transferred group in the transition state. These and prior results are consistent with a strong electrostatic interaction between the active site bimetallo Zn2+ cluster and one of the nonbridging oxygen atoms on the transferred group. We suggest that the lower charge density of this oxygen atom on a transferred sulfuryl group accounts for a large fraction of the decreased stabilization of the transition state for its reaction relative to phosphoryl transfer.
Introduction
The explosion of structural and sequence information over the past decades has revealed extensive evolutionary relationships between enzymes that form ‘families’ and ‘superfamilies’.1, 2 Much recent work in enzymology has investigated these relationships from a functional perspective and has yielded insights into changes in and around active sites that allow enzymes with the same overall fold and often with active site residues in common to catalyze different chemical transformations.1-10
An enzymatic attribute that has been emphasized in functional work is ‘catalytic promiscuity’ – the ability of an enzyme to catalyze, at a low level, a reaction other than its cognate reaction that is maintained via selective pressure.11-17 Catalytic promiscuity is thought to provide an important foothold for the evolution of new enzymes as follows. Under new selective pressures and following a gene duplication event, the promiscuous enzyme can provide a head start in evolving a specialized and highly functional new enzyme by optimizing the promiscuous activity. The advantage arises because fewer mutations would be needed to reach a threshold that would provide a selective advantage and allow natural selection to guide the remainder of the optimization process, relative to an enzyme lacking the promiscuous activity. Catalytic promiscuity thus has implications for understanding evolution and evolutionary relationships and may provide lessons for the design of enzymes with new, useful activities.14 Furthemore, studies of evolutionarily related enzymes have revealed how common mechanistic features can be utilized in different chemical transformations and how the presence or absence of additional catalytic groups can result in different reaction outcomes.12, 14, 17 Here we take advantage of another potential impact of catalytic promiscuity: its use to elucidate the underlying mechanisms that provide the enormous rate enhancements and reaction specificities of enzymes via direct comparisons between cognate and promiscuous reactions.
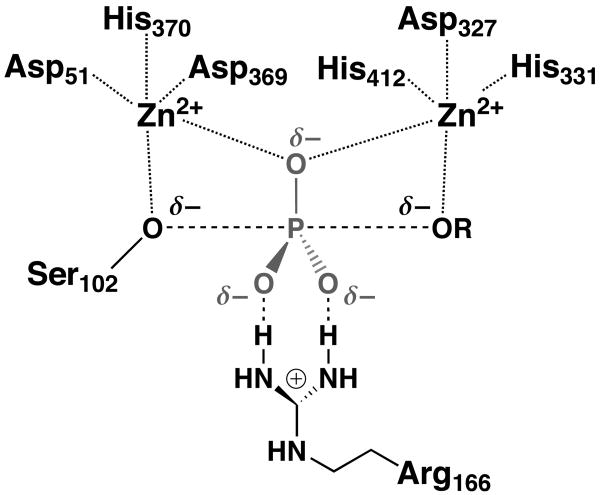
The primary, cognate activity of E. coli alkaline phosphatase (AP) is phosphate monoester hydrolysis.18 AP carries out phosphoryl transfer in a two-step reaction via a covalent enzyme-phosphate intermediate.18 Structural and mechanistic work has led to the following model for active site interactions (Scheme 1).19, 20 The nucleophilic serine residue is positioned adjacent to one of two Zn2+ atoms in the bimetallocluster. The other Zn2+ atom interacts with the leaving group oxygen atom, and one of the nonbridging phosphoryl oxygen atoms is situated between the two Zn2+ atoms in the transition state. Arg166 at the active site interacts with the other two nonbridging phosphoryl oxygen atoms.
Scheme 1.
In addition to its cognate phosphatase activity, AP has promiscuous activities toward sulfate monoesters, phosphate diesters, and phosphite.21-23 A comparison of the phosphatase and sulfatase reactions is particularly intriguing. In solution, phosphate and sulfate monoester hydrolysis reactions occur with similar rate constants,21, 24 and both reactions proceed through loose transition states.25-32 Furthermore, the structural properties of phosphate and sulfate monoesters are highly similar (Table 1). Thus, the observation that AP has some detectable sulfatase activity may not be surprising. However, AP's catalytic proficiency is dramatically different for the two reactions, with catalysis of the cognate phosphatase reaction 109-fold more proficient than the sulfatase reaction (Table 2). Given the similarity of the two types of substrates (Scheme 2), how does AP so robustly distinguish between them?
Table 1.
Comparison of Properties for Phosphate and Sulfate Monoesters.a
| Phosphate Monoesters | Sulfate Monoesters | |
|---|---|---|
| P(S)-OR Bond Length (Å) | 1.56 ± 0.02 | 1.60 ± 0.01 |
| P(S)-O Bond Length (Å) | 1.52 ± 0.02 | 1.46 ± 0.01 |
| kw (M-1s-1)b | 3 × 10-11 | 9 × 10-12 |
| Transition Statec | Loose | Loose |
| Charge | −2 | −1 |
Bond lengths are for ethyl phosphate and ethyl sulfate.69-71 A comparison of bond lengths for the p-nitrophenyl esters was not possible because no structure is available for pNPS.
Table 2.
Comparison of Catalytic Proficiency of AP-catalyzed Reactions of pNPP and pNPS.a
| Enzyme | pNPP | pNPS | Proficiency Ratio (P/S) |
|---|---|---|---|
| wt AP | 1.1 × 1018 | 1.1 × 109 | 1.0 × 109 |
| R166S AP | 3.3 × 1015 | 1.6 × 107 | 2.1 × 108 |
Catalytic proficiencies are defined as [(kcat/KM)/kw], the ratio of the bimolecular rate constants for the enzymatic and corresponding solution hydrolysis reactions for the p-nitrophenyl esters at 25 °C. Rate constants for enzymatic reactions21, 22, 33, 35 and solution reactions21, 24, 33 are from literature sources. The catalytic proficiency for the reaction of R166S AP with pNPS was determined at 30 °C, and control reactions indicated that there was no significant effect of temperature on the catalytic proficiency between 25 and 30 °C.33 The proficiency ratio is the catalytic proficiency for phosphate monoester hydrolysis divided by the catalytic proficiency for sulfate monoester hydrolysis.
Scheme 2.
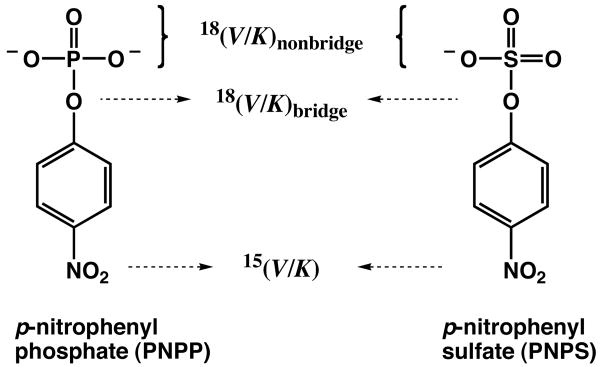
Substrates used in this study and sites of isotope labeling.
A prominent difference between phosphate and sulfate monoesters is the amount of total charge. Phosphate monoesters carry two negative charges while sulfate esters have only a single negative charge (Scheme 2). Prior work with AP revealed a strong correlation of the catalytic proficiency for several substrates with the amount of charge on the nonbridging oxygen atom that would be situated between the Zn2+ ions in the transition state.33 The linear dependence of the free energy of transition state stabilization (relative to the uncatalyzed reaction) with a slope of 31 kcal/mol/unit charge (R2 = 0.98) suggested an important electrostatic contribution to catalysis arising from interactions between the Zn2+ ions and a nonbridging oxygen atom (Scheme 1).
The correlation described above was obtained with the R166S mutant of AP to avoid differential effects from interactions of the other nonbridging atoms with Arg166 (Scheme 1).33 Indeed, these interactions have been demonstrated to provide preferential catalysis for the phosphate ester reaction over the sulfate ester reaction. Nevertheless, Arg166 is responsible for only a small fraction of the discrimination between phosphate and sulfate ester substrates observed in wild type (wt) AP, and R166S AP still discriminates 108-fold between phosphate and sulfate ester substrates (Table 2). Mutation of Arg166 to Ser decreases the rate of p-nitrophenyl phosphate (pNPP) hydrolysis by ∼102-fold.22, 34 However, this decrease does not reflect the full contribution of Arg166 because the chemical step is not rate-determining in the reaction of wt AP with pNPP.35 For alkyl phosphate substrates for which the chemical step is rate-determining, removal of Arg166 decreases the hydrolysis rate by ∼104-fold.36 In contrast, mutation of Arg166 to Ser has a ∼102-fold effect on hydrolysis of p-nitrophenyl sulfate,21, 33 indicating that Arg166 provides a ∼102-fold preference for phosphate monoesters over sulfate monoesters when chemistry is rate-determining. Thus, other active site interactions must be responsible for the large reactivity difference.
To further probe the interactions of AP with the leaving group and nonbridging oxygen atoms of phosphate and sulfate ester substrates, we have compared heavy atom isotope effects for these reactions in solution and at the AP active site. Kinetic isotope effect (KIE) measurements for the enzymatic reactions reported herein are combined with previous measurements for the corresponding solution reactions to make these comparisons. The results suggest that while active site interactions at the leaving group oxygen atom are similar for phosphate and sulfate esters, the interactions with the nonbridging oxygen atom differ significantly, consistent with the prior electrostatic model for discrimination.
Results and Discussion
Traditionally, the contributions of various active site groups to catalysis have been probed by site-directed mutagenesis, but interpretation of these experiments in terms of quantitative contributions to catalytic proficiency is fraught with difficulties.37-45 Instead, we have taken a distinct approach, comparing two of the activities of AP – the cognate phosphate monoesterase activity and the promiscuous sulfate monoesterase activity. We consider, in turn, interactions of AP with the leaving group and with the nonbridging oxygen atoms. These comparisons have allowed us to determine catalytic features that may have different effects on the two reactions. Specifically, we compare our measurements of the heavy atom KIEs for AP-catalyzed hydrolysis of p-nitrophenyl sulfate (pNPS) and p-nitrophenyl phosphate (pNPP) to those obtained previously for the corresponding solution reactions. The KIEs were determined by the competitive method and are therefore effects on V/K (i.e., kcat/KM) for the enzymatic reactions.46 Effects on V/K reflect differences between the enzymatic transition state and the ground state in which the enzyme and substrate are free in solution.
Analysis of interactions with the leaving group oxygen atom
AP, because of its open active site that presumably evolved to generate inorganic phosphate from a wide array of phosphorylated substrates,18, 19 is an ideal enzyme for determining linear free energy relationships (LFERs). The value of βlg determined from a LFER measures the sensitivity of the transition state to charge buildup at the position of bond cleavage and can reflect the extent of bond cleavage in the transition state.47, 48 Previous work showed that the values of βlg for reactions of substituted phenyl phosphorothioates49 and substituted phenyl sulfates are identical within error to each other for both nonenzymatic and AP-catalyzed reactions.50, 51 However, the enzymatic βlg values were less negative than those for the reactions in solution. There is considerable evidence suggesting that the decreased Brønsted slopes for the enzymatic reactions do not originate from a more compressed transition state; rather, the values obtained are in line with expectations for reactions with a stronger nucleophile than in the solution reactions (serine oxide vs. water) and an electrostatic interaction of the leaving group oxygen atom with an active site Zn2+ atom.35, 51 Nevertheless, there are assumptions in and limitations to the analysis of βlg to distinguish contributions from bond cleavage and electrostatic interactions.47, 48
To further probe active site catalytic interactions with the leaving group oxygen atom and to test whether differential interactions at this position might account for the greatly reduced catalytic proficiency of AP as a sulfatase, we determined the 18O leaving group heavy atom isotope effects for AP-catalyzed hydrolysis of pNPS and pNPP (see Methods). For the pNPP reaction it was necessary to use the R166S variant of AP, as chemistry is not rate-determining for the reaction of wt AP with pNPP and kinetic isotope effects are suppressed.35, 52 Removal of Arg166, which interacts with two of the nonbridging oxygen atoms (Scheme 1), substantially slows catalysis but does not affect the value of βlg for reactions of both alkyl phosphates and phenyl phosphorothioates, suggesting that this residue does not perturb interactions with the leaving group.36, 53 Wt AP was used to determine KIEs for enzymatic pNPS hydrolysis, as chemistry is rate-determining for this substrate and it was not practical to measure KIEs for the reaction of R166S AP with pNPS due to the extremely large enzyme concentrations and long reaction times that would be required to observe significant product formation.
Table 3 lists the results of the enzymatic isotope effects for the leaving group oxygen atoms (18(V/K)bridge) of pNPS and pNPP and the values for the corresponding nonenzymatic reactions (18kbridge). As noted previously, the nonenzymatic isotope effects of ∼2% (1.02) for pNPP and pNPS suggest substantial bond cleavage in the transition state,46, 52, 54 consistent with the body of work on phosphoryl and sulfuryl transfer that supports a loose transition state for these reactions.25-32
Table 3.
Kinetic Isotope Effects for Solution and Enzymatic Reactions of pNPP and pNPSa
| Enzymatic Reactions b | Substrate | 15(V/K) | 18(V/K) bridge | 18(V/K)nonbridge |
| wt AP33 | pNPP2- | 1.0003 (2) | 1.0003 (4) | 0.9982 (1) |
| R166S AP | pNPP2- | 1.0007 (1) | 1.0091 (6) | 0.9925 (11) |
| wt AP | pNPS1- | 1.0019 (4) | 1.0084 (10) | 0.9946 (4) |
| Uncatalyzed Reactions | Substrate | 15k | 18kbridge | 18knonbridge |
| At 95 °C52 | pNPP2- | 1.0028 (2) | 1.0189 (5) | 0.9994 (5) |
| At 85 °C52 | pNPS1- | 1.0026 (1) | 1.0210 (10) | 0.9951 (3) |
Standard errors for the last decimal place(s) are in parenthesis. Data for the reaction of wt AP with pNPP2- and for solution reactions are from previous work.52, 54 Data for all other reactions were obtained for this work. KIEs for wt AP-catalyzed pNPP2- hydrolysis are close to unity because the chemical step is not rate-determining.35, 52
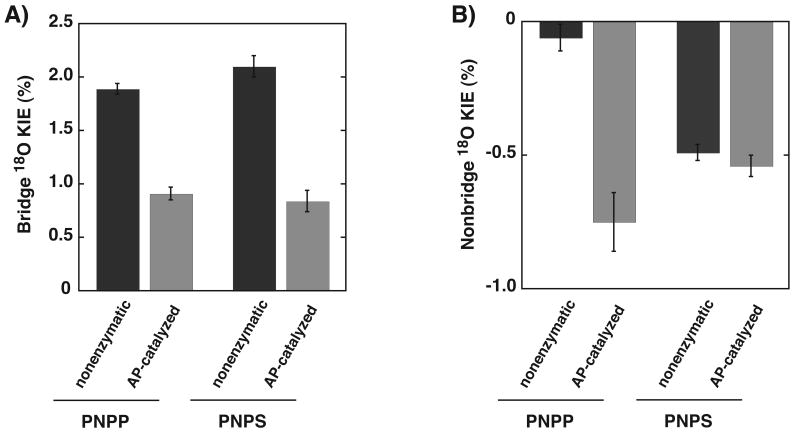
The isotope effect for R166S AP-catalyzed hydrolysis of pNPP is reduced relative to that in solution, with a value of ∼1% (Table 3). As linear free energy relationships give values of βlg consistent with the same extent of bond cleavage for nonenzymatic and AP-catalyzed reactions of phosphate, phosphorothioate, and sulfate monoesters,35, 36, 50, 51, 53 we suggest that the reduced isotope effect results from transition state interactions between the leaving group oxygen and the active site Zn2+ ion (Scheme 1). A similar decrease in this isotope effect is observed in reactions in which the leaving group oxygen is protonated in the transition state,46 but the effect reported herein probably has a different origin. Leaving group stabilization by interaction with a zinc ion does not result in a new covalent bond, so that no new vibrational modes are introduced. However, coordination to zinc may stiffen bending modes, contributing an inverse effect to lower the observed isotope effect. Regardless of the origin, here we emphasize that the isotope effect is reduced to the same extent for the sulfatase reaction catalyzed by AP (Table 3 and Figure 1A). These results suggest that AP has the same catalytic interactions with the leaving group in its phosphatase and sulfatase reactions. Thus, differential interactions with other portions of the substrates are presumably responsible for the large difference in AP's catalytic efficiency for these reactions.
Figure 1.
Comparison of KIEs for nonenzymatic and AP-catalyzed reactions of pNPP and pNPS. Values for the 18O bridge KIEs are plotted in (A) and values for the 18O nonbridge KIEs are plotted in (B). Isotope effects are expressed as a percentage difference from unity [KIE% = (KIEobs –1)*100]. Values for nonenzymatic reactions are shown in black and values for AP-catalyzed reactions are shown in grey.
Analysis of interactions with the nonbridging oxygen atoms
In the transition state model for the AP-catalyzed phosphoryl transfer, one of the nonbridging phosphoryl oxygen atoms is situated between the two Zn2+ atoms of the bimetallocluster (Scheme 1). Evidence in support of this model comes from x-ray structures with covalently and noncovalently bound phosphate and with a bound pentacoordinate vanadate transition state analog.19, 20, 55
Functional data support an important energetic interaction of this oxygen atom with the active site bimetallocluster. R166S AP exhibits a remarkable correlation between the amount of charge on this oxygen atom and the observed rate enhancement. The correlation holds for six substrates investigated and gives a slope of 31 kcal/mol per unit charge (R2 = 0.98).33 Thus, AP appears to be enormously sensitive to electrostatic interactions in the active site, leading to the suggestion that the enzyme and its bimetallocluster establish a region of extraordinarily high positive charge density that preferentially stabilizes transition states with higher negative charge density on the oxygen atom that is situated between the two Zn2+ ions.
Nevertheless, there are many other bimetalloenzymes that preferentially hydrolyze substrates that have less charge situated between the two metal ions.10, 56, 57 It is possible that other features of the AP active site lead to a strong electrostatic interaction energy at this position, such as the nearby Mg2+ ion that is present in AP but absent in other members of the AP superfamily.10, 19, 58, 59 It is also possible that there is an alternative explanation for the observed correlation that is unrelated to local electrostatics.
To probe interactions with the nonbridging oxygen atom we determined the KIEs for the AP-catalyzed reactions of pNPP and pNPS with 18O substitutions at the nonbridging positions (Scheme 2) and compared these values to those obtained for the corresponding nonenzymatic reactions (Table 3). There is a significant difference for the nonenzymatic and AP-catalyzed reactions of pNPP (Figure 1B). The 18O nonbridge KIE for pNPP hydrolysis becomes much more inverse, from −0.06% for the nonenzymatic hydrolysis reaction to −0.75% for the AP-catalyzed reaction. As described above in the case of the bridging oxygen, the interaction between an oxygen atom and the Zn2+ bimetallo site does not result in new covalent bonds, so that no new vibrational modes are introduced, and the large inverse effect probably arises from a strong interaction with the active site Zn2+ ions that stiffens or freezes some bending and torsional modes.60 In contrast to the isotope effect for the pNPP reaction, the isotope effect for the pNPS reaction is the same, within error, for the enzymatic and nonenzymatic reactions, providing no evidence for a special or strong interaction of the sulfuryl group with the Zn2+ ions (Figure 1B). The pNPS KIEs are consistent with a similar transition state on and off the enzyme and an absence of strongly perturbing interactions with the Zn2+ bimetallocluster that stiffens bending and torsional vibrational modes of the nonbridging S-O bonds.
The different effect of AP on the nonbridge KIEs for phosphoryl versus sulfuryl transfer reactions supports a model in which there are differential active site interactions. As there is evidence that both reactions occur in the same active site in AP and require the presence of Ser102 and Arg166,61 we suggest that the effect arises due to differing strengths of the interaction between a nonbridging oxygen atom and the active site Zn2+ ions. Indeed, this interpretation of the KIE data supports a previously proposed model in which the high positive charge density in the AP active site leads to stronger interactions with a nonbridging oxygen atom of highly negatively charged substrates, such as phosphate monoesters, relative to less negatively charged substrates, such as sulfate monoesters.33, 62
Alternative models to account for the differing behavior of the nonbridge KIEs for pNPP and pNPS have been considered as follows. One possibility is that the different behavior originates because the data for pNPP were obtained with R166S AP, whereas the data for pNPS were obtained with wt AP. However, one might have anticipated a greater inverse contribution to the KIE with Arg166 present, contrary to the observed effects. Furthermore, removal of the active site arginine residue in protein tyrosine phosphatases has no significant effect on the value of the 18O nonbridge KIE.63 Another possibility is that because the nonbridge KIE for pNPS hydrolysis in solution is already significantly inverse (−0.5%), interactions with Zn2+ ions in the AP active site cannot provide any additional inverse contribution. Physically, this model suggests that vibrational modes for the nonbridging S-O bonds are already stiffened or frozen in the transition state for solution hydrolysis and that the enzymatic transition state does not provide any additional tightening of the vibrational modes. However, there is no theoretical basis for assuming that −0.5% represents an upper limit in the magnitude of a secondary 18O KIE, and secondary 18O KIEs that are significantly more inverse than −0.5% have been observed.64 Nevertheless, this model cannot be explicitly ruled out because we lack a complete, quantitative description of the behavior of the vibrational modes that give rise to the observed KIEs even for the reactions in solution.
The proposal that the bimetallo site of AP provides electrostatic discrimination to allow preferential catalysis of phosphate monoester hydrolysis immediately raises the question of how the bimetallo site in AP is adapted for this preference, given that many other bimetallo enzymes act preferentially on substrates with less charge than phosphate monoesters.10, 56, 57 Furthermore, there are enzymes in the AP superfamily known as arylsulfatases that preferentially catalyze sulfate ester hydrolysis. Although there are several conserved features between the AP and arylsulfatase active sites, there are also prominent differences. In particular, the AP active site contains three metal ions, two of which directly contact the substrate,19 whereas arylsulfatases contain only a single metal ion and have a lysine residue in place of one of the missing metal ions.58 Nevertheless, there are several other residues in the arylsulfatase active site that could potentially bear a positive charge,58 making it difficult to assess the differences in active site electrostatics between AP and arylsulfatase by comparison of the structures alone.
KIEs for arylsulfatase-catalyzed reactions may offer additional insight into the differences between the AP and arylsulfatase active sites. The simplest expectation for arylsulfatase-catalyzed pNPS hydrolysis would be that there are strong active site interactions with the transferred sulfuryl group, leading to an inverse contribution to 18(V/K)nonbridge similar to that observed for AP-catalyzed pNPP hydrolysis. KIEs for pNPS hydrolysis catalyzed by two different arylsulfatases have been determined.65 The values of 18(V/K)nonbridge are small and normal (0.09% to 0.24%), distinct from the small inverse effects observed for uncatalyzed hydrolysis and AP-catalyzed hydrolysis. The small normal aryl sulfatase values could reflect differences in nucleophilic participation and/or weaker electrostatic interactions with the sulfuryl group in the aryl sulfatase transition state than in the AP transition state. Further experimental work is needed before conclusions can be drawn about the comparative nature and strength of interactions in the active sites of arylsulfatase versus AP.
Continued comparative structural and functional studies on evolutionarily related enzymes in the AP superfamily will reveal more about how these similar active sites discriminate between phosphate ester, sulfate ester, and other substrates. In addition to comparisons within the AP superfamily, comparisons with unrelated enzymes that catalyze phosphoryl and sulfuryl transfer reactions may ultimately help identify features of the AP active site that make important contributions to its electrostatic behavior and reveal common mechanistic strategies that underlie the remarkable specificities and catalytic proficiencies of enzymes.
Methods
Materials
The plasmids for expression of wt and R166S AP and the phoA- strain of E. coli (SM547) were provided by Evan Kantrowitz.34 Natural abundance and isotopically labeled salts of pNPP and pNPS were synthesized as previously described.52, 54
Alkaline Phosphatase Purification
Wild type and R166S AP were purified as previously described.22
Isotope Effect Determinations
18O KIEs were measured using isotope ratio mass spectrometry by the remote label method, using the nitrogen atom as a reporter for isotopic fractionation at the bridge or nonbridge oxygen atoms. The experimental procedures used to measure these isotope effects were similar to those previously reported.46
Isotope effects on the alkaline phosphatase reaction with pNPS
Reactions were carried out at 35 °C using 100 μmoles of pNPS in 1.0 mL of 0.1 M MOPS buffer, pH 7.0, containing 10 μM alkaline phosphatase. Reactions were allowed to proceed for 10 to 21 days, giving fractions of reaction ranging from 17 to 35%. A side-by-side control reaction under identical conditions but without enzyme showed only a negligible amount of nonenzymatic hydrolysis of pNPS (≪1% of the total). The reactions were then diluted with water to a volume of 20 mL. To determine the fraction of reaction, two aliquots were removed. One was assayed for p-nitrophenol from the absorbance at 400 nm of a sample added to 3 mL of 1 N NaOH. The other aliquot was made 1 N in HCl and heated for 12 hours at 80 °C, which control experiments showed resulted in complete hydrolysis of pNPS. This aliquot was then similarly assayed for p-nitrophenol, and the ratio of the two assays gave the fraction of enzymatic hydrolysis at the time the reaction was stopped.
The original 20 mL aqueous reaction mixture was titrated to pH 4 with HCl, and p-nitrophenol was extracted with ether (3 × 20 mL). The ether extracts were dried over MgSO4, and the solvent removed by rotary evaporation. The remaining pNPS in the aqueous layer was hydrolyzed by addition of 2.2 mL of concentrated HCl and heating for 12 h at 80 °C. The p-nitrophenol thus released was isolated by ether extraction as described. The p-nitrophenol samples were purified by sublimation before analysis using a Europa 20-20 isotope ratio mass spectrometer interfaced with an ANCA-NT combustion system.
Isotope effects on the R166S alkaline phosphatase reaction with pNPP
Reactions were carried out at 25 °C with 100 μmoles of pNPP in 2.5 mL of 0.5 M Tris·HCl buffer, pH 8, containing 0.5 μM enzyme. At partial completion, reactions were stopped by lowering the pH to 4 and diluting to 20 mL with ice water, and two aliquots were removed. One aliquot was added to a solution of Tris·HCl at pH 9 containing excess wild type alkaline phosphatase to hydrolyze remaining pNPP. The second aliquot was assayed for p-nitrophenol immediately, and the other was similarly assayed after 24 hours, from the absorbance at 400 nm after addition of a sample to a cuvette containing 1 N NaOH. The ratio of the two assays gave the fraction of enzymatic hydrolysis. A side-by-side control reaction under identical conditions without enzyme showed only a negligible amount of nonenzymatic hydrolysis of pNPP (≪1% of the total).
The p-nitrophenol product at partial reaction was isolated from the original 20 mL reaction mixture as described for the pNPS reaction. Residual pNPP in the aqueous layer was hydrolyzed by titration to pH 9 and overnight reaction after addition of commercial alkaline phosphatase. Subsequently, p-nitrophenol was isolated by titration to pH 4 and ether extractions as described above.
Calculation of isotope effects
Isotope effects were calculated from the isotopic ratios at partial reaction in the product (Rp), in the residual substrate (Rs), and in the starting material (Ro).
| (1) |
| (2) |
Equations 1 or 2 were used to calculate the observed isotope effect either from Rp and Ro or from Rs and Ro, respectively, at the measured fraction of reaction.66 The observed isotope effects from experiments to determine 18O isotope effects were corrected for the 15N effect and for incomplete levels of isotopic incorporation in the starting material as previously described.67 The independent calculation of each isotope effect using Rp and Ro and using Rs and Ro from Eqs. 1 and 2, respectively, provided an internal check of the results. These values agreed within experimental error, and were averaged to give the values reported in Table 3.
Acknowledgments
This work was supported by a grant to D.H. from the NIH (GM64798) and to A.H. from the NIH (GM47297). I.N.H. was supported in part by an American Heart Association Predoctoral Fellowship. J.G.Z. was supported in part by a Hertz Foundation Graduate Fellowship. We thank members of the Herschlag and Hengge labs for comments on the manuscript. I.N.H. acknowledges valuable interactions with Doug Rees and members of his laboratory.
Abbreviations
- AP
alkaline phosphatase
- wt
wild type
- LFER
linear free energy relationship
- KIE
kinetic isotope effect
- pNPP
p-nitrophenyl phosphate
- pNPS
p-nitrophenyl sulfate
- pNPPS
p-nitrophenyl phosphorothioate
References
- 1.Gerlt JA, Babbitt PC. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 2.Orengo CA, Thornton JM. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 3.Todd AE, Orengo CA, Thornton JM. J Mol Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong RN. Biochemistry. 2000;39:13625–13632. doi: 10.1021/bi001814v. [DOI] [PubMed] [Google Scholar]
- 5.Whitman CP. Arch Biochem Biophys. 2002;402:1–13. doi: 10.1016/S0003-9861(02)00052-8. [DOI] [PubMed] [Google Scholar]
- 6.Anantharaman V, Aravind L, Koonin EV. Curr Opin Chem Biol. 2003;7:12–20. doi: 10.1016/s1367-5931(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 7.Allen KN, Dunaway-Mariano D. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Seibert CM, Raushel FM. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 9.Glasner ME, Gerlt JA, Babbitt PC. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Zalatan JG, Fenn TD, Brunger AT, Herschlag D. Biochemistry. 2006;45:9788–9803. doi: 10.1021/bi060847t. [DOI] [PubMed] [Google Scholar]
- 11.Jensen RA. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien PJ, Herschlag D. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 13.Palmer DRJ, Garrett JB, Sharma V, Meganathan R, Babbitt PC, Gerlt JA. Biochemistry. 1999;38:4252–4258. doi: 10.1021/bi990140p. [DOI] [PubMed] [Google Scholar]
- 14.Penning TM, Jez JM. Chem Rev. 2001;101:3027–3046. doi: 10.1021/cr000049n. [DOI] [PubMed] [Google Scholar]
- 15.Copley SD. Curr Opin Chem Biol. 2003;7:265–272. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 16.James LC, Tawfik DS. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 17.Khersonsky O, Roodveldt C, Tawfik DS. Curr Opin Chem Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Coleman JE. Annu Rev Biophys Biomol Struct. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 19.Kim EE, Wyckoff HW. J Mol Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 20.Holtz KM, Stec B, Kantrowitz ER. J Biol Chem. 1999;274:8351–8354. doi: 10.1074/jbc.274.13.8351. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien PJ, Herschlag D. J Am Chem Soc. 1998;120:12369–12370. [Google Scholar]
- 22.O'Brien PJ, Herschlag D. Biochemistry. 2001;40:5691–5699. doi: 10.1021/bi0028892. [DOI] [PubMed] [Google Scholar]
- 23.Yang KC, Metcalf WW. Proc Natl Acad Sci USA. 2004;101:7919–7924. doi: 10.1073/pnas.0400664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng H, Nikolic-Hughes I, Wang JHH, Deng H, O'Brien PJ, Wu L, Zhang ZY, Herschlag D, Callender R. J Am Chem Soc. 2002;124:11295–11306. doi: 10.1021/ja026481z. [DOI] [PubMed] [Google Scholar]
- 25.Kirby AJ, Jencks WP. J Am Chem Soc. 1965;87:3209–3216. [Google Scholar]
- 26.Kirby AJ, Varvoglis AG. J Am Chem Soc. 1967;89:415–423. [Google Scholar]
- 27.Thatcher GRJ, Kluger R. Adv Phys Org Chem. 1989;25:99–265. [Google Scholar]
- 28.Hengge AC. Adv Phys Org Chem. 2005;40:49–108. [Google Scholar]
- 29.Benkovic SJ, Benkovic PA. J Am Chem Soc. 1966;88:5504–5511. [Google Scholar]
- 30.Fendler EJ, Fendler JH. J Org Chem. 1968;33:3852–3859. [Google Scholar]
- 31.Hopkins A, Day RA, Williams A. J Am Chem Soc. 1983;105:6062–6070. [Google Scholar]
- 32.Bourne N, Hopkins A, Williams A. J Am Chem Soc. 1985;107:4327–4331. [Google Scholar]
- 33.Nikolic-Hughes I, O'Brien PJ, Herschlag D. J Am Chem Soc. 2005;127:9314–9315. doi: 10.1021/ja051603j. [DOI] [PubMed] [Google Scholar]
- 34.Chaidaroglou A, Brezinski DJ, Middleton SA, Kantrowitz ER. Biochemistry. 1988;27:8338–8343. doi: 10.1021/bi00422a008. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien PJ, Herschlag D. Biochemistry. 2002;41:3207–3225. doi: 10.1021/bi012166y. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien PJ, Herschlag D. J Am Chem Soc. 1999;121:11022–11023. [Google Scholar]
- 37.Knowles JR. Science. 1987;236:1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- 38.Wells JA. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 39.Mildvan AS, Weber DJ, Kuliopulos A. Arch Biochem Biophys. 1992;294:327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- 40.Mark AE, Vangunsteren WF. Journal of Molecular Biology. 1994;240:167–176. doi: 10.1006/jmbi.1994.1430. [DOI] [PubMed] [Google Scholar]
- 41.Plapp BV. Meth Enzymol. 1995;249:91–119. doi: 10.1016/0076-6879(95)49032-9. [DOI] [PubMed] [Google Scholar]
- 42.Licata VJ, Ackers GK. Biochemistry. 1995;34:3133–3139. doi: 10.1021/bi00010a001. [DOI] [PubMed] [Google Scholar]
- 43.Dill KA. J Biol Chem. 1997;272:701–704. doi: 10.1074/jbc.272.2.701. [DOI] [PubMed] [Google Scholar]
- 44.Kraut DA, Carroll KS, Herschlag D. Annu Rev Biochem. 2003;72:517–571. doi: 10.1146/annurev.biochem.72.121801.161617. [DOI] [PubMed] [Google Scholar]
- 45.Albeck S, Unger R, Schreiber G. J Mol Biol. 2000;298:503–520. doi: 10.1006/jmbi.2000.3656. [DOI] [PubMed] [Google Scholar]
- 46.Hengge AC. Acc Chem Res. 2002;35:105–112. doi: 10.1021/ar000143q. [DOI] [PubMed] [Google Scholar]
- 47.Williams A. Acc Chem Res. 1984;17:425–430. [Google Scholar]
- 48.Williams A. Adv Phys Org Chem. 1992;27:1–55. [Google Scholar]
- 49.Phenyl phosphorothioate monoesters were used instead of phenyl phosphate monoesters because the chemical step is not rate-determining for AP-catalyzed phenyl phosphate hydrolysis.35
- 50.Hollfelder F, Herschlag D. Biochemistry. 1995;34:12255–12264. doi: 10.1021/bi00038a021. [DOI] [PubMed] [Google Scholar]
- 51.Nikolic-Hughes I, Rees DC, Herschlag D. J Am Chem Soc. 2004;126:11814–11819. doi: 10.1021/ja0480421. [DOI] [PubMed] [Google Scholar]
- 52.Hengge AC, Edens WA, Elsing H. J Am Chem Soc. 1994;116:5045–5049. [Google Scholar]
- 53.Holtz KM, Catrina IE, Hengge AC, Kantrowitz ER. Biochemistry. 2000;39:9451–9458. doi: 10.1021/bi000899x. [DOI] [PubMed] [Google Scholar]
- 54.Hoff RH, Larsen P, Hengge AC. J Am Chem Soc. 2001;123:9338–9344. doi: 10.1021/ja0163974. [DOI] [PubMed] [Google Scholar]
- 55.Murphy JE, Stec B, Ma L, Kantrowitz ER. Nat Struct Biol. 1997;4:618–622. doi: 10.1038/nsb0897-618. [DOI] [PubMed] [Google Scholar]
- 56.Strater N, Lipscomb WN, Klabunde T, Krebs B. Angew Chem Int Ed Engl. 1996;35:2024–2055. [Google Scholar]
- 57.Wilcox DE. Chem Rev. 1996;96:2435–2458. doi: 10.1021/cr950043b. [DOI] [PubMed] [Google Scholar]
- 58.Hanson SR, Best MD, Wong CH. Angew Chem Int Ed Engl. 2004;43:5736–5763. doi: 10.1002/anie.200300632. [DOI] [PubMed] [Google Scholar]
- 59.Galperin MY, Jedrzejas MJ. Proteins. 2001;45:318–324. doi: 10.1002/prot.1152. [DOI] [PubMed] [Google Scholar]
- 60.Because the KIEs report on changes in stretching, bending, and torsional vibrational modes, it is difficult to draw conclusions about the nonbridging P-O bond order in the transition state. While it is possible that the increased inverse effect on the nonbridge KIE reflects changes in the nature of the transition state in the AP active site relative to that in solution, prior LFER studies provide no indication of such a change.35, 36, 50, 51, 53 We therefore suggest that the increased inverse effect in the enzymatic reaction arises from the interaction between a nonbridging oxygen atom and the Zn2+ ions.
- 61.See O'Brien and Herschlag, 1998,21 and Table 2.
- 62.We have also measured the 18O nonbridging isotope effect for AP-catalyzed p-nitrophenyl phosphorothioate (pNPPS) hydrolysis. The observed KIE of −2% is significantly more inverse than the value of 1% observed for the nonenzymatic hydrolysis reaction.64, 68 This observation support the conclusion that there is a strong interaction between the bimetallo site and a nonbridging oxygen atom for good substrates of AP, such as phosphate and phosphorothioate monoesters. The KIEs for AP-catalyzed pNPPS hydrolysis will be discussed extensively elsewhere.64
- 63.Hoff RH, Wu L, Zhou B, Zhang ZY, Hengge AC. J Am Chem Soc. 1999;121:9514–9521. [Google Scholar]
- 64.Zalatan JG, Catrina IE, Mitchell R, Grzyska PK, O'Brien PJ, Herschlag D, Hengge AC. 2007 doi: 10.1021/ja072196+. Manuscript in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibby SG, Younker JM, Hengge AC. J Phys Org Chem. 2004;17:541–547. [Google Scholar]
- 66.Bigeleisen J, Wolfsberg M. Adv Chem Phys. 1958;1:15–76. [Google Scholar]
- 67.Caldwell SR, Raushel FM, Weiss PM, Cleland WW. Biochemistry. 1991;30:7444–7450. doi: 10.1021/bi00244a011. [DOI] [PubMed] [Google Scholar]
- 68.Catrina IE, Hengge AC. J Am Chem Soc. 2003;125:7546–7552. doi: 10.1021/ja0340026. [DOI] [PubMed] [Google Scholar]
- 69.Jarvis JAJ. Acta Cryst. 1953;6:327–330. [Google Scholar]
- 70.Truter MR. Acta Cryst. 1958;11:680–685. [Google Scholar]
- 71.McDonald WS, Cruickshank DW. Acta Cryst. 1971;B27:1315–1319. [Google Scholar]
- 72.Czyryca PG, Hengge AC. Biochim Biophys Acta. 2001;1547:245–253. doi: 10.1016/s0167-4838(01)00191-1. [DOI] [PubMed] [Google Scholar]