Abstract
Fibroblasts, when plated on the extracellular matrix protein fibronectin (FN), rapidly spread and form an organized actin cytoskeleton. This process is known to involve both the central α5β1 integrin-binding and the C-terminal heparin-binding regions of FN. We found that within the heparin-binding region, the information necessary for inducing organization of stress fibers and focal contacts was located in a 29–amino acid segment of FN type III module 13 (III13). We did not find a cytoskeleton-organizing role for repeat III14, which had previously been implicated in this process. Within III13, the same five basic amino acids known to be most important for heparin binding were also necessary for actin organization. A substrate of III13 alone was only weakly adhesive but strongly induced formation of filopodia and lamellipodia. Stress fiber formation required a combination of III13 and III7–11 (which contains the integrin α5β1 recognition site), either as a single fusion protein or as separate polypeptides, and the relative amounts of the two binding sites appeared to determine whether stress fibers or filopodia and lamellipodia were the predominant actin structures formed. We propose that a balance of signals from III13 and from integrins regulates the type of actin structures assembled by the cell.
INTRODUCTION
Cells encountering the extracellular matrix protein fibronectin (FN)1 adhere, spread, form an organized cytoskeleton, and activate biochemical pathways that allow cell cycle progression and prevent apoptosis (for review, see Zigmond, 1996; Assoian, 1997; Frisch and Ruoslahti, 1997; Schwartz, 1997). It is clear that the interaction of receptors of the integrin family with FN, although largely responsible for the adhesion of cultured cells to FN, is not sufficient to produce the entire cellular response. In particular, cytoskeletal organization and the formation of sites of cytoskeleton–extracellular matrix connections called focal contacts depend not only on the α5β1 integrin-binding RGD site in type III modules 9 and 10 but also on a second region, the C-terminal heparin-binding domain. Fibroblasts attached to FN fragments that contain an integrin-binding site but lack the heparin-binding domain fail to spread and form focal contacts (Izzard et al., 1986; Woods et al., 1986), and NIH3T3 cells on a similar substrate only partially activate focal adhesion kinase (Guan et al., 1991). The heparin-binding domain is also required for maximal motility of several cell types (Yoneda et al., 1995; Kapila et al., 1997).
The actin-organizing elements within the heparin-binding domain, which consists of type III repeats 12–14, are not well defined, and a cell surface receptor responsible for actin organization has not been identified. The heparin-binding domain has been shown to harbor binding activity for proteoglycans, molecules covalently modified with long chains of sulfated disaccharides structurally similar to heparin, including the heparan sulfate proteoglycan (HSPG) syndecan-1 (Saunders and Bernfield, 1988; Elenius et al., 1990) and the chondroitin sulfate proteoglycan CD44 (Jalkanen and Jalkanen, 1992). Based on sequence predictions, five basic peptides derived from clusters of basic residues in III12–III14 have been found to bind heparin and to support adhesion and spreading in various cell types (McCarthy et al., 1988; Iida et al., 1992; Mooradian et al., 1992, 1993; Huebsch et al., 1995). However, most of these peptides have not been tested in the context of a folded domain. Three of them are derived from module III14, which by itself binds heparin only weakly at physiological ionic strength (Ingham et al., 1993). Mutational studies of larger recombinant FN fragments have localized the principal heparin-binding site to III13 (Barkalow and Schwarzbauer, 1991) and, in particular, to a cluster of six basic residues in III13 (Busby et al., 1995).
Experiments with the heparin-binding peptides and recombinant FN fragments have led to different conclusions as to the importance of heparin-binding domain elements in cytoskeletal organization. Yoneda et al. (1995) demonstrated that recombinant FN fragments encompassing type III repeats 8–10 followed by III13 could support spreading and focal contact formation in HT1080 human fibrosarcoma cells, whereas similar recombinant fragments incorporating III12 or III14 in place of III13 could not. The effects of III13 were only observed when it was in the same molecule as the integrin-binding region; mixed substrates of III8–10 and III13 were ineffective. By contrast, Woods et al. (1993) demonstrated that a heparin-binding peptide from III14, with amino acid sequence PRARI, had potent focal contact-inducing activity when applied in soluble form to human embryo fibroblasts already adherent to the cell-binding domain of FN, whereas four other heparin-binding peptides from elsewhere in the heparin-binding domain, including III13, had substantially weaker activity. Similarly, Huebsch et al. (1995) found that endothelial cells formed stress fibers on coverslips coated with the PRARI peptide but not with the other four heparin-binding peptides, and Huhtala et al. (1995) observed focal contact formation in fibroblasts on mixtures of the cell-binding domain of FN and PRARI peptides. Again, the PRARI peptide has not been tested in the context of a folded domain. Thus, the reasons for the conflicting data concerning both the location of the focal contact-inducing site and the constraints on the molecular context in which this site is presented are not clear. In the present study we sought to clarify both of these issues. Using bacterially expressed FN fragments, we have localized the actin-organizing site in FN to the same set of basic amino acids that bind heparin in III13 (Busby et al., 1995) and have found that cells form filopodia and lamellipodia within minutes of contacting this site. We have failed to identify a function for intact III14 or for the PRARI sequence contained within III14.
MATERIALS AND METHODS
Cells and Reagents
MG-63 (American Type Culture Collection [ATCC], Rockville, MD; CRL-1427), Rat1 (Freeman et al., 1973), and normal rat kidney (NRK) cells (ATCC CRL-6509) were maintained in Dulbecco’s modified Eagle’s medium (DMEM)-HG with 10% FBS (Hyclone, Logan, UT). NIH3T3 cells (ATCC CRL-1658) were maintained in DMEM-HG containing 10% calf serum (JRH Biosciences, Lenexa, KS). The 120-kDa fragment of FN was purified from a chymotryptic digest of human plasma FN (New York Blood Center, New York, NY) by passage of the digest over gelatin-Sepharose and heparin-Sepharose columns (Pharmacia, Piscataway, NJ) followed by size fractionation on Sephacryl S-200 (Pharmacia).
Recombinant FN Fragments
Clones were constructed by standard molecular biological methods (Sambrook et al., 1989; Ausubel et al., 1991). The expression vector pGH for production of fusion proteins containing an N-terminal GST domain and a C-terminal His6 tag was derived from pGEX-2T (Pharmacia) by insertion of an adaptor encoding a His6 tag (5′-GATCCCCCGGGCATCACCATCACCATCACT-3′ annealed to 5′-AATTAGTGATGGTGATGGTGATGCCCGGGG-3′) between the BamHI and EcoRI sites of pGEX-2T (Figure 1A). The vector pGH.PL was generated by insertion of a short linker with NsiI, XbaI, and KpnI sites between the BamHI and SmaI sites of pGH (Figure 1A). Fusion clones containing rat FN type III repeats 7–15 (pGH.F7–15) were generated by cloning a 2.4-kb HincII fragment from a rat plasma FN cDNA (Guan et al., 1990) into the SmaI site of pGH (Figure 1B). pGH.F7–11, a clone containing type III repeats 7–11, was obtained by insertion of a 1.4-kb BamHI–PstI fragment from pGH.F7–15 into pGH.PL digested with BamHI and NsiI. Clones containing type III repeats 7–11 followed by an additional type III repeat (e.g., pGH.F7–11,13; Figure 1B) were generated by amplification of the additional type III repeat by the PCR, digestion with XbaI and KpnI, and insertion into the corresponding sites in pGH.F7–11. Clones containing a single domain 13 or 14 were constructed by cloning a single-domain cassette into the XbaI and KpnI sites of pGH.PL. The following primers were used for amplification of type III repeats from rat FN (lower case indicates added cloning site; upper case indicates FN sequence): for III12, FN12N (5′-gctctagaGCCATTCCTGCGCC-3′) and FN12C (5′-ggggtaccTCCAGAGTCGTGAC-3′); for III13, FN13N (5′-gctctagaAATGTCAGCCCTCCAA-3′) or alternately FN13NB (5′-gctctagaAATGTCAGCCCACCA-3′) and FN13C (5′-ggggtaccGTGGAGGCATCAATGAC-3′); and for III14, FN14N (5′-gctct-agaGCCATTGATGCCCCA-3′) and FN14C (5′-ggggtaccGTCTTTTTCCTCCCAAT-3′). Rat and human III13 were equivalent in activity in preliminary experiments (our unpublished results), and human III13 (from Busby et al., 1995) was used in the F7–11,13 construct described in this paper.
Figure 1.
GST–His6 vectors and FN fusion proteins. (A) Polycloning regions of the vectors pGH and pGH.PL. DNA sequences, shown grouped into codons, begin after the last codon of the GST coding region and end at the TAA stop codon that formed part of the EcoRI site of pGEX-2T. Restriction sites and the sequence encoding the His6 tag are indicated. (B) Schematic drawing of FN and recombinant GST-FN-His6 fusion proteins. Small rectangles, FN type I repeats; open circles, FN type II repeats; squares, FN type III repeats. Important FN sites are denoted by filled symbols: circles, RGD sequence; arrowhead pointing up, heparin-binding site; arrowhead pointing down, PRARI; shaded area, V region.
Cassettes containing rat type III repeats EIIIB and III10 were obtained from GST fusion clones described by Peters et al. (1995) by digestion with EcoRI and BamHI (partial with BamHI for EIIIB and partial with EcoRI for III10) and ligation into the BamHI and EcoRI sites of pBluescript II SK (Stratagene, La Jolla, CA). Single-domain GST–His6 fusions were generated by cloning XbaI–EcoRV fragments from these cassettes into the XbaI and SmaI sites of pGH.PL, and pGH.F7–11 clones containing these cassettes were constructed by cloning SpeI–MluI fragments from the resulting clones into pGH.F7–11 digested with XbaI and MluI.
Mutant III13 domain cassettes were generated by PCR amplification of maltose-binding protein fusions to human III13 (Busby et al., 1995) using primers FN13N or FN13NB and FN13C. Double mutant combinations were made by PCR amplification of two overlapping fragments from each of the single mutants, using the primer sets FN13NB and FN12/13-12 (5′-GGTGGTCTCAGTAG-3′) for the 5′ portion and FN12/13-11 (5′-CTACTGAGACCACC-3′) and FN13C for the 3′ portion. The purified products, which overlap in a 14-bp region between codons 13 and 17, were mixed, and a full-length PCR product was amplified with primers FN13NB and FN13C. The structures of these clones and all others generated by PCR or site-directed mutagenesis (below) were confirmed by DNA sequencing.
Chimeras between rat III12 and human III13 were made by PCR of III12 and III13 templates to generate fragments with overlapping ends at the III12–III13 junction, which were then mixed and reamplified with flanking primers. Primers were as follows, with sequences from III12 in lower case, from III13 in upper case, and from both in italics: B/C strand junction (see Figure 6A for domain structure), 12/13-1 (5′-TGAGACGATCACTGGCTaccgagtg-3′) and its complement, 12/13-2; C/D strand junction, 12/13-3 (5′-ggagaagacaggaCCAATCCAGAGAACCATC-3′) and its complement, 12/13-4; and E/F strand junction, 12/13-5 (5′-GTCAGAAGCTACACCATctcagggctcatggtgg-3′) and its complement, 12/13-6. Chimera structures are shown in Figure 6B. 12(13AB+loop) was generated by amplifying a III13 template with FN13N and 12/13-2 and a III12 template with 12/13-1 and FN 12C and then mixing the two purified products and amplifying with FN13N and FN12C. 12(13AB+loop,DE) was constructed in three stages. First, a III13 template was amplified either with FN13N and 12/13-2 to generate fragment A or with 12/13-3 and 12/13-6 to generate fragment D, and a III12 template was amplified either with 12/13-1 and 12/13-4 to generate fragment H or with 12/13-5 and FN12C to generate fragment E. Second, fragments A and H were mixed and amplified with 12/13-3 and FN12C, and fragments D and E were mixed and amplified with FN13N and 12/13-4. Third, the resulting products were mixed and amplified with FN13N and FN12C to yield the full-length chimera. Other chimeras were built by a similar strategy.
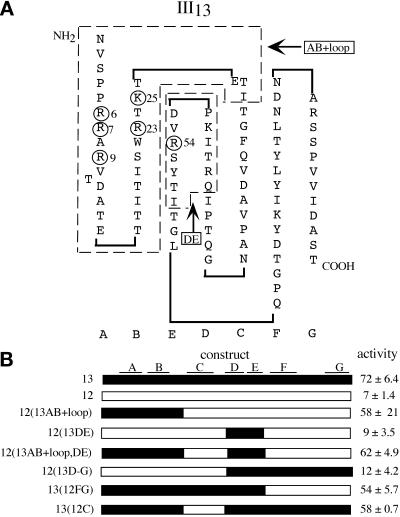
Figure 6.
The N-terminal 29 amino acids of repeat III13 confer stress fiber–inducing activity on repeat III12. (A) Schematic drawing of repeat III13, based on the crystal structure of repeat III10 (Dickinson et al., 1994) and confirmed by the recently solved crystal structure of the heparin-binding domain (Sharma et al., 1999). Beta strands A–G are shown below the domain. The heparin-binding amino acids R6, R7, R9, R23, K25, and R54 are circled (Busby et al., 1995). Dashed lines show the parts of III13 used in III12/III13 chimeras, designated AB + loop and DE. (B) Schematic drawing of III12/III13 chimeras, showing regions of III13 in black and III12 in white. Beta strands are drawn above. The percentages of cells with organized stress fibers when plated on F7–11, III12/III13 chimeras at 9 μg/ml (mean ± SD) are shown at right.
The R6T/R7 M double mutation in rat FN was derived from clone MSV-SF-diS RR>TM, a gift from Dr. Jean Schwarzbauer (Princeton University, Princeton, NJ) (Barkalow and Schwarzbauer, 1991). A 1.05-kb BglII–SacI fragment from this clone was used to replace the equivalent sequence in pGH.F7–15, and the resulting mutant III13 cassette was PCR amplified and cloned into pGH.F7–11,13 as described above. A clone with rat III13 preceding domains III7–III11, called pGH.F13,7–11, was constructed by ligation of a fragment of pGH.F7–11 digested with BamHI, blunted with Klenow fragment, and digested with MluI into pGH.F13 digested with MluI and SmaI.
Site-directed mutagenesis of the PRARI sequence in III14 was carried out with the mutagenic oligonucleotide 5′-GGCAGGCACCCAGXGCCAGXATTACTGGCTACATT-3′, in which equal mixtures of A and C were included at the sites marked X to generate a mixture of arginine and serine codons at the two positions. Mutagenesis using uridine misincorporation (Sambrook et al., 1989) was performed on a pBluescript II SK plasmid containing a 143-nucleotide SacI–BamHI FN fragment, and a double mutant SacI BamHI fragment was ligated together with 4.1-kb MluI–SacI and 3.1-kb MluI–BamHI fragments of pGH.F7–15 to generate the mutant plasmid pGH.F7–15(14SS). The structure of the reassembled mutant III14 region was confirmed by DNA sequencing.
Production of Fusion Proteins
Expression of fusion proteins in Escherichia coli strain DH5α grown to late log phase in Luria–Bertani medium and 100 μg/ml ampicillin was induced with 0.33–1 mM isopropyl-β-d-thiogalactopyranoside (Boehringer Mannheim, Indianapolis, IN) for 4 h. Cells were harvested by centrifugation and stored at −80°C in PBS containing 2 mM PMSF (Sigma, St. Louis, MO). Bacterial pellets were thawed in water at 4–8°C, and the buffer was adjusted to 50 mM sodium phosphate, pH 8.0, 10 mM imidazole, 300 mM NaCl, 2 mM PMSF, 1:100 aprotinin stock (Sigma), and 12.5 μg/ml leupeptin (Sigma). Pellets were incubated with lysozyme (2 mg/ml; Sigma) for 30 min at 4°C, lysed by sonication, brought to 1% Triton X-100 (Sigma), and centrifuged at 28,000 × g for 20 min. Lysates were incubated with TALON resin (Clontech, Palo Alto, CA) for 1 h at room temperature, followed by washing in 10 vol of wash buffer 1 (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 10 mM imidazole), 10 vol of wash buffer 2 (wash buffer 1 with 10% glycerol), and 20 vol of wash buffer 1. Proteins were eluted with 6 vol of elution buffer (wash buffer 1 adjusted to 150 mM imidazole) and dialyzed against CAPS buffer (20 mM 3-[cyclohexylamino]-1-propanesulfonic acid, pH 11, 150 mM NaCl, and 2 mM EDTA). In several experiments, proteins were then bound to glutathione-agarose columns (Sigma) in PBS containing 0.2% Tween 20 and protease inhibitors, washed in PBS and 2 mM PMSF, and eluted in 10 mM glutathione, 50 mM Tris, pH 8.0, and 2 mM PMSF. The second purification was usually omitted, because it reduced protein yield without apparent improvement in protein specific activity.
Actin Assembly Assays
Substrates were prepared by coating 12-mm glass coverslips (Fisher, Scientific, Pittsburgh, PA) in a 24-well plate with 40-μl drops of FN fusion proteins in CAPS buffer overnight at 4°C. Coverslips were washed three times in PBS, blocked for 2 h at 37°C with 2 mg/ml BSA (Calbiochem, San Diego, CA) that had been treated at 70°C for 2 h, and washed three times in PBS before plating cells. Substrates to be treated with heparin were incubated with 100 μg/ml heparin (from porcine intestinal mucosa; Sigma) for 1 h at 37°C before plating cells, and cells were plated in the presence of heparin. Near-confluent MG-63 cells were treated with 2.5 μg/ml cycloheximide (CHX; Sigma) for 2 h, released from their dishes with versene (0.02% EDTA in buffered saline), washed in serum-free DMEM-HG containing 2.5 μg/ml CHX (SFM/CHX), and allowed to recover in SFM/CHX for 30 min at 37°C. Five thousand cells per well were added, and the plates were brought momentarily to 188 × g in a swinging bucket centrifuge to begin adhesion. Cells were incubated for 2 h at 37°C, washed gently three times with PBS, fixed for 10 min in 4% paraformaldehyde in PBS, washed, permeabilized for 15 min in 0.5% NP-40 or IGEPAL CA-630 (Sigma) in PBS, washed, and stained for 30 min in rhodamine– or Texas Red–conjugated phalloidin (Molecular Probes, Eugene, OR; or Sigma). Coverslips were coded, mounted on slides in Gelvatol (Monsanto, St. Louis, MO) with 1,4-diazabicyclo [2.2.2]octane (Sigma), and scored under epifluorescence optics on a Zeiss (Thornwood, NY) Axiophot microscope. Samples were scored blind, with at least 100 cells per coverslip scored as having either well-organized or poorly organized actin filaments. To avoid the influence of cell–cell contacts, only isolated cells were scored. Some samples were stained with mAbs to vinculin (VIN-11-5; Sigma), phosphotyrosine (PY99; Santa Cruz Biotechnology, Santa Cruz, CA), talin (clone 8d4; Sigma), or paxillin (clone 349; Transduction Laboratories, Lexington, KY) and rhodamine- or FITC-conjugated anti-mouse antibody (Tago, Camarillo, CA).
Actin assembly in Rat1 cells was assayed according to the procedure of Clark et al. (1998), similar to the protocol described for MG-63 cells, except that Rat1 cells were starved in serum-free DMEM-HG for 16 h before each experiment, released from dishes with 0.00625% trypsin and EDTA instead of EDTA alone, and washed in 0.5 mg/ml soybean trypsin inhibitor in SFM (Fluka, Milwaukee, WI). CHX was omitted from these experiments, because Rat1 cells spread poorly in the presence of the drug.
Adhesion Assays
Cell adhesion to glass coverslips coated with 80-μl drops of substrates was measured by plating 12,500 cells per coverslip, starting adhesion with a brief spin at 188 × g, and gentle washing and fixation as described above after 15 min at 37°C. Cells were permeabilized, stained with 25 ng/ml DAPI (Sigma) and TRITC-phalloidin (for scoring filopodia) in PBS for 30 min at 37°C, and the coverslips were washed and mounted as described above. DAPI-stained cells were counted in video images of five fields for each sample by Macintosh computer using Scion Image, a derivative of the public domain NIH Image program (developed at the National Institutes of Health, Bethesda, MD, and available on the Internet at http://rsb.info.nih.gov/nih-image/).
Type III Structural Analysis
Folding of type III repeats was measured by monitoring the ratio of fluorescence at 350 nm to that at 320 nm with excitation at 280 nm as the samples were slowly heated and cooled, as described in Busby et al. (1995). Type III repeats were purified from GST–type III–His6 fusions in E. coli lysates by binding to TALON columns as described above, followed by washing with thrombin buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 2.5 mM CaCl2) and cleavage of GST with 1.3% (wt/wt) thrombin (Sigma) for 1.5 h at 37°C. Type III repeats were eluted from TALON columns as described above, and residual uncleaved protein and free GST were removed by passage of the eluate over a glutathione-agarose column, followed by dialysis against Tris-buffered saline (150 mM NaCl and 20 mM Tris, pH 7.4).
RESULTS
Heparin Interferes with Stress Fiber Formation on FN
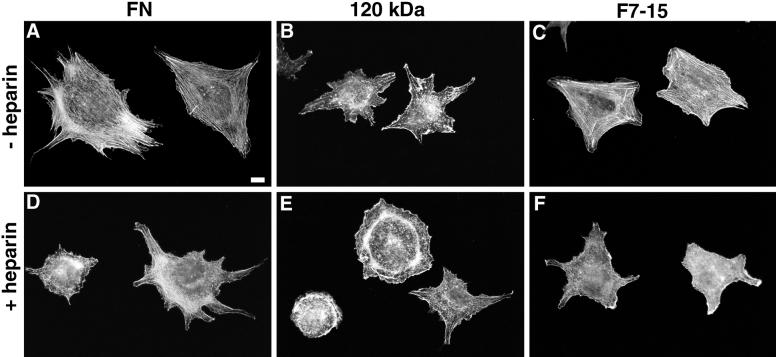
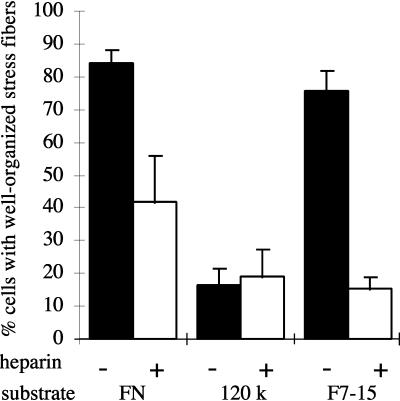
Previous published work suggested a role for the FN heparin-binding domain in spreading and focal contact formation. To determine whether the heparin-binding function of this domain (and hence its likely ability to bind HSPGs) is involved in organizing cellular morphology, we tested the role of heparin as a competitor for endogenous HSPGs in inducing cytoskeletal organization in MG-63 cells, a human osteosarcoma cell line. Cells plated on FN for 2 h in the absence of heparin spread well and formed an extensive array of parallel actin stress fibers (Figure 2A), whereas cells plated on FN in the presence of 100 μg/ml heparin were more poorly spread and had few stress fibers (Figure 2D). Instead, these cells contained short, irregular actin filaments extending upward from the ventral surface of the cell. This morphology resembled that of cells plated on the 120-kDa fragment of FN, which lacks the heparin-binding domain (Figure 2B). Heparin did not appear to affect the actin organization in cells plated on the 120-kDa FN fragment (Figure 2E). A quantitative index of cytoskeletal organization, the percentage of cells able to form well-organized stress fibers (Figure 3), showed that the morphological differences shown in Figure 2 are representative of the population. NIH3T3 and NRK cells responded to heparin, FN, and the 120-kDa domain in a manner similar to MG-63 cells (our unpublished results). The ability of heparin to inhibit stress fiber formation suggested that the heparin-binding function of the heparin-binding domain is involved in inducing stress fiber formation, or that the heparin-binding site is sufficiently close to the actin-organizing site to allow competition.
Figure 2.
Actin organization in MG-63 cells on FN and recombinant fragments in the presence or absence of heparin. Cells were plated for 2 h on 20 μg/ml FN (A and D), 10 μg/ml 120-kDa fragment of FN (B and E), which contains the central cell-binding domain but not the heparin-binding domain, or 10 μg/ml F7–15 (C and F), a GST–FN–His6 fusion containing both the cell-binding and heparin-binding domains. Rhodamine–phalloidin-stained cells without heparin (A–C) or with 100 μg/ml heparin (D–F) applied to substrates before plating and to cells during incubation. Cells in A and C are representative of cells with well-organized stress fibers in quantitative stress fiber assays. Bar, 10 μm.
Figure 3.
Heparin inhibits stress fiber formation in MG-63 cells on FN or a recombinant FN fragment. Cells were plated for 2 h on FN, 120-kDa fragment, or F7–15 as described in Figure 2. The percentage of cells with well-organized stress fibers (see Figure 2, A and C) was scored (mean ± SD for duplicate samples).
Bacterially Expressed FN Fragments Can Support Stress Fiber Formation Equivalent to That Supported by Intact FN
To determine the precise location of the site in the heparin-binding domain active in stress fiber and focal contact formation, we developed a set of bacterially expressed FN fusion proteins, in which sequences in the heparin-binding domain could be easily manipulated. We produced proteins with an N-terminal GST domain and a C-terminal His6 tag to allow two types of affinity purification (Figure 1). The fusion F7–15 contained FN type III repeats 7–15, which include sites for α5β1 integrin binding (the sequence RGD in repeat III10 and the synergy site in repeat III9 needed for high-affinity binding) and the heparin-binding domain, type III repeats 12–14. MG-63 cells spread well and formed stress fibers to the same extent on human pFN and bacterially expressed F7–15 (Figures 2C, 3, and 4A′), as did NIH3T3 cells (our unpublished results). Cells plated on F7–15 in the presence of 100 μg/ml heparin formed few stress fibers in a manner similar to cells on FN in the presence of heparin (Figures 2F and 3). Cells adhering to the fusion protein F7–11, which contained type III repeats 7–11 but lacked the heparin-binding domain, appeared similar to cells on the 120-kDa proteolytic fragment of FN: cells adhered to this protein but formed few well-organized stress fibers or focal contacts (Figures 2B and 4, B and B′). These observations demonstrated that recombinant F7–15 produced in bacteria contains all of the heparin-sensitive FN elements needed for cytoskeletal organization, presumably within type III repeats 12–15.
Figure 4.
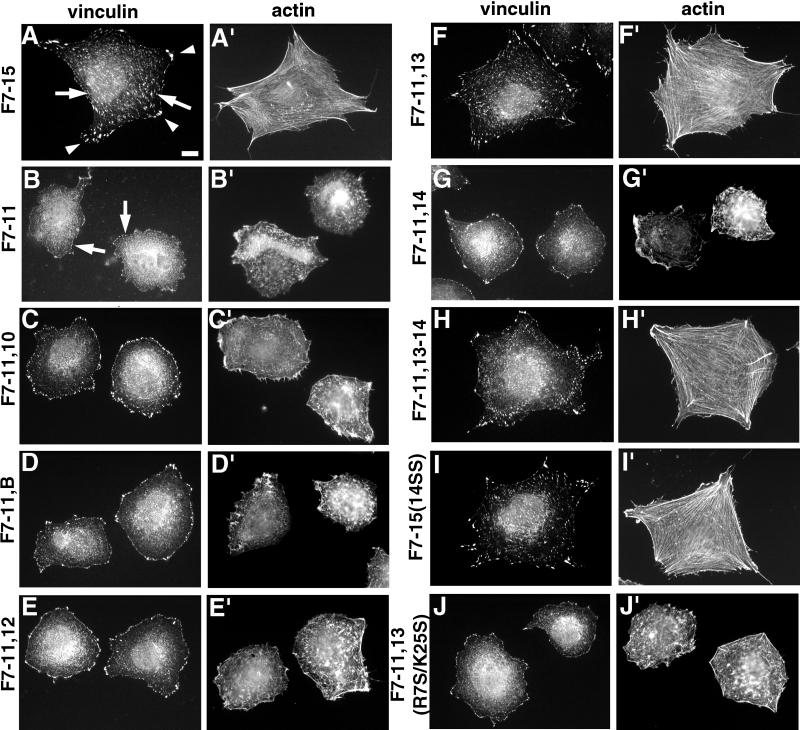
Repeat III13, together with repeats III7–11, supports full stress fiber and focal contact formation. Cells were plated for 2 h on a 7.5 μg/ml concentration of each substrate and then fixed and stained for vinculin (A–J) and actin (A′–J′). (A and A′) F7–15; (B and B′) F7–11; (C and C′) F7–11,10; (D and D′) F7–11, EIIIB; (E and E′) F7–11,12; (F and F′) F7–11,13; (G and G′) F7–11,14; (H and H′) F7–11,13–14; (I and I′) F7–15(14SS); (J and J′) F7–11,13(R7S/K25S) (see text below). Arrowheads in A, representative focal contacts at cell apices; arrows in A, representative internal focal contacts; arrows in B, representative peripheral vinculin patches in cells with poor actin organization. Cells in A, F, H, and I are representative of cells scored as having well-organized focal contacts. Bar, 10 μm.
FN Repeat III13 Contains the Stress Fiber– and Focal Contact–inducing Activities of the FN Heparin-binding Domain
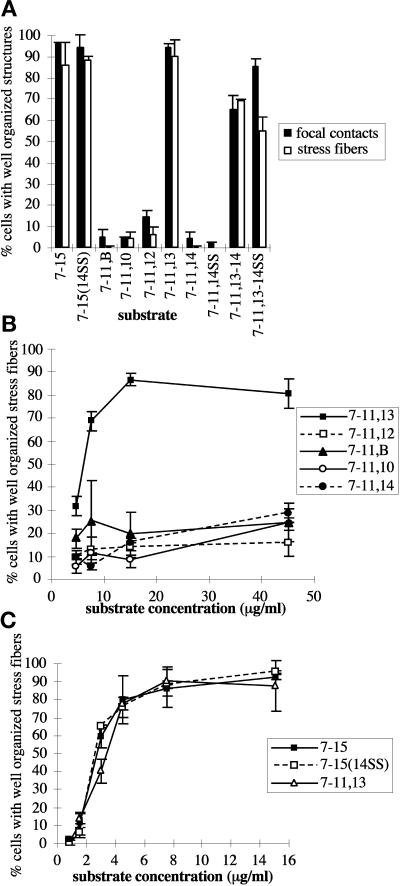
Although several peptides representing sequences throughout the heparin-binding domain can bind heparin in solid-phase assays (McCarthy et al., 1988; Iida et al., 1992; Mooradian et al., 1992), experiments with intact FN type III repeats have identified a region of noncontiguous sequence in III13 that accounts for the heparin-binding activity of the heparin-binding domain (Busby et al., 1995). Consistent with this observation, Yoneda et al. (1995) showed that fusions of FN repeats III8–10 to III13 were sufficient for induction of focal contacts in HT1080 fibrosarcoma cells, whereas fusions of III8–10 to III12 or III14 were not. We confirmed this observation with GST-His6 fusions. F7–11,13 was able to support formation of stress fibers and focal contacts comparable with those observed in cells on F7–15 or rat pFN (Figures 4, A, A′, F, and F ′, and 5, A and C).
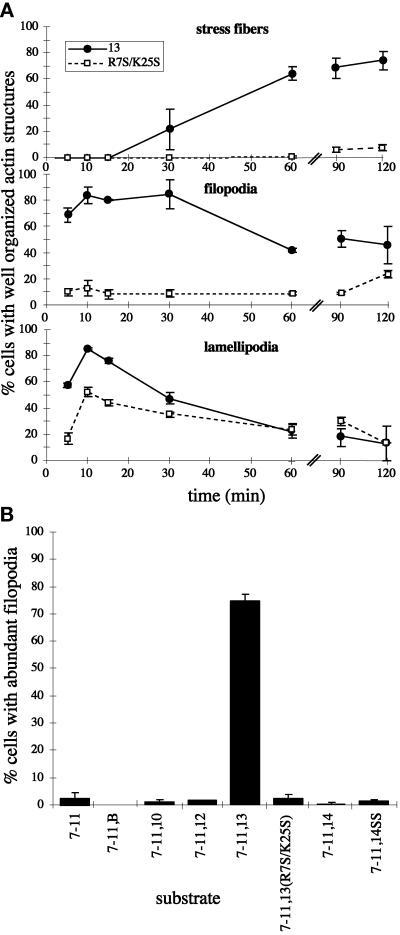
To determine whether the stress fiber–inducing ability of III13 in F7–11,13 is unique to III13, we tested several other type III repeats in the context of similar fusion proteins. GST-His6 fusions of III7–11 to III10, EIIIB, III12, or III14 did not support stress fiber or focal contact formation, in contrast with F7–11,13 (Figures 4 and 5). Cells plated on all of these fusion proteins appeared identical to cells plated on F7–11: cells were rounded and poorly spread, and their actin was present in disorganized short filaments (Figure 4). Vinculin was present in small patches outlining the cells, as were paxillin and phosphotyrosine. F7–11 fusions to type III repeats other than III13 appeared generally inactive even when coated on coverslips at 45 μg/ml, 5- to 10-fold higher than the minimum concentration of F7–11,13 required for significant stress fiber induction (Figure 5B). This minimum concentration and the concentration at which maximal stress fiber induction occurred varied up to twofold between experiments, but the relative activities of the fusion proteins reported in this paper remained consistent (our unpublished results). Rat1 cells responded to type III repeat fusions in a manner similar to that of MG-63 cells (our unpublished results).
Figure 5.
III13 is unique in its stress fiber– and focal contact–inducing ability and is as potent as the entire heparin-binding domain. MG-63 cells were plated on the GST–F7–11, type III–His6 fusions for 2 h, and the proportions of cells with well-organized stress fibers and vinculin-containing focal contacts were scored (mean ± SD for duplicate samples). (A) Stress fiber– and focal contact–organizing activities of F7–11,13 but not other F7–11, type III repeat fusions, are similar to those of F7–15. Cells were plated on 7.5 μg/ml fusion protein for 2 h and scored for distribution of vinculin (black bars) and actin (open bars). (B) Dose dependence of stress fiber–organizing activity of F7–11, type III repeat fusions. 7–11,13 (▪), 7–11,12 (□), 7–11,B (▴), 7–11,10 (○), and 7–11,14 (●) were tested for stress fiber–inducing activity over a range of concentrations. (C) Mutation of the PRARI site in III14 does not alter stress fiber induction by F7–15. F7–15 (▪), F7–15(14SS) (□), and F7–11,13 (▵) were tested for stress fiber–organizing activity over a range of concentrations.
The spatial relationship of III13 to the integrin-binding site of the recombinant FN fragments did not appear to affect its stress fiber induction ability. Constructs in which III13 was fused to the N terminus of F7–11 (F13,7–11) or to the C terminus of F7–11 (F7–11,13) were equally potent in stress fiber induction assays, and mixed substrates of F7–11 and F13 also induced stress fiber formation (see below).
Because the experiments of Woods et al. (1993), Huebsch et al. (1995), and Huhtala et al. (1995) showed that peptides containing a sequence from III14, PRARI, were particularly potent inducers of cytoskeletal organization, and because domain III14 slightly enhances the heparin binding of III13 (Ingham et al., 1993), we examined the role of III14 further. Despite the enhancement of heparin binding in the III13–14 pair, the presence of III14 did not appear to enhance the stress fiber–inducing function of III13. Stress fibers and focal contacts in cells plated on 7.5 μg/ml F7–11,13–14 appeared similar to those in cells on F7–11,13 (Figures 4, F, F′, H, and H′, and 5A). Together with the observation that F7–11,14 had little or no stress fiber or focal contact-inducing activity (Figures 4, G and G′, and 5, A and B), these data suggest that, at least in the context of these fusion proteins, III14 does not play a significant role in MG-63 cytoskeletal organization.
We also examined the role of the PRARI sequence in the context of an intact III14 domain. Because heparin and heparan sulfate binding elsewhere in FN depend on basic amino acids (Busby et al., 1995), we mutated the two arginine residues in the III14 PRARI sequence to serine and tested the resulting III14 mutant, 14SS (which did not appear to alter the structure of III14; see below), in the context of III7–15 and III7–11,13–14. Stress fibers in cells plated on mutant F7–15(14SS) appeared identical to those in cells on wild-type F7–15 (Figures 4, A′ and I′, and 5A) over a range of concentrations (Figure 5C), and focal contacts on mutant and wild-type F7–15 at 7.5 μg/ml were also indistinguishable (Figure 4, A and I). Likewise, the stress fiber–inducing activity of F7–11,13–14 appeared unaffected by the mutation (Figure 5A). These results indicated that an intact PRARI sequence is not necessary for stress fiber induction, at least in the presence of a wild-type III13.
Basic Residues in III13 Required for Heparin Binding Are Also Required for Stress Fiber and Focal Contact Induction
We attempted to identify the active site in III13 by replacing portions of the inactive repeat III12 with corresponding portions of repeat III13 and assaying the resulting F7–11,12/13 chimeras for stress fiber–inducing activity. Because heparin was able to block this function of III13, we focused on six basic residues (arginines 6, 7, 9, 23, and 54 and lysine 25) whose individual mutations were shown previously to diminish heparin binding (Busby et al., 1995). According to their model, these residues, located on β-strands A, B, and E and adjacent loops, form a “cationic cradle” on one face of the III13 module (Figure 6A). The recently solved x-ray crystal structure of III13 confirms this model (Sharma et al., 1999). Chimeras containing 29 amino acids of III13 sequence from the N terminus to the start of strand C (including all heparin-binding residues except the least critical one, arginine 54) replacing the equivalent sequence of III12, called 12(13AB+loop), were nearly as active as wild-type III13 in stress fiber induction, whereas chimeras lacking this sequence were inactive (Figure 6B). Additional III13 sequence in the chimeras (containing arginine 54 in the E strand) did not alter stress fiber–inducing activity, suggesting that all critical residues are within this 29–amino acid region (or that III12 sequences can replace key III13 sequences outside of this region).
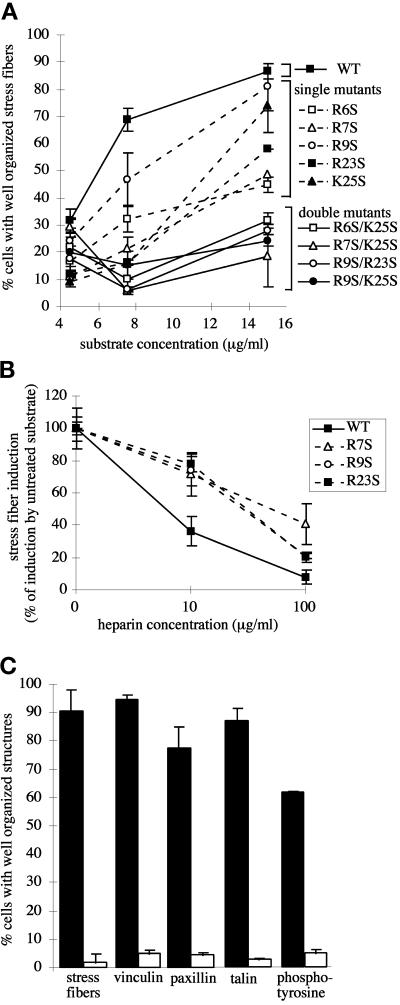
To test the contribution of individual basic residues in the III13 heparin-binding site, we used mutations shown by Busby et al. (1995) to eliminate solution phase heparin binding nearly completely but to leave the structure of III13 intact. We constructed variants of F7–11,13 using the human III13 domain (identical to the rat III13 domain in 85 of 89 residues) containing the single amino acid substitutions R6S, R7S, R9S, R23S, and K25S (Busby et al., 1995). MG-63 stress fiber formation was impaired to varying degrees on each fusion protein containing a mutant domain III13 (Figure 7A). The R9S mutation caused only a slight reduction in stress fiber–inducing function of F7–11,13, whereas R6S, R7S, and R23S caused major reductions. The K25S mutation caused an intermediate level of impairment. The impairment of all single mutants was dependent on substrate concentration (Figure 7A), and at high concentrations, all could support wild-type levels of stress fiber formation (our unpublished results).
Figure 7.
Heparin-binding site mutations reduce stress fiber– and focal contact–inducing activity of III13 and confer partial resistance to heparin. (A) Effects of heparin-binding site mutations on stress fiber formation. MG-63 cells were plated for 2 h on wild-type and mutant F7–11,13 substrates over a range of concentrations, and stress fiber formation was scored (mean ± SD of duplicate samples). ▪, wild-type F7–11,13. Single mutants are indicated by dashed lines: □, R6S; ▵, R7S; ○, R9S; ▪, R23S; ▴, K25S. Double mutants are indicated by solid lines: □, R6S/K25S; ▵, R7S/K25S; ○, R9S/R23S; ●, R9S/K25S. (B). Effects of heparin on stress fiber formation. MG-63 cells were plated for 2 h on 15 μg/ml F7–11,13 fusions carrying wild-type (▪), R7S (▵), R9S (○), or R23S (▪) mutant III13.Substrates were pretreated with 0, 10, or 100 μg/ml heparin, which remained present throughout the experiment. For each substrate, stress fiber induction is shown normalized to induction in the absence of heparin. (C) The R7S/K25S mutation in III13 reduces focal contact organization. MG63 cells plated for 2 h on F7–11,13 (black bars) or F7–11,13(R7S/K25S) (open bars) were fixed and stained for localization of actin, vinculin, paxillin, talin, and phosphotyrosine, and the percentage of cells (±SD) with well-organized focal contacts containing each protein was scored.
Derivatives of F7–11,13 carrying either R7S, R9S, or R23S were partially resistant to the effects of heparin: whereas stress fiber formation on wild-type F7–11,13 was strongly reduced in the presence of 10 μg/ml heparin, stress fiber formation on the single mutant substrates was only partially reduced (Figure 7B). At 100 μg/ml heparin, stress fiber formation on all substrates except F7–11,13(R7S), the most severely impaired of these mutants in the absence of heparin, was virtually eliminated. The inverse relationship between heparin sensitivity and stress fiber induction is consistent with a colocalization of the heparin-binding and stress fiber–inducing sites, with soluble heparin binding to the shared site more weakly than a putative stress fiber-inducing receptor.
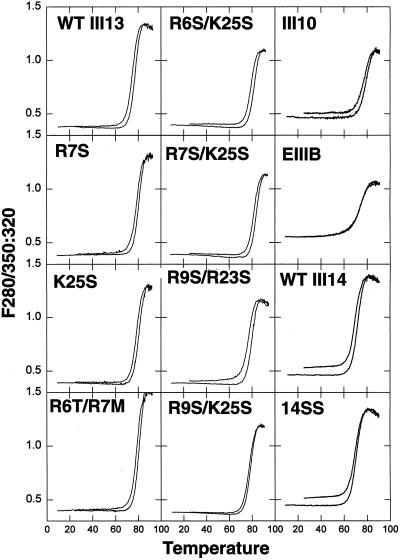
The single point mutations tested reduce but do not eliminate binding of III13 to solid-phase heparin (Busby et al., 1995), suggesting that a substrate of mutant F7–11,13 protein may still retain the ability to bind heparin-like molecules such as cell surface HSPGs, especially at high coating concentrations. To determine whether both heparin-binding and stress fiber–inducing activity could be further reduced, we constructed double-mutant combinations of heparin-binding residue mutations. We measured the thermal stability of purified III13–His6 domains containing these mutations to determine the effects of the mutations on domain structure (Figure 8). Mutant and wild-type domains showed reversible unfolding at 75–85°C, indicating that the domains were stable and unaffected by the presence of the double mutations. Other type III repeats that lacked stress fiber–inducing activity (EIIIB, III10, and wild-type and mutant III14) also showed normal melting profiles.
Figure 8.
Thermal stability of type III repeats lacking stress fiber–inducing activity is normal. Purified mutant III13–His6 and III14–His6 domains showed melting profiles similar to that of their wild-type counterparts, indicating that they are properly folded. Melting properties of purified type III–His6 fusion proteins were determined by heating samples at 1°C/min and measuring the ratio of fluorescence intensity at 350 nm to that at 320 nm with excitation at 280 nm. The upper curve for each sample was obtained during cooling.
All of the double mutant combinations tested in the context of F7–11,13 (R6S/K25S, R7S/K25S, R9S/R23S, and R9S/K25S) were severely defective in stress fiber induction, even at 15 μg/ml (Figure 7A). Cells plated on the F7–11,13 double mutants had short, disorganized actin filaments resembling those on F7–11,12 or other F7–11, inactive type III repeat fusions or heparin-treated intact FN (Figures 2 and 4). Cells plated on F7–11,13(R7S/K25S) also showed poorly organized focal contacts, as judged by the distribution of vinculin, phosphotyrosine, paxillin, and talin (Figures 4J and 7C). We also tested the double mutation R6T/R7 M, described by Barkalow and Schwarzbauer (1991), and found that in the context of either F7–15 or F7–11,13, this mutation abolished the stress fiber– and focal contact–inducing activity of III13 (our unpublished results). The reduction in stress fiber induction caused by single and double III13 mutations generally correlated with the reduction in the heparin-Sepharose binding strength of single mutant III13 domains (Busby et al., 1995) and F7–11,13 fusions (our unpublished results).
Taken together, these results indicated that the stress fiber– and focal contact–inducing activities of domain III13 arise from a portion of the protein coincident with the region required for heparin binding. No single amino acid was essential for stress fiber induction, but mutation of any one of the positively charged residues that contribute to heparin binding caused a measurable decrease in this function, and simultaneous mutation of any two of them nearly abolished it. The largest effects were observed with the residues R6, R7, and R23, located near the center of the cationic cradle (Figure 6A).
III13 Plays Only a Minor Role in Cell Adhesion
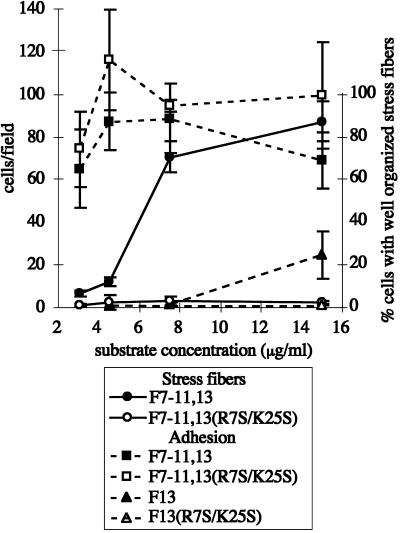
To determine whether the reduction in stress fiber induction caused by the R7S/K25S double mutation was merely a consequence of reduced adhesion to the mutant F7–11,13 protein, we measured the adhesion of cells to glass coverslips coated with fusion proteins over a range of concentrations (Figure 9). We found that, over the range of coating concentrations tested (3–15 μg/ml), F7–11,13(R7S/K25S) supported adhesion similar to that supported by wild-type F7–11,13 (Figure 9). Adhesion did not vary significantly over this range, indicating that wild-type III13 heparin-binding function does not contribute to adhesion to the F7–11,13 molecule. Furthermore, stress fiber induction varied significantly over this concentration range, suggesting that the contribution of III13 to stress fiber induction is not simply a function of the adhesiveness of the protein. We obtained similar results with the set of five single-mutant F7–11,13 constructs (our unpublished results). At or above 15 μg/ml, GST–III13–His6 protein did support a low level of cell adhesion, however, which was eliminated by the R7S/K25S mutation (Figure 9), indicating that an intact heparin-binding, stress fiber–inducing site is weakly adhesive.
Figure 9.
Differences in stress fiber–inducing activity between wild-type and mutant III13 domains do not result from differences in adhesivity. Cells adhering to varying concentrations of F7–11,13 (▪), F7–11,13(R7S/K25S) (□), F13 (▴) or F13(R7S/K25S) (▵) on glass coverslips for 15 min were fixed, stained with DAPI, and counted. Stress fiber formation was scored in a second set of cells adhering for 2 h to glass coverslips coated with F7–11,13 (●) or F7–11,13(R7S/K25S) (○).
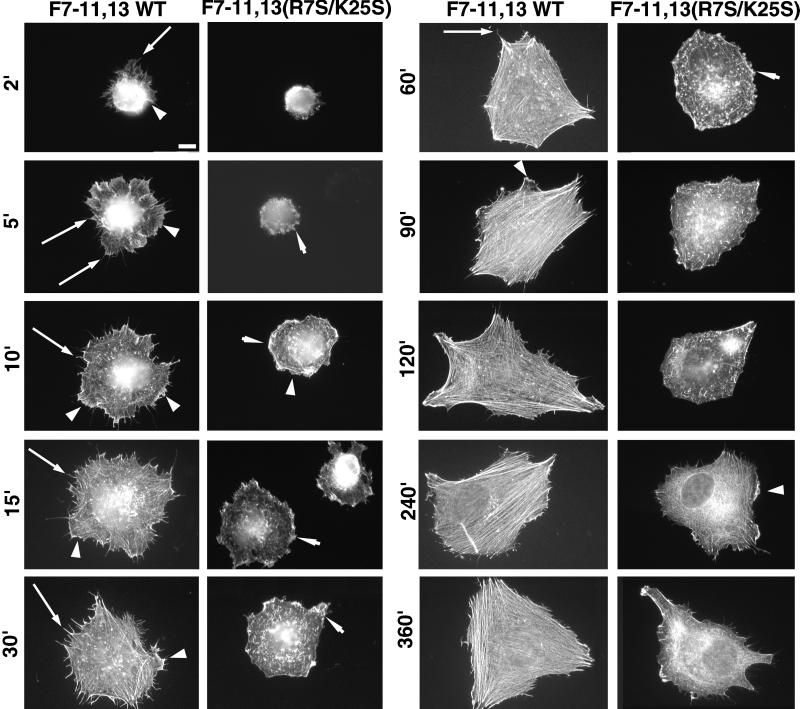
III13 Function Is Required for Early Events in Cytoskeletal Assembly
We investigated the role of III13 in the formation of actin structures over time by plating cells on F7–11,13 fusions containing either wild-type or R7S/K25S mutant III13 and fixing cells over the course of 6 h (Figures 10 and 11). MG-63 cells spreading on wild-type F7–11,13 extended large numbers of filopodia as early as 2 min after plating, and by 5 min, the cell peripheries had broad lamellipodia with ruffles. At 10–15 min after plating, the filopodia extended radially out from the center of the spreading cells, and lamellae occupied the space between the filopodia. Beginning at 30 min, the actin appeared to begin to reorganize from radially directed filopodia to circumferentially oriented parallel bundles, which became oriented along a small number of axes of the cell. Lamellipodia appeared to fill in the spaces between filopodia, and by 60–90 min, only a few prominent radially directed filopodia remained extending out from the apices of the cells.
Figure 10.
Time course of actin assembly of MG-63 cells plated on 7.5 μg/ml F7–11,13 or F7–11,13(R7S/K25S). Cells were fixed at the times indicated and stained with TRITC-phalloidin. Long arrows, representative filopodia; arrowheads, representative lamellipodia extending parallel to the substratum; short arrows, representative ruffles perpendicular to the substratum. Cells shown for wild-type F7–11,13 at 2–30 min are representative of cells scored as having abundant filopodia in quantitative assays. Cells shown for F7–11,13 at 2–15 min are representative of cells with abundant lamellipodia. Bar, 10 μm.
Figure 11.
Wild-type III13 function is necessary during early and late events of actin assembly. (A) Cells were plated on 7.5 μg/ml F7–11,13 or F7–11,13(R7S/K25S) and fixed at intervals and stained with TRITC-phalloidin. The percentages of cells with well-organized stress fibers, abundant filopodia (>10 per cell), or abundant lamellipodia (>25% of the cell perimeter occupied by actin-rich sheets) were scored (±SD, in duplicate samples). ●, F7–11,13; □, F7–11,13(R7S/K25S). (B) Formation of filopodia on recombinant FN fragments. The percentages of cells with abundant filopodia were scored in cells plated for 15 min on 15 μg/ml F7–11,type III repeat substrates.
The profound inhibition of stress fiber induction caused by the R7S/K25S mutation, initially observed at 2 h after plating, persisted until at least 6 h (Figure 10). A similarly profound inhibition of filopodia formation was evident as early as 2 min after plating. Cells on F7–11,13(R7S/K25S) remained rounded with few or no filopodia, although they did spread slowly and develop a few long stress fiber–like actin filaments by 6 h (Figures 10 and 11). Filopodia also failed to form in cells plated on fusions of F7–11 to type III repeats 10, 12, 14, or EIIIB (Figure 11B). The R7S/K25S mutation also impaired lamellipodia formation. Cells spreading on F7–11,13(R7S/K25S) had abundant membrane ruffles at the edge of spreading cells but relatively few large flat lamellipodia extending along the substrate. The effects of the R7S/K25S mutation on actin assembly in Rat1 cells were similar to those observed for MG-63 (our unpublished results).
III13 Induces Formation of Filopodia and Lamellipodia, and Mixtures of III13 and F7–11 Can Induce Formation of Stress Fibers
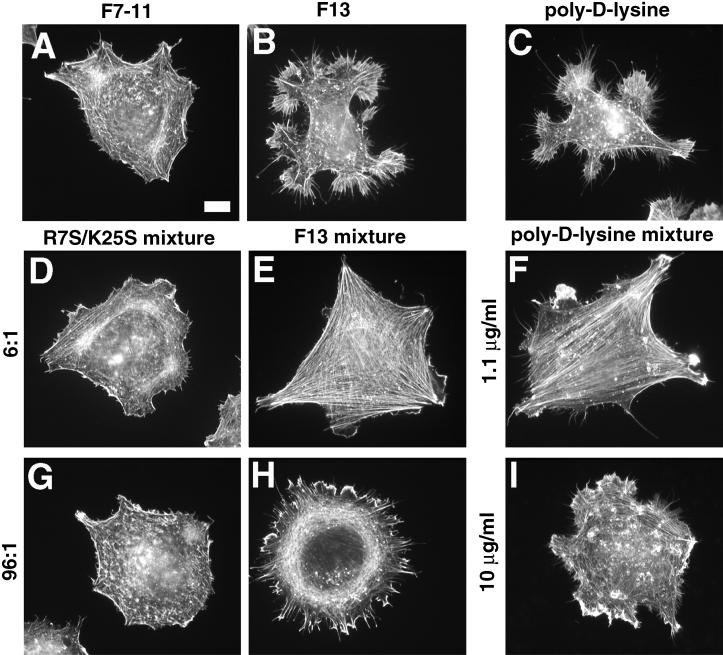
Although F13 is only weakly adhesive, we observed that cells adherent to F13 for 2 h developed extensive arrays of filopodia and lamellipodia, in contrast both with the disorganized actin in cells on F7–11 (Figures 4B′ and 12, A and B) and with the stress fibers observed in cells on F7–11,13 (Figure 4, F and F′). This observation indicated that F13 could induce actin organization in the absence of F7–11 but suggested that an additional signal from F7–11 was required to generate stress fibers. To determine whether these two stimuli could be provided by separate polypeptides, we coated coverslips with mixtures of F13 and F7–11 in varying proportions. Cell morphology varied with the proportion of the two polypeptides (Figures 12, E and H, and 13). At F13:F7–11 molar ratios between 1.5:1 and 6:1, cells adopted a polygonal shape with well-ordered stress fibers and few filopodia, similar to cells on F7–11,13 (Figures 4, F and F′, and 12E). At higher ratios, cells remained well spread but became less polygonal in shape, formed fewer stress fibers, and instead formed large numbers of filopodia and lamellipodia (Figures 12H and Figure 13). The range of molar ratios required for optimal stress fiber formation varied slightly between experiments. In some experiments, a molar ratio of 1:1 was not sufficient to induce stress fiber formation (our unpublished results), similar to the observations reported by Yoneda et al. (1995). The mutation R7S/K25S abolished the effects of wild-type F13 (Figures 12, D and G, and 13).
Figure 12.
GST–III13–His6 or poly-d-lysine alone can support cell adhesion and extension of filopodia and lamellipodia and can induce stress fibers in a mixed substrate with F7–11. Cells were stained with TRITC-phalloidin after 2 h on 4.5 μg/ml F7–11 alone (A), 206 μg/ml GST–III13–His6 alone (B), 1.1 μg/ml poly-d-lysine alone (C), or the following mixed substrates: 4.5 μg/ml F7–11 together with mutant GST–III13(R7SK25S)–His6 at 12.9 μg/ml (D; molar ratio, 6 GST–III13(R7SK25S)–His6:1 F7–11) or 206 μg/ml (G; 96:1 molar ratio), 4.5 μg/ml F7–11 with wild-type GST–III13–His6 at 12.9 μg/ml (E; 6:1) or 206 μg/ml (H; 96:1), or 4.5 μg/ml F7–11 together with poly-d-lysine at 1.1 μg/ml (F) or 10 μg/ml (I). Poly-d-lysine samples were tested in a separate experiment from wild-type and mutant III13 samples. Bar, 10 μm.
Figure 13.
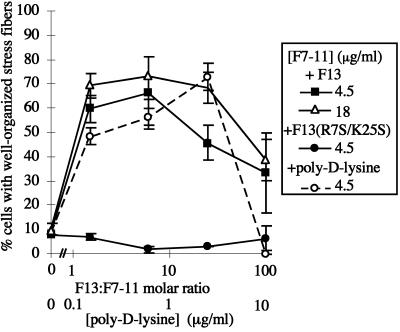
Stress fiber induction depends on a balance of F7–11 and cationic substrates. Cells were plated on coverslips coated with mixtures of 18 μg/ml F7–11 with varying concentrations of F13 (▵) or on 4.5 μg/ml F7–11 with varying concentrations of F13 (▪), F13(R7S/K25S) (●), or poly-d-lysine (○), and stress fiber formation after 2 h was scored.
Stress fiber formation on mixed substrates of F13 and F7–11 appeared to depend primarily on the molar ratio of the two proteins rather than on the absolute concentration of either protein. For example, a concentration of F13 (206 μg/ml) that caused cells to form primarily filopodia and lamellipodia in a mixture with 4.5 μg/ml F7–11 (96:1 molar ratio) gave rise primarily to cells with stress fibers when the F7–11 concentration was increased to 18 μg/ml (molar ratio, 24:1; Figure 13). A fourfold increase in F7–11 concentration generally produced a small increase in the proportion of cells with organized stress fibers but did not alter the range of molar ratios that produced an optimal response (Figure 13).
We observed that MG-63 cells plated on poly-d-lysine substrates formed filopodia and lamellipodia resembling those formed on F13 (Figure 12C). To determine whether poly-d-lysine, which might resemble the cluster of basic amino acids at the heparin-binding site of III13, was also able to stimulate stress fiber formation, we plated cells on mixed substrates of poly-d-lysine and F7–11. Like F13, poly-d-lysine was able to induce formation of either stress fibers or filopodia and lamellipodia, depending on its concentration relative to that of F7–11 (Figures 12, F and G, and 13).
DISCUSSION
We have found that the stress fiber–, focal contact–, and filopodia–inducing activities of the extracellular matrix molecule FN are contained within a short sequence at the N terminus of FN type III repeat 13 previously identified as the major heparin-binding site (Barkalow and Schwarzbauer, 1991; Busby et al., 1995). Heparin interfered with stress fiber formation in several cell types: MG-63 osteosarcoma cells, NIH3T3 fibroblasts, and NRK cells. Using recombinant GST–FN–His6 fusion proteins containing type III repeats 7–11 (which include the high-affinity integrin α5β1 binding site composed of the RGD sequence in repeat III10 and the synergy site in III9) and any of several type III repeats, we have found that III13 is unique among the type III repeats tested in its ability to induce stress fibers and focal contacts. Several non-RGD–containing FN type III repeats from FN and from other proteins have been shown to support adhesion and stress fiber and focal adhesion formation when β1 integrins are activated by Mn2+ or activating antibodies (Chi-Rosso et al., 1997). However, in the absence of exogenous integrin-activating reagents in our experiments, stress fiber and focal contact induction appeared highly specific for III13. When substituted for the equivalent sequence of the inactive III12 repeat, a 29–amino acid III13 sequence containing five arginine or lysine residues previously shown to be critical for solution phase heparin binding (Busby et al., 1995) provided most of the activity of the intact III13 domain. Mutations in any of these five basic residues reduced the ability of III13 to induce actin assembly, and all of the double-mutant combinations tested eliminated actin-organizing activity nearly completely. Taken together, these data map the actin-organizing and heparin-binding activities of FN to the same key amino acids.
Point mutations that partially reduced stress fiber activity also partially reduced the ability of heparin to interfere with stress fiber formation, further underscoring this close association. Among these, a central core of three III13 residues, R6, R7, and R23, is most critical for both stress fiber induction and heparin binding. The crystal structure of III13 (Sharma et al., 1999) indicates that these three residues are in fact closely spaced on the surface of III13, forming the center of a cationic cradle able to interact electrostatically with the negatively charged sulfate groups on heparin (Busby et al., 1995). Mutations in R9 and K25, which are located slightly farther from the center of the cationic cradle, had relatively weak effects on stress fiber induction, but the strong effects of double-mutant combinations involving R9 and K25 (including the R9S/K25S combination) clearly demonstrated a contribution of these sites to the activity of the domain. We also found that poly-d-lysine could substitute for III13 in stress fiber induction, further indicating the importance of clustered basic amino acids in this process. Because molecules structurally similar to heparin, such as the heparan sulfate chains attached to the core proteins of HSPGs, are likely binding partners for III13, our findings implicate HSPGs as likely to be involved in actin assembly. This is consistent with the observation that elimination of cell surface heparan sulfate by mutation or enzymatic cleavage reduces focal contact formation (LeBaron et al., 1988; Woods et al., 1993). Our data indicate that III13 binds to the cell surface via an HSPG (Bloom and Hynes, unpublished data). The HSPG syndecan-4 is localized to focal contacts (Woods and Couchman, 1994; Baciu and Goetinck, 1995) and can activate PKC (Oh et al., 1997, 1998), which has been implicated in focal contact formation (Woods and Couchman, 1992). Furthermore, antibodies to syndecan-4 can synergize with integrin occupancy to induce stress fibers and focal contacts (Saoncella et al., 1999), suggesting that syndecan-4 may be among the relevant HSPGs.
We did not find a function for III14 in actin organization, in contrast to the results of Woods et al. (1993), Huebsch et al. (1995), and Huhtala et al. (1995), who identified the III14 peptide PRARI as a potent inducer of focal contacts and stress fibers. In our experiments, III14 had no effect in the context of an F7–11,14 fusion, and mutation of the arginines in PRARI to serines in a manner analogous to the III13 heparin-binding mutations had no effect on the function of F7–15 (which still contained an intact III13). The differences between our findings and those of Woods et al. (1993), Huhtala et al. (1995), and Huebsch et al. (1995) may result from differences between receptors and signaling systems expressed by the cultured osteosarcoma cells and Rat1 fibroblasts used in this study and by the primary fibroblasts and primary endothelial cells and cell lines used in the other studies. Even with the MG-63 cell line used in this study, we have observed that different batches of serum used during cell growth can lead to significant quantitative differences in the response to III13, suggesting that the responsive machinery is highly sensitive to environmental conditions (our unpublished results). Alternatively, it is possible that the PRARI peptide (but not the PRARI sequence within the globular III14 domain) can either adopt a conformation that mimics another ligand or can present cells with a high density of basic residues similar to that found in III13. We cannot rule out the possibility that the activity of the PRARI sequence does not depend on the basic property of the two arginine residues. However, zebrafish FN has the sequence PRSTI in this position (Collodi, personal communication; GenBank number AF081128), suggesting that the second basic residue is not required. By contrast, all six amino acids of the cationic cradle are completely conserved in all known FNs, including those from zebrafish, Xenopus (DeSimone et al., 1992), the newt Pleurodeles (Clavilier et al., 1993), chicken (Norton and Hynes, 1987), rat (Schwarzbauer et al., 1983), and human (Kornblihtt et al., 1984).
We found that the heparin-binding site in III13 is a weak adhesive ligand but a potent inducer of filopodia and lamellipodia. When cells are exposed to both III7–11 and III13, III13 induces stress fibers and focal contacts as well, and the degree to which cells form stress fibers instead of radially directed filopodia can be manipulated by altering the relative amounts of III7–11 and III13. The two FN domains can be supplied in separate polypeptides or in different relative positions on the same polypeptide, suggesting that III13 acts by engaging a separate signaling mechanism rather than by affecting FN structure or accessibility of the RGD site. We propose that a balance of signals emanating from cell interactions with III13 and with the RGD–synergy region determines the type of actin structures formed by the cell. Stress fiber and focal contact formation resulting from engagement of both integrins and proteoglycans has been observed for melanoma cells attached to a mixture of the CS1 peptide (which binds integrin α4β1) and a mAb against the chondroitin sulfate proteoglycan NG2 (Iida et al., 1995). Interestingly, NG2 is localized to a subset of filopodia formed on poly-l-lysine (Lin et al., 1996).
We observed III13-dependent formation of filopodia in MG-63 and Rat1 cells within 2 min of adhesion to an F7–11,13 substrate. The recent demonstration that the rapid appearance of filopodia and lamellipodia in NIH3T3 cells on FN results from activation of the rho family GTPases cdc42 and rac (Price et al., 1998) suggests that III13, acting via its putative HSPG receptor, may be an activator of cdc42 and rac. Furthermore, our observation that relatively high ratios of III13 to III7–11 drive the cell to form filopodia at the expense of stress fibers is reminiscent of the observations of Kozma et al. (1995), who found that microinjection of wild-type cdc42 into Swiss 3T3 cells produced a similar shift toward filopodia. It is interesting to note that members of the syndecan family, a set of transmembrane HSPGs that could be III13 receptors, bind intracellularly to the PDZ domains of a recently described protein, syntenin (Grootjans et al., 1997), and of CASK/LIN-2, a multidomain protein that both binds to the actin-binding protein 4.1 and contains a guanylate kinase domain, which itself could potentially regulate the activity of small GTPases (Cohen et al., 1998). A plausible model is that III13–syndecan-4 interactions activate primarily cdc42 and rac, whereas III9/10–integrin interactions activate a different spectrum of rho family GTPases (Clark et al., 1998), and that the balance among these inputs provided by different domains of FN yields the typical cytoskeletal organization induced by this extracellular matrix protein.
ACKNOWLEDGMENTS
We are grateful to Jane Trevithick for construction of the clones pGH and pGH.F7–15 and to Shelesa Brew for thermal stability analysis of type III repeats. We thank members of the Hynes laboratory, Philip Gotwals, David McClay, and the developers of NIH Image for sharing materials and ideas. Oligonucleotide synthesis and DNA sequencing were performed at the Howard Hughes Medical Institute–Massachusetts Institute of Technology Biopolymers Laboratory. This work was supported by grants RO1 CA-17007 from the National Cancer Institute (to R.O.H.) and RO1 HL-21791 from the National Heart, Lung, and Blood Institute (to K.C.I.) and by the Howard Hughes Medical Institute. L.B. was supported by a fellowship from the Cancer Research Institute. R.O.H. is a Howard Hughes Medical Institute Investigator.
Abbreviations used:
- CHX
cycloheximide
- CSPG
chondroitin sulfate proteoglycan
- DMEM-HG
Dulbecco’s modified Eagle’s medium with 4.5 g/l glucose
- FN
fibronectin
- HSPG
heparan sulfate proteoglycan
- NRK
normal rat kidney
- SFM
serum-free DMEM-HG
REFERENCES
- Assoian RK. Anchorage-dependent cell cycle progression. J Cell Biol. 1997;136:1–4. doi: 10.1083/jcb.136.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- Baciu PC, Goetinck PF. Protein kinase C regulates the recruitment of syndecan-4 into focal contacts. Mol Biol Cell. 1995;6:1503–1513. doi: 10.1091/mbc.6.11.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkalow FJ, Schwarzbauer JE. Localization of the major heparin-binding site in fibronectin. J Biol Chem. 1991;266:7812–7818. [PubMed] [Google Scholar]
- Busby TF, Argraves WS, Brew SA, Pechik I, Gilliland GL, Ingham KC. Heparin binding by fibronectin module III-13 involves six discontinuous basic residues brought together to form a cationic cradle. J Biol Chem. 1995;270:18558–18562. doi: 10.1074/jbc.270.31.18558. [DOI] [PubMed] [Google Scholar]
- Chi-Rosso G, Gotwals PJ, Yang J, Ling L, Jiang K, Chao B, Baker DP, Burkly LC, Fawell SE, Koteliansky VE. Fibronectin type III repeats mediate RGD-independent adhesion and signaling through activated β1 integrins. J Biol Chem. 1997;272:31447–31452. doi: 10.1074/jbc.272.50.31447. [DOI] [PubMed] [Google Scholar]
- Clark EA, King WG, Brugge JS, Symons M, Hynes RO. Integrin-mediated signals regulated by members of the rho family of GTPases. J Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavilier L, Riou JF, Shi DL, DeSimone DW, Boucaut JC. Amphibian Pleurodeles waltl fibronectin: cDNA cloning and developmental expression of spliced variants. Cell Adhes Commun. 1993;1:83–91. doi: 10.3109/15419069309095683. [DOI] [PubMed] [Google Scholar]
- Cohen AR, Wood DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol. 1998;142:129–138. doi: 10.1083/jcb.142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone DW, Norton PA, Hynes RO. Identification and characterization of alternatively spliced fibronectin mRNAs expressed in early Xenopus embryos. Dev Biol. 1992;149:357–369. doi: 10.1016/0012-1606(92)90291-n. [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Veerapandian B, Dai X-P, Hamlin RC, Xuong N, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J Mol Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Elenius K, Salmivirta M, Inki P, Mali M, Jalkanen M. Binding of human syndecan to extracellular matrix proteins. J Biol Chem. 1990;265:17837–17843. [PubMed] [Google Scholar]
- Freeman AE, Gilden RV, Vernon ML, Wolford RG, Hugunin PE, Huebner RJ. 5-Bromo-2′-deoxyuridine potentiation of transformation in rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci USA. 1973;70:2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Ruoslahti E. Integrins and anoikis. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Dürr J, David G. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci USA. 1997;94:13683–13688. doi: 10.1073/pnas.94.25.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JL, Trevithick JE, Hynes RO. Retroviral expression of alternatively spliced forms of rat fibronectin. J Cell Biol. 1990;110:833–847. doi: 10.1083/jcb.110.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JL, Trevithick JE, Hynes RO. Fibronectin/integrin interaction induces tyrosine phosphorylation of a 120-kDa protein. Cell Regul. 1991;2:951–964. doi: 10.1091/mbc.2.11.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch JC, McCarthy JB, Diglio CA, Mooradian DL. Endothelial cell interactions with synthetic peptides from the carboxyl-terminal heparin-binding domains of fibronectin. Circ Res. 1995;77:43–53. doi: 10.1161/01.res.77.1.43. [DOI] [PubMed] [Google Scholar]
- Huhtala P, Humphries MJ, McCarthy JB, Tremble P, Werb Z, Damsky C. Cooperative signaling by α5β1 and α4β1 integrins regulates metalloproteinase gene expression in fibroblasts adhering to fibronectin. J Cell Biol. 1995;129:867–879. doi: 10.1083/jcb.129.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida J, Meijne AM, Spiro RC, Roos E, Furcht LT, McCarthy JB. Spreading and focal contact formation of human melanoma cells in response to the stimulation of both melanoma-associated proteoglycan (NG2) and α4β1 integrin. Cancer Res. 1995;55:2177–2185. [PubMed] [Google Scholar]
- Iida J, Skubitz AP, Furcht LT, Wayner EA, McCarthy JB. Coordinate role for cell surface chondroitin sulfate proteoglycan and α4β1 integrin in mediating melanoma cell adhesion to fibronectin. J Cell Biol. 1992;118:431–444. doi: 10.1083/jcb.118.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham KC, Brew SA, Migliorini MM, Busby TF. Binding of heparin by type III domains and peptides from the carboxy terminal hep-2 region of fibronectin. Biochemistry. 1993;32:12548–12553. doi: 10.1021/bi00097a035. [DOI] [PubMed] [Google Scholar]
- Izzard CS, Radinsky R, Culp LA. Substratum contacts and cytoskeletal reorganization of BALB/c 3T3 cells on a cell-binding fragment and heparin-binding fragments of plasma fibronectin. Exp Cell Res. 1986;165:320–336. doi: 10.1016/0014-4827(86)90586-0. [DOI] [PubMed] [Google Scholar]
- Jalkanen S, Jalkanen M. Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J Cell Biol. 1992;116:817–825. doi: 10.1083/jcb.116.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila YL, Niu J, Johnson PW. The high affinity heparin-binding domain and the V region of fibronectin mediate invasion of human oral squamous cell carcinoma cells in vitro. J Biol Chem. 1997;272:18932–18938. doi: 10.1074/jbc.272.30.18932. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, Vibe-Pedersen K, Baralle FE. Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J. 1984;3:221–226. doi: 10.1002/j.1460-2075.1984.tb01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBaron RG, Esko JD, Woods A, Johansson S, Höök M. Adhesion of glycosaminoglycan-deficient chinese hamster ovary cell mutants to fibronectin substrata. J Cell Biol. 1988;106:945–952. doi: 10.1083/jcb.106.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XH, Grako KA, Burg MA, Stallcup WB. NG2 proteoglycan and the actin-binding protein fascin define separate populations of actin-containing filopodia and lamellipodia during cell spreading and migration. Mol Biol Cell. 1996;7:1977–1993. doi: 10.1091/mbc.7.12.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JB, Chelberg MK, Mickelson DJ, Furcht LT. Localization and chemical synthesis of fibronectin peptides with melanoma adhesion and heparin binding activities. Biochemistry. 1988;27:1380–1388. doi: 10.1021/bi00404a044. [DOI] [PubMed] [Google Scholar]
- Mooradian DL, McCarthy JB, Cameron DJ, Skubitz APN, Furcht LT. Rabbit corneal epithelial cells adhere to two distinct heparin binding synthetic peptides derived from fibronectin. Invest Ophthalmol Vis Sci. 1992;33:3034–3040. [PubMed] [Google Scholar]
- Mooradian DL, McCarthy JB, Skubitz AP, Cameron DJ, Furcht LT. Characterization of FN-C/H-V, a novel synthetic peptide from fibronectin that promotes rabbit corneal epithelial cell adhesion, spreading, and motility. Invest Ophthalmol Vis Sci. 1993;34:153–164. [PubMed] [Google Scholar]
- Norton PA, Hynes RO. Alternative splicing of chicken fibronectin in embryos and in normal and transformed cells. Mol Cell Biol. 1987;7:4297–4307. doi: 10.1128/mcb.7.12.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh ES, Woods A, Couchman JR. Syndecan-4 proteoglycan regulates the distribution and activity of protein kinase C. J Biol Chem. 1997;272:8133–8136. doi: 10.1074/jbc.272.13.8133. [DOI] [PubMed] [Google Scholar]
- Oh ES, Woods A, Lim ST, Theibert AW, Couchman JR. Syndecan-4 proteoglycan cytoplasmic domain and phosphatidylinositol 4,5-bisphosphate coordinately regulate protein kinase C activity. J Biol Chem. 1998;273:10624–10629. doi: 10.1074/jbc.273.17.10624. [DOI] [PubMed] [Google Scholar]
- Peters JH, Trevithick JE, Johnson P, Hynes RO. Expression of the alternatively spliced EIIIB segment of fibronectin. Cell Adhes Commun. 1995;3:67–89. doi: 10.3109/15419069509081278. [DOI] [PubMed] [Google Scholar]
- Price LS, Leng J, Schwartz MA, Bokoch GM. Activation of rac and cdc42 by integrins mediates cell spreading. Mol Biol Cell. 1998;9:1863–1871. doi: 10.1091/mbc.9.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saoncella S, Echtermeyer F, Denhez F, Nowlen JK, Mosher DF, Robinson SD, Hynes RO, Goetinck PF. Syndecan-4 signals cooperatively, and in a Rho-dependent manner, with integrins in the assembly of focal adhesions and actin stress fibers. Proc Natl Acad Sci USA. 1999;96:2805–2810. doi: 10.1073/pnas.96.6.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders S, Bernfield M. Cell surface proteoglycan binds mouse mammary epithelial cells to fibronectin and behaves as a receptor for interstitial matrix. J Cell Biol. 1988;106:423–430. doi: 10.1083/jcb.106.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA. Integrins, oncogenes, and anchorage independence. J Cell Biol. 1997;139:575–578. doi: 10.1083/jcb.139.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer JE, Tamkun JW, Lemischka IR, Hynes RO. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983;35:421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI. Crystal structure of a heparin and integrin binding segment of human fibronectin. EMBO J. 1999;18:1468–1479. doi: 10.1093/emboj/18.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Couchman JR. Protein kinase C involvement in focal adhesion formation. J Cell Sci. 1992;101:277–290. doi: 10.1242/jcs.101.2.277. [DOI] [PubMed] [Google Scholar]
- Woods A, Couchman JR. Syndecan-4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Mol Biol Cell. 1994;5:183–192. doi: 10.1091/mbc.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Couchman JR, Johansson S, Höök M. Adhesion and cytoskeletal organization of fibroblasts in response to fibronectin fragments. EMBO J. 1986;5:665–670. doi: 10.1002/j.1460-2075.1986.tb04265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, McCarthy JB, Furcht LT, Couchman JR. A synthetic peptide from the COOH-terminal heparin-binding domain of fibronectin promotes focal adhesion formation. Mol Biol Cell. 1993;4:605–613. doi: 10.1091/mbc.4.6.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda J, Saiki I, Igarashi Y, Kobayashi H, Fujii H, Ishizaki Y, Kimizuka F, Kato I, Azuma I. Role of the heparin-binding domain of chimeric peptides derived from fibronectin in cell spreading and motility. Exp Cell Res. 1995;217:169–179. doi: 10.1006/excr.1995.1076. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Signal transduction and actin filament organization. Curr Opin Cell Biol. 1996;8:66–73. doi: 10.1016/s0955-0674(96)80050-0. [DOI] [PubMed] [Google Scholar]