Abstract
PrP binds copper in the highly conserved, unstructured N-terminal half of the protein. The octarepeat region consists of 4 tandem repeats of PHGGGWGQ and binds four equivalents of copper at full occupancy. Adjacent to the octarepeats are two additional histidines that may also bind copper. We recently showed that when the octarepeat region is titrated with Cu2+, the copper binding mode depends on the number of equivalents of copper bound. In addition to copper, other metals have been associated with PrP, however zinc is the only metal other than copper that induces PrP endocytosis, inhibits fibril formation and promotes inter-molecular interactions. In this work we show that even large excesses of zinc (> 1mM) are unable to displace copper from either the octarepeat region or the full-length protein. However, EPR reveals that physiologically relevant levels of zinc significantly alter the distribution of copper among the available binding modes. Diethyl pyrocarbonate (DEPC) modification and Mass Spectrometry is used to identify the octarepeat region as the zinc binding site and to confirm that the affinity of PrP for zinc is ~200 μM. PrP can simultaneously bind both copper and zinc by shifting to binding modes that minimize the ratio of histidines to copper.
The Transmissible Spongiform Encephalopathies (TSE) are a unique class of neurodegenerative diseases where the transmissible agent (the prion) consists of misfolded protein,1 designated PrPSC. The normal cellular form (PrPC) is expressed throughout the body, but mainly in the brain. The physiological role of PrPC is not known, but growing evidence points to a function related to copper binding.2 PrP has been shown to bind copper in vivo and although PrP knockout mice flourish, they show increased signs of oxidative stress. Cellular studies show that copper induces PrP endocytosis.3, 4
PrP binds copper in the highly conserved, unstructured N-terminal half of the protein. The octarepeat region, PrP(60–91), consists of 4 tandem repeats of PHGGGWGQ and binds four equivalents of copper at full occupancy.5 Adjacent to the octarepeats are two additional histidines (H96 and H111) that may also bind copper (the so-called “non-octarepeat” copper binding sites).6 We recently showed that when the octarepeat region is titrated with Cu2+, the copper binding mode depends on the precise molar ratio of copper to protein.7 The first equivalent of copper coordinates in a multi-histidine mode involving 3–4 imidazole side chains and we identify this mode by its CW Electron Paramagnetic Resonance (EPR) spectrum as Component 3. With additional copper, Component 3 decreases and is replaced by Component 2 (2 histidines per copper) and then four equivalents of component 1 (one copper per octarepeat), at saturation.8
In addition to copper, other metals have been associated with PrP9, with zinc (Zn2+) having the next highest affinity.10 Zinc is also the only metal other than copper that induces PrP endocytosis.3, 4 Zinc, like copper, also inhibits fibril formation (synthetic prions)11 and promotes inter-molecular interactions.12 Some have suggested that in vivo PrP may actually bind zinc rather than copper given the abundance of available zinc in the brain, with peak levels up to 300 μM in the synaptic cleft of glutaminergic neurons.13 While peak copper levels in the synaptic cleft may be as high as 100 – 250 μM,14 the basal level in Cerebral Spinal Fluid (CSF) is micromolar, with most exchangeable copper bound by amino acids and peptides. In this work we show that even large excesses of zinc are unable to displace copper from either the octarepeat region or the full-length protein. However, EPR reveals that physiologically relevant levels of zinc significantly alter the distribution of copper among the available binding modes. Diethyl pyrocarbonate (DEPC) modification10 and mass spectrometry are used to identify the octarepeat region as the zinc binding site and to confirm that PrP-Zn2+ dissociation constant is ~200 μM, reflecting an affinity significantly lower than that for copper.
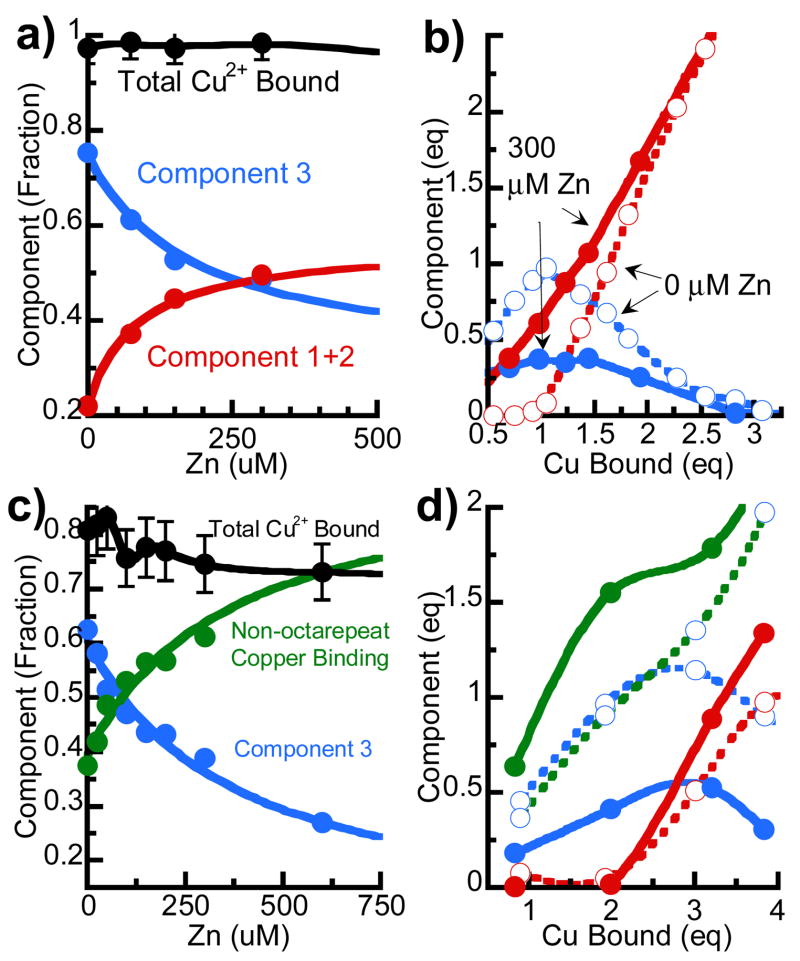
First we employ a direct competition between copper and zinc for binding sites in the octarepeat region, with the copper binding monitored by X-band EPR. Zn2+ is diamagnetic and therefore has no EPR signal; likewise, unbound Cu2+ at pH 7.4 exists as anti-ferromagnetically coupled hydroxides and is EPR silent. Component analysis of the copper-PrP EPR spectrum, developed by our lab, is used to determine both the concentration and the binding mode.8 Figure 1a shows the analysis of the EPR spectra from a zinc titration of PrP(23–28, 57–91) with copper held constant at one equivalent. At physiologically relevant zinc levels (< 1 mM), the total amount of bound copper is unchanged. However, a decomposition of the spectra into component spectra shows that the copper binding mode shifts dramatically with zinc concentration, from mostly Component 3 in the absence of zinc, to a majority of Component 1 (Component 2 remains a minor component throughout, and is summed with Component 1 for simplicity). When these spectral changes are fit to saturation curves (solid lines), they indicate a Zn2+ affinity of order 10−4 M (Supporting Information). Alternatively, Figure 1b shows a copper titration in the absence of zinc or with zinc held at 300 μM, to match the anticipated maximal synaptic concentration. The influence of zinc is the greatest with < 2 equivalents of bound copper; with > 3 equivalents of bound copper, zinc has little effect. When these techniques were applied to peptides encompassing the non-octarepeat copper binding sites, PrP(90–114), no change in the EPR spectrum was evident. Additionally, Ca2+ and Cd2+ were substituted for zinc; Ca2+ showed no effect while Cd2+ showed an effect qualitatively similar to Zn, but with weaker (~10x) affinity.
Figure 1. Effect of zinc on the distribution of the components of the copper EPR spectrum.
a)PrP(23–28,57–91) w/1 eq copper titrated with zinc b) PrP(23–28,57–91) with and without 300 μM Zn titrated with copper c) recombinant PrP(23–231) w/2 eq copper titrated with zinc d) PrP(23–231) with and without 300 μM Zn titrated with copper. Total copper bound (black), Component 3 (blue), 1+2 (red) and non-octarepeat (green). Copper and zinc (solid), copper only (dotted).
Similar experiments were also performed with full-length recombinant protein from Syrian Hamster, SHaPrP(23–231). SHaPrP(23–231) with 2 equivalents of copper was titrated with Zn2+. (Note, that the extra copper equivalent relative to the peptide experiment is added to partially populate the non-octarepeat sites.) The spectra showed only component 3 and non-octarepeat binding, with the amounts of each as a function of zinc shown in Figure 1c. While the zinc binding affinity derived from the fit saturation curves is slightly less than for the octarepeat peptide, (300 μM vs. 200 μM, See Supporting Information), the change in copper distribution is even more significant. As with the octarepeat peptide, the presence of zinc shifts copper away from the multi-histidine component 3 binding mode. Figure 1d shows that the binding mode, to which the copper is diverted, depends on how many equivalents of copper are bound. With less than two copper equivalents, it is the high copper affinity non-octarepeat binding sites that show increased copper. At higher copper levels, when the non-octarepeat sites are saturated, the distribution change occurs in the octarepeat region, favoring component 1.
To test zinc binding affinity in the absence of copper, we used DEPC modification and mass spectrometry. Peptides spanning the octarepeat region, PrP(60–91), and the non-octarepeat copper binding sites, PrP(90–114), were allowed to react with zinc. Then DEPC was added and allowed to react for one minute. The reaction was then quenched with imidazole. The reaction products were separated and quantified by reverse-phase HPLC and identified by ESI-MS (see Supporting Information). DEPC modifies the imidizole side chain of histidines to give a characteristic change in mass; however, histidines that are involved in metal binding will be protected from such modification. When PrP(60–91), (PHGGGWGQ)4, is titrated with ZnCl2, the amount of unmodified peptide goes up and the amount of peptide with 4 modifications goes down, giving a saturable binding curve with a Kd of 200 μM (Supporting Information). Similar experiments with peptides spanning the non-octarepeat copper binding region PrP(90–114) and peptides with 1–3 octarepeats show no change in DEPC modification distribution with the addition of zinc. Together, these show that zinc is bound exclusively by the octarepeat region, with four octarepeats necessary to bind one equivalent of Zn2+.
These findings show that Zn2+ binding to the PrP octarepeat domain is possible with reported synaptic zinc concentrations. As summarized in Figure 2, when copper levels are low, PrP can simultaneously bind both copper and zinc. At higher copper levels it accommodates the zinc by shifting to binding modes (Components 1 and 2) that minimize the ratio of histidines to copper. But when no rearrangement can accommodate both zinc and the available copper, it is the zinc that is displaced, not the copper. This is true even at millimolar zinc concentrations.
Figure 2. Models representing metal binding in the N-terminal domain of PrP.
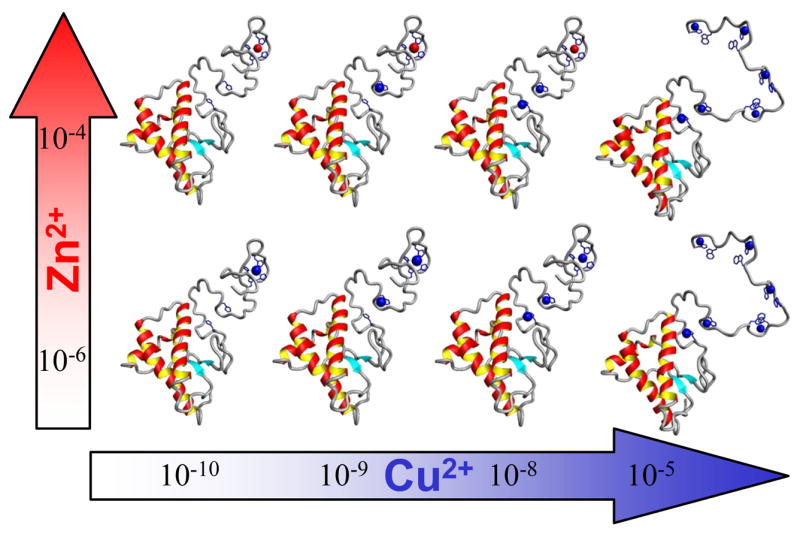
Top row (High Zinc); Zinc (red) is bound by the octarepeat region (left) while non-octarepeat sites (H96 and H111) are available for copper binding (blue, middle). Copper at high concentration will displace zinc from octarepeats to form up to 4 eq of Component 1 (right). Bottom row (Low Zinc); Copper (blue) is bound by the octarepeats in Component 3 when copper is low (left), with increasing copper loads the non-octarepeat sites (middle). High copper (right column) results in Component 1 copper binding by the octarepeats. Approximate molar metal concentrations are shown in the arrows. Octarepeat structures based on data from Chattopadhyay et al.7
Our results indicate that zinc may effect changes in two different ways: either directly by being bound by PrP or indirectly by changing the copper binding mode. A shift in binding mode may explain how both copper and zinc are able to stimulate PrP endocytosis. Unlike copper, zinc is redox inactive; however, zinc can change the overall Cu2+ redox properties of the protein by shifting copper coordination from the redox accessible component 3 binding mode to redox inactive component 1 and non-octarepeat binding modes. If so, proposed reductase properties based on component 3 binding may not be relevant in vivo. Finally, there are implications for experimental design; in vivo studies on either metal must consider the basal levels of the other. In vitro results for each metal individually may not extrapolate to the combination.
Supplementary Material
Experimental parameters and details for peptide and protein production, DEPC/MS footprinting, and EPR. HPLC chromatograms and spectra for ESI-MS, and EPR.
Acknowledgments
This work was supported by NIH Grant GM 65790
References
- 1.Prusiner SB. Proc Natl Acad Sci U S A. 1998;95(23):13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millhauser GL. Acc Chem Res. 2004;37(2):79–85. doi: 10.1021/ar0301678. [DOI] [PMC free article] [PubMed] [Google Scholar]; Millhauser GL. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pauly PC, Harris DA. J Biol Chem. 1998;273(50):33107–10. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 4.Perera WS, Hooper NM. Curr Biol. 2001;11(7):519–23. doi: 10.1016/s0960-9822(01)00147-6. [DOI] [PubMed] [Google Scholar]
- 5.Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, Antholine WE, Olmstead MM, Vrielink A, Gerfen GJ, Peisach J, Scott WG, Millhauser GL. Biochemistry. 2002;41(12):3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, Gerfen GJ, Peisach J, Millhauser GL. Biochemistry. 2003;42(22):6794–803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay M, Walter ED, Newell DJ, Jackson PJ, Aronoff-Spencer E, Peisach J, Gerfen GJ, Bennett B, Antholine WE, Millhauser GL. J Am Chem Soc. 2005;127(36):12647–56. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter ED, Chattopadhyay M, Millhauser GL. Biochemistry. 2006;45(43):13083–92. doi: 10.1021/bi060948r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leach SP, Salman MD, Hamar D. Anim Health Res Rev. 2006;7(1–2):97–105. doi: 10.1017/S1466252307001181. [DOI] [PubMed] [Google Scholar]
- 10.Qin K, Yang Y, Mastrangelo P, Westaway D. J Biol Chem. 2002;277(3):1981–90. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 11.Bocharova OV, Breydo L, Salnikov VV, Baskakov IV. Biochemistry. 2005;44(18):6776–87. doi: 10.1021/bi050251q. [DOI] [PubMed] [Google Scholar]
- 12.Kenward AG, Bartolotti LJ, Burns CS. Biochemistry. 2007;46(14):4261–71. doi: 10.1021/bi602473r. [DOI] [PubMed] [Google Scholar]
- 13.Watt NT, Hooper NM. Trends Biochem Sci. 2003;28(8):406–10. doi: 10.1016/S0968-0004(03)00166-X. [DOI] [PubMed] [Google Scholar]
- 14.Kardos J, Kovacs I, Hajos F, Kalman M, Simonyi M. Neurosci Lett. 1989;103:139–144. doi: 10.1016/0304-3940(89)90565-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental parameters and details for peptide and protein production, DEPC/MS footprinting, and EPR. HPLC chromatograms and spectra for ESI-MS, and EPR.