Abstract
Lacticin 481 synthetase (LctM) is a bifunctional enzyme that undertakes dehydration and cyclization in the structural region of the pre-lacticin peptide (LctA) to introduce three thioether rings and one dehydrobutyrine residue. The order and timing of these events has been investigated employing high resolution ESI-FTMS based tandem MS/MS techniques and chemical derivatization. LctM demonstrates highly processive behavior as seen by MS analysis of the reaction course of dehydration. Furthermore, cyclization is not tightly coupled to dehydration and follows at a later stage of the enzymatic reaction.
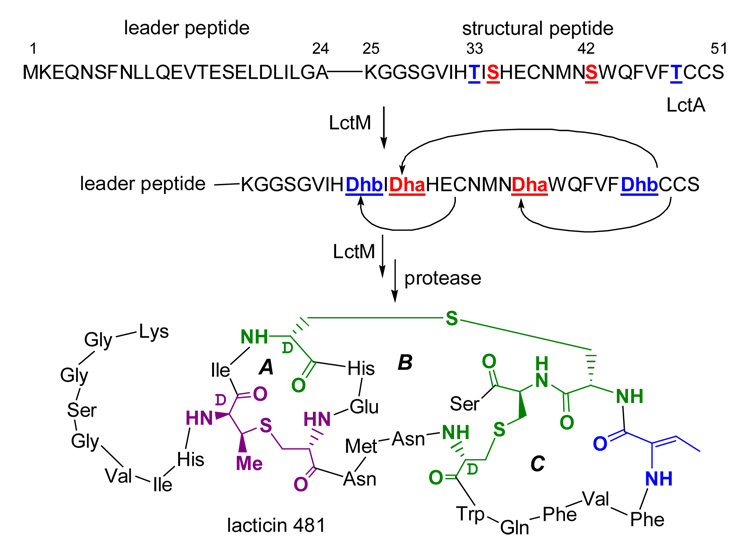
Lantibiotics are gene encoded peptide antimicrobials that contain the unusual residues lanthionine (Lan) and/or methyllanthionine (MeLan) (e.g., Scheme 1).1 The formation of these residues involves the initial dehydration of specific Ser and Thr residues in the pre-lantibiotic peptide and the subsequent stereoselective Michael-type addition of cysteine thiols to the newly formed dehydroalanine (Dha) and dehydrobutyrine (Dhb) residues, respectively (Figure 1A). A family of bifunctional enzymes (LanM) undertake both processes for the class II lantibiotics such as lacticin 481 (Scheme 1).
Scheme 1.
Figure 1.
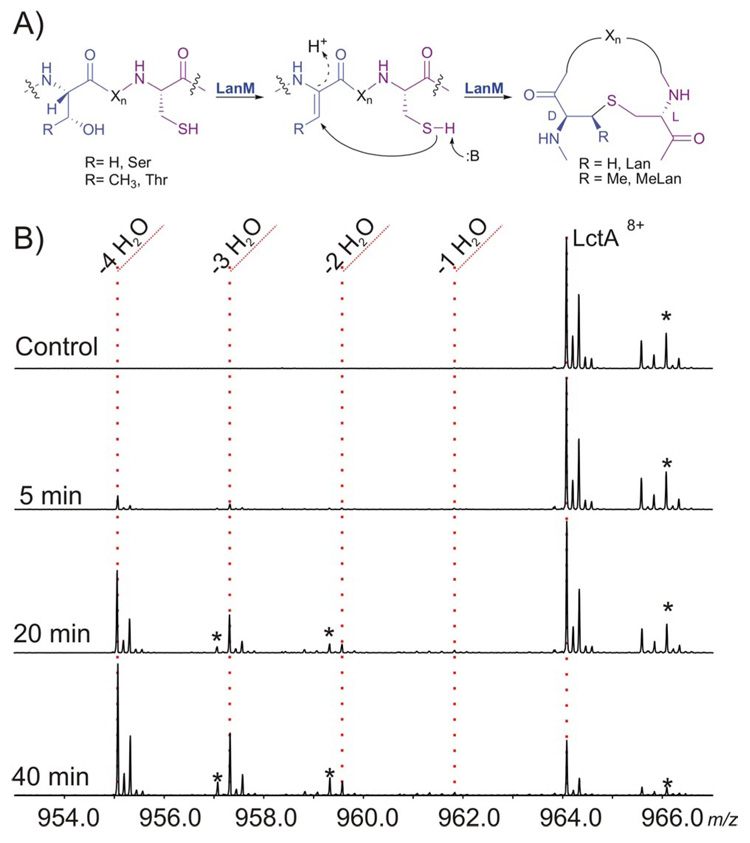
A) LanM catalyzed formation of (Me)Lan. B) Time dependent ESI/FT mass spectra of the assay of His6-LctA with LctM. * = oxidized products M+(O).
The activity of the lacticin 481 synthetase, LctM, was recently reconstituted in vitro.2 This synthetase catalyzes four dehydration and three cyclization reactions in the pre-lacticin peptide, LctA (Scheme 1). Chemical derivatization of the products and intermediates observed in time-dependent assays along with analysis by Fourier-Transform mass spectrometry (FTMS) analysis has now afforded a first glimpse of the order and timing of both the dehydration and cyclization reactions.
Conventional analysis of the products of the enzymatic modification process by MALDI-MS or ESI-MS is hampered by several experimental obstacles. A single dehydration (Δm = −18 Da) is difficult to distinguish from oxidation of a doubly dehydrated product (Δm = −36+16), and the potential of disulfide bond formation further clouds the interpretation (Supporting Figure 1). Therefore it is difficult to distinguish putative dehydrated intermediates from oxidized dead-end products. To overcome this hurdle, high resolution MS was used and the N-terminally His-tagged substrate, His6-LctA, was isolated from cells grown in isotopically depleted media (99.9% 12C and 99.99% 14N).3 This facilitated the precise identification of intermediates vs. oxidation artifacts. The time-course of conversion of the precursor peptide to the completely dehydrated product (M – 4 H2O) was observed by FTMS using a 1:900 enzyme to substrate ratio (Figure 1B).
Interestingly, during the course of this assay very little ion intensity was observed from the build-up of partially dehydrated species. For instance, neither the M – H2O nor the M – 2 H2O species were observed at the 5 min time-point (Figure 1B), whereas the M – 4 H2O species was clearly produced in a time-dependent manner. The data in Figure 1 is consistent with a processive mode of action and suggests that the dehydration of all four Ser/Thr residues in His6-LctA can occur prior to its dissociation from LctM. Processive enzymes, such as nucleotide polymerases, endocellulases, and some protein kinases, remain bound to their polymeric substrate throughout multiple rounds of catalysis.4–6 Distributive enzymes, however, dissociate from their substrate after each catalytic event, demonstrating a build-up of discrete intermediates, which was not observed for LctM.
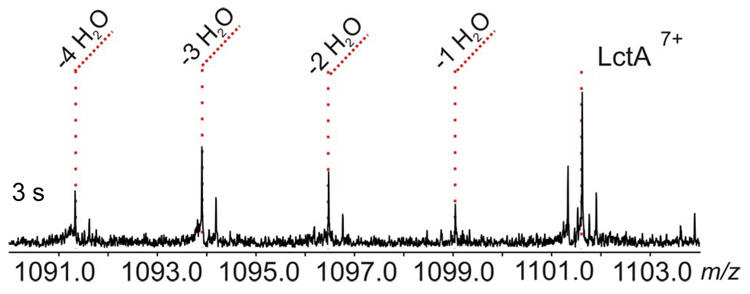
In order to test this hypothesis, rapid-quench assays were conducted with a 1:1 enzyme to substrate ratio under single-turnover like conditions. While no significant reaction was seen within 50 ms, after 3 s the product, substrate, and all partially dehydrated intermediates were observed with similar abundances (Figure 2 and Figure S3). The data indicates that under these conditions the intermediates can be trapped on the enzyme, further supporting a processive model during steady state turnover.7
Figure 2.
Assay product of LctM with His6-LctA at 1:1 enzyme to substrate ratio. The 3 s time-point exhibits the presence of all possible dehydrated forms of LctA (7+ charge state).
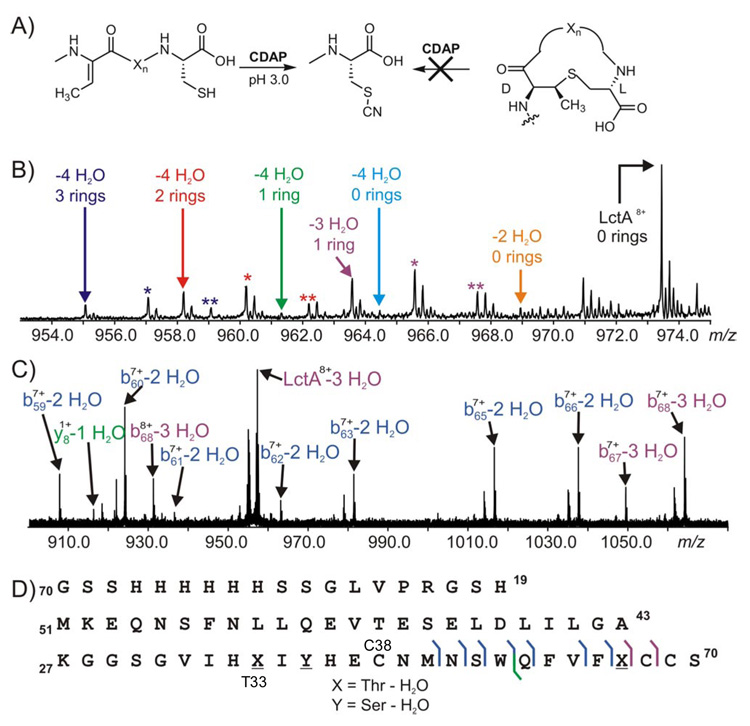
The timing of cyclization of the dehydrated species was interrogated to determine the relative order of the dehydration and cyclization events. All three Cys residues in His6-LctA are involved in thioether formation in the mature product (Scheme 1). Ring formation in dehydrated substrates does not lead to a change in mass of the cyclized product, thus precluding direct MS analysis to identify (Me)Lan formation. However, selective chemical derivatization of free thiols in uncyclized products permits assignment of (Me)Lan ring number by high resolution MS (Figure 3A).8 In order to count thioether rings in assay products and intermediates, assays were conducted for 20 min, 40 min, and 2 h. All species present in an assay mixture were co-purified by HPLC under acidic conditions to prevent non-enzymatic cyclization and subjected to thiol cyanylation at pH 3 with excess 1-cyano-4-dimethylaminopyridinium tetrafluoroborate (CDAP) prior to FTMS analysis.9
Figure 3.
ESI/FT MS/MS interrogation of thioether formation in His6-LctA during LctM catalysis. A) Expected cyanylation products. B) Cyanylation of the assay products after 40 min. Colored asterisks depict oxidation products, M+(O), of the identically colored parent ions. C) MS/MS analysis of the LctA-3 H2O species from Figure 1B (40 min). D) Fragment ion map of the LctA-3 H2O species.
Upon cyanylation of the 40 min assay product (Figure 1B, final time point) with CDAP, the M – 4 H2O species is a mixture of all possible ring states (Figure 3B).10 The presence of a maximum of three uncyclized cysteines in the M – 4 H2O species clearly indicates that each LctM catalyzed dehydration is not coupled to a subsequent cyclization. The M – 3 H2O species contains 1 ring (cyanylated twice) and the M – 2 H2O species is cyanylated at all Cys residues (Figure 3B) and therefore has no (Me)Lan rings, which supports our conclusion. In contrast, the 2 h time point shows near quantitative conversion to a final product with 4 dehydrations and 3 lanthionine rings (Figures S5 and S6).
To localize the residues that underwent modification in the M – 3 H2O and M – 2 H2O species, they were interrogated by tandem mass spectrometry (MS/MS) using infrared photodissociation. The M – 3 H2O ion from the 40 min time point of Figure 1B was isolated by SWIFT11 and dissociated. The fragment ions observed in Figure 3C localized the three dehydrations in the M – 3 H2O species to Thr33, Ser35, and Thr48 (Figure 3D), whereas Ser42 remained unmodified. Due to low ion abundance, no single major species could be deduced from the fragmentation ion pattern of the M – 2 H2O species. The incompletely dehydrated species observed in our assays may be due to intermediates that dissociate from the enzyme,12 or to the enzyme skipping a residue (Ser42) for dehydration as seen in the case of the M – 3 H2O peptide. By virtue of the abundant fragment ions from cleavage of backbone bonds in the B and C ring regions (Figure 3D, bottom right), the one lanthionine ring in M – 3 H2O species from the 40 min time point involves residues Thr33 and Cys38.
Based on the results presented here we conclude that the bifunctional enzyme LctM is highly processive in its dehydrative function. Furthermore, the dehydration process precedes the cyclization to form (Me)Lan residues as seen by the presence of an uncyclized yet completely dehydrated species in our experiments, as well as other partially cyclized products. Finally, the skipping of residues normally targeted for dehydration is observed to some extent in a putatively off-pathway activity of LctM which accounts for the small amounts of M – 3 H2O and possibly M – 2 H2O species observed. Structural interrogation of the “on-pathway” intermediates trapped by rapid quench are underway to assess the directionality and regioselectivity of LctM’s dehydration activity.
Supplementary Material
detailed experimental procedures for substrate preparation and FTMS-based assays.
Acknowledgment
This work was supported by NIH Grants to WAV (GM 58822) and to NLK (GM 67725).
References
- 1.Chatterjee C, Paul M, Xie L, van der Donk WA. Chem. Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 2.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 3.Marshall AG, Senko MW, Li W, Li M, Dillon S, Guan S, Logan TM. J. Am. Chem. Soc. 1997;119:433–434. [Google Scholar]
- 4.Breyer WA, Matthews BW. Protein Sci. 2001;10:1699–1711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Proc. Nat. Acad. Sci. USA. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott MP, Miller WT. Biochemistry. 2000;39:14531–14537. doi: 10.1021/bi001850u. [DOI] [PubMed] [Google Scholar]
- 7.Although unlikely, a model that cannot be unequivocally ruled out with the current data would invoke a much reduced Km for partially dehydrated substrates and increased kcat for each consecutive dehydration step. This model can be tested using the isotope trapping method (Rose IA. Methods Enzymol. 1980;64:47–59. doi: 10.1016/s0076-6879(80)64004-x.), and experiments along these lines are underway.
- 8.Kelleher NL, Hendrickson CL, Walsh CT. Biochemistry. 1999;38:15623–15630. doi: 10.1021/bi9913698. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Watson JT. Protein Sci. 1997;6:391–398. doi: 10.1002/pro.5560060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDAP treatment of the assay mixtures at 0 min, 20 min, 40 min, and 2 h time-points gave rise to the spectra shown in Figure S5.
- 11.Marshall AG, Wang TCL, Ricca TL. J. Am. Chem. Soc. 1985;107:7893–7897. [Google Scholar]
- 12.The possibility that (partial) cyclization of the M - 3 H2O species prevented dehydration at Ser42 cannot be ruled out from the results obtained here. However, the conversion of some of the partially processed species to the completely processed peptide does occur in a slower process that likely represents a minor distributive pathway for LctM action like that in other processive enzymes such as DNA polymerase beta:Osheroff WP, Jung HK, Beard WA, Wilson SH, Kunkel TA. J. Biol. Chem. 1999;274:3642–3650. doi: 10.1074/jbc.274.6.3642.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
detailed experimental procedures for substrate preparation and FTMS-based assays.