Abstract
HypA is an accessory protein and putative metallochaperone that is critical for supplying nickel to the active site of NiFe hydrogenases. In addition to binding Ni(II), HypA is known to contain a Zn site that has been suggested to play a structural role. X-ray absorption spectroscopy has been used to show that the Zn site changes structure upon binding nickel, from a S3(O/N)-donor ligand environment to an S4-donor ligand environment. This provides a potential mechanism for discriminating Ni(II) from other divalent metal ions. The Ni(II) site is shown to be a six-coordinate complex composed of O/N-donors including two histidines. As such, it resembles the nickel site in UreE, a nickel metallochaperone involved in nickel incorporation into urease.
The use of reactive and potentially toxic transition metals in enzyme active sites depends on proteins that can acquire metals from the environment (e.g., permeases), transport them inside cells and incorporate them into apoenzymes (e.g., metallochaperones), and control their intracellular concentration (e.g. transcriptional regulators), often with great specificity for the cognate metal.1 How proteins achieve this specificity is largely unknown. However, competition for ligands between metals and changes in ligand environments would provide a mechanism for discriminating different metals. Several accessory proteins have been implicated in the highly choreographed incorporation of nickel in the bimetallic active site of NiFe hydrogenases,2 including HypA,2–4 HypB,3, 5, 6 and SlyD.7 HypA is critical for supplying Ni to the active site of NiFe hydrogenases.2, 3 In addition to binding Ni(II), HypA is known to contain a Zn site that has been suggested to play a structural role in E. coli4 as has its homolog, HybF.8 Herein we show that the Zn site of H. pylori HypA (HpHypA) undergoes a structural change in response to Ni binding, suggesting that it has a role in the specific binding of nickel.
H. pylori hypA was cloned, expressed and purified as described in the supporting information. The purification procedure was a modification of that previously described by Mehta et al.3 Most notably, 1 mM DTT was included in all stages of the purification, but removed before the addition of nickel. The protein eluted from gel-filtration at a molecular weight consistent with a monomer. Metal content was determined by ICP-AE after treating metallated samples with Chelex and buffer exchanging ten times to remove any trace metal that was not tightly bound. Urea denaturation experiments indicated the protein unfolded cooperatively (see supporting information). The addition of stoichiometric Ni(II) had only a slight effect on the stability. Nickel titration of the nickel-free protein monitored by Tyr fluorescence showed a decrease in signal that reached a minimum at 1:1 Ni(II):HypA stoichiometry.
XAS data were collected on frozen protein solutions in Tris-buffer (20 mM Tris with 100 mM NaCl, pH 7.2, no DTT) held at ~50 K using a He displex cryostat. Protein solutions were prepared aerobically. Data were collected at the National Sychrotron Light Souce (NSLS) under ring conditions of 2.8 GeV and 120–300 mA using a sagitally focusing Si(111) double-crystal monochromator, and employing a 13-channel Ge fluorescence detector (Canberra). The data presented are an average of 5 or more scans. XAS data was analyzed using EXAFS1239 and phase and amplitude parameters obtained from FEFF8.10 UV-vis data were collected on a Hewlett Packard diode-array UV-vis spectrometer and 1H NMR measurements were performed on a Bruker 400 MHz NMR spectrometer.
The ‘apo’ form of HpHypA purifies with one equivalent of Zn per subunit. Addition of Ni(II) results in stoichiometric binding as previously reported.3 Studies on E.coli HypA and HybF have shown that the Zn ion is likely coordinated by two CXXC motifs,8 while the N-terminal MHE motif is likely involved in the coordination of the Ni(II) ion (Figure 2).3,8
Figure 2.
Sequence alignment for HpHypA and EcHypA. Conserved histidines and carboxylate residues are highlighted in red, the two conserved CXXC motifs in yellow and the conserved MHE N-terminus in grey.
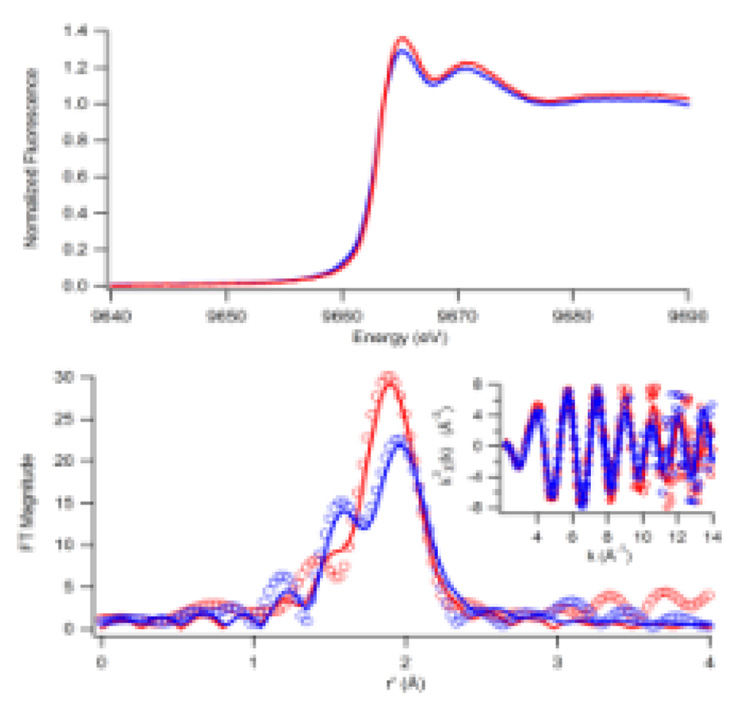
Remarkably, Zn K-edge XAS spectra show that in the apo-protein the Zn site is coordinated by 3 sulfur atoms and one N/O scatterer (Figure 1). This differs from that reported for EcHypA; however, that data is from a protein sample in which the zinc ion has been substituted with a cobalt ion and assumes that the coordination environment of the two species is the same. There are also four additional Cys residues in EcHypA that may coordinate the Zn ion (Figure 2). Upon coordination of Ni, the EXAFS spectrum of the Zn site changes; the N/O scatterer is replaced by a fourth S-donor ligand. The XANES spectra differ in the relative intensities of the two peaks immediately after the edge. The greater relative intensity of the first peak in the HpHypA+ Ni spectrum is consistent with an increase in sulfur coordination at the Zn site.11
Figure 1.
Zn K-edge XAS of HpHypA without a second metal (blue) and with Ni(II) (red). Top: The XANES spectra show a small shift in the relative intensities of the first two peaks. Bottom: The Fourier-transformed EXAFS spectra (FT window = 2–14 Å−1, uncorrected for phase shifts, bottom) show that in the Ni-free sample a second peak corresponding to an N/O-donor is present (data shown as circles), unfiltered exafs data is shown in the inset. Fits calculated for the apo-protein 1N/O @ 2.03(3) Å (σ2 = 0.002(2) Å2) + 3S @ 2.34(1) Å (σ2 = 0.003(1) Å2), GOF = 0.88, and for the protein with Ni added, 4S @ 2.32(1) Å (σ2 = 0.003(1) Å2), GOF = 0.78, are shown as solid lines (see supporting information).
In the FT-EXAFS spectra for the Zn sites, a second peak corresponding to N/O scatterers is clearly visible in the apo-protein compared to the Ni-loaded protein (Figure 1).
The titration of free sulfhydryl groups with DTNB shows that the ‘apo’-protein containing only Zn has approximately one free cysteine (0.88(15)) that is absent for the Ni–loaded protein (See supporting information for experimental procedure). This result is consistent with the XAS data.
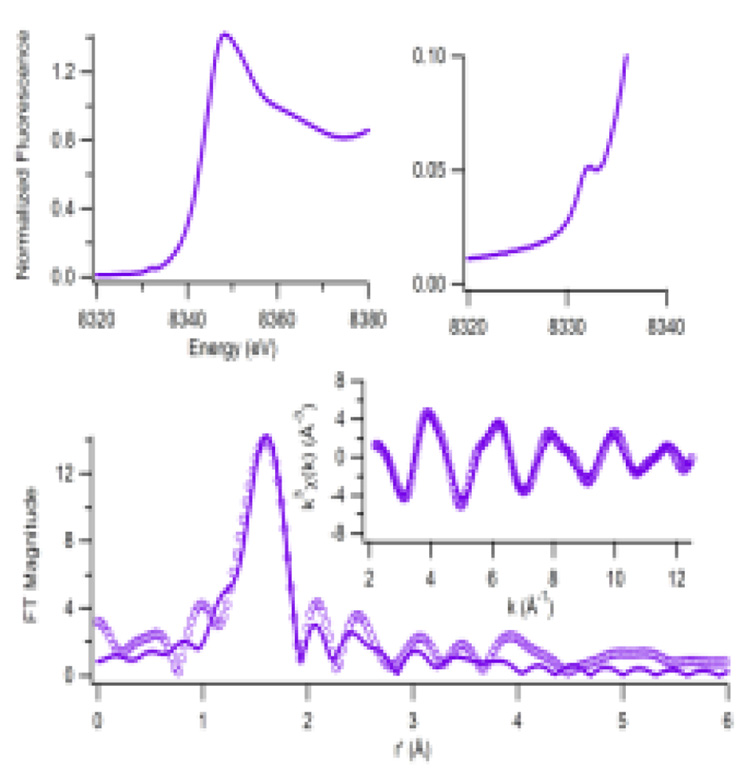
The Ni K-edge XANES data for HpHypA is consistent with a 6-coordinate Ni site (Figure 3). The XANES spectrum exhibits a pre-edge peak near 8332 eV (peak area = 3.1(6) × 10−2 eV) with no evidence of additional features, indicating a 6-coordinate site.12 The EXAFS spectrum is best fit with a 6 N/O-ligand donor environment. Using multiple scattering parameters, 1–2 histidine ligands can be accommodated. The histidine residue H2, has been previously implicated as a Ni ligand, consistent with this analysis.3
Figure 3.
Ni K-edge XAS of HpHypA. The Ni XANES spectrum (top left) shows a small peak associated with a 1s→3d transition (top right). Bottom: The Fourier-transformed EXAFS spectra (circles (FT window = 2–12.5 Å−1, uncorrected for phase shifts,) and selected fits (solid line) calculated for the Ni center (3N/O @ 2.04(1) Å (σ2 = 0.003(1) Å2) + 3N/O @ 2.15(3) Å (σ2 = 0.010(6) Å2) with one of the N/O @ 2.04 Å being fit as an imidazole using multiple scattering parameters, GOF = 0.59) and for the Cu site (4N/O @ 1.97(1) Å (σ2 = 0.005(1) Å2) with one of the being fit as an imidazole, GOF = 0.44 (See supporting information).
The coordination environment is similar to that of another structurally characterized nickel metallochaperone, UreE, that has also been shown to bind Ni in an 6-coordinate environment with 6 N/O donors including two histidines.13, 14 There is no structural Zn site in UreE, but a similar conformational change may still exist by which the disordered C-terminus in the apo-protein participates in binding the Ni ion via a conserved HXH sequence.
The 6-coordinate Ni site found for HpHypA is also consistent with Evans (NMR) susceptibility15 data that determined the number of unpaired electrons for the Ni site (1.9 ± 0.2), demonstrating the presence of a high-spin (S = 1) Ni (II) ion.
The XAS data presented here clearly show that the Zn site in HpHypA rearranges upon coordination of a second metal ion. Preliminary studies suggest that this rearrangement is dependent on the nature of the second metal ion, and may help provide a mechanism for distinguishing between otherwise similar divalent metal ions. The possibilities for this rearrangement include metal-dependent ligand exchange and/or protein conformational changes that are specific to the metal bound and affect the Zn site structure. Protein conformations providing different ligand environments for different metals have been documented in other metal-trafficking proteins.16
Supplementary Material
Tables for XAS data, details of analysis and unfiltered EXAFS spectra, data for urea melt experiments and fluorescence titrations, and details of protein over-expression, sample preparation sulfhydryl titrations and Evan’s method experiment. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgement
This work was supported by the National Institutes of Health Grant GM69696 (M.J.M). XAS data collection at the National Synchrotron Light Source at Brookhaven National Laboratory was supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences. Beamline X9B at NSLS is supported in part by the NIH.
References
- 1.Finney LA, O'Halloran TV. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 2.Hube M, Blokesch M, Bock A. J. Bacteriol. 2002;184:3879–3885. doi: 10.1128/JB.184.14.3879-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta N, Olson JW, Maier RJ. J. Bacteriol. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atanassova A, Zamble DB. J. Bacteriol. 2005;187:4689–4697. doi: 10.1128/JB.187.14.4689-4697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson JW, Mehta NS, Maier RJ. Mol. Microbiol. 2001;39:176. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 6.Leach MR, Sandal S, Sun H, Zamble DB. Biochemistry. 2005;44:12229–12238. doi: 10.1021/bi050993j. [DOI] [PubMed] [Google Scholar]
- 7.Zhang JW, Butland G, Greenblatt JF, Emili A, Zamble DB. J. Biol. Chem. 2005;280:4360–4366. doi: 10.1074/jbc.M411799200. [DOI] [PubMed] [Google Scholar]
- 8.Blokesch M, Rohrmoser M, Rode S, Boeck A. J. Bacteriol. 2004;186:2603–2611. doi: 10.1128/JB.186.9.2603-2611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padden KM, Krebs JF, Trafford KT, Yap GPA, Rheingold AH, Borovik AS, Scarrow RC. Chem. Mater. 2001;13:4305. [Google Scholar]
- 10.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Phys. Rev. B. 1998;58:7565. [Google Scholar]
- 11.Clark-Baldwin K, Tierney DL, Govindaswamy N, Gruff ES, Kim C, Berg J, Koch SA, Penner-Hahn JE. J. Am. Chem. Soc. 1998;120:8401–8409. [Google Scholar]
- 12.Colpas GJ, Maroney MJ, Bagyinka C, Kumar M, Willis WS, Suib SL, Mascharak PK, Baidya N. Inorg. Chem. 1991;30:920–928. [Google Scholar]
- 13.Colpas GJ, Brayman TG, McCracken J, Pressler MA, Babcock GT, Ming L-J, Colangelo CM, Scott RA, Hausinger RP. J. Biol. Inorg. Chem. 1998;3:150–160. [Google Scholar]
- 14.Stola M, Musiani F, Mangani S, Turano P, Safarov N, Zambelli B, Ciurli S. Biochemistry. 2006;45:6495–6509. doi: 10.1021/bi0601003. [DOI] [PubMed] [Google Scholar]
- 15.Evans DF. J. Chem. Soc. 1959:2003. [Google Scholar]
- 16.Pennella MA, Giedroc DP. BioMetals. 2005;18:413–428. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables for XAS data, details of analysis and unfiltered EXAFS spectra, data for urea melt experiments and fluorescence titrations, and details of protein over-expression, sample preparation sulfhydryl titrations and Evan’s method experiment. This material is available free of charge via the Internet at http://pubs.acs.org.