Abstract
Antisense oligonucleotides are powerful tools for the in vivo regulation of gene expression. We have characterized the intracellular distribution of fluorescently tagged phosphorothioate oligodeoxynucleotides (PS-ONs) at high resolution under conditions in which PS-ONs have the potential to display antisense activity. Under these conditions PS-ONs predominantly localized to the cell nucleus where they accumulated in 20–30 bright spherical foci designated phosphorothioate bodies (PS bodies), which were set against a diffuse nucleoplasmic population excluding nucleoli. PS bodies are nuclear structures that formed in cells after PS-ON delivery by transfection agents or microinjection but were observed irrespectively of antisense activity or sequence. Ultrastructurally, PS bodies corresponded to electron-dense structures of 150–300 nm diameter and resembled nuclear bodies that were found with lower frequency in cells lacking PS-ONs. The environment of a living cell was required for the de novo formation of PS bodies, which occurred within minutes after the introduction of PS-ONs. PS bodies were stable entities that underwent noticeable reorganization only during mitosis. Upon exit from mitosis, PS bodies were assembled de novo from diffuse PS-ON pools in the daughter nuclei. In situ fractionation demonstrated an association of PS-ONs with the nuclear matrix. Taken together, our data provide evidence for the formation of a nuclear body in cells after introduction of phosphorothioate oligodeoxynucleotides.
INTRODUCTION
The modulation of gene expression by antisense oligonucleotides is emerging as a promising new concept for treatment of human diseases (Sharma and Narayanan, 1995; Agrawal, 1996; Crooke and Bennett, 1996). In addition, antisense molecules prove to be a valuable tool in basic research applications (e.g., Coats et al., 1996; Luger et al., 1996). Antisense oligonucleotides are short polymers comprised of 10–25 modified DNA or RNA nucleotides that are complementary to sequences in their target RNA. Their hybridization to the target sequence is proposed to interfere with RNA intermediary metabolism and/or to induce RNA degradation, resulting in a decreased expression of the target gene (for review see Crooke, 1996). So far, the lead chemistry for antisense oligonucleotides has been the phosphorothioate modification in which a nonbridging oxygen atom in the phosphate backbone is replaced by sulfur. The focus on this modification is due to its acceptable properties in terms of stability, uptake, selectivity, toxicology, and the support of ribonuclease (RNase) H activity (Crooke, 1997). Furthermore, these compounds can be easily and cost-effectively synthesized in bulk quantities. As a consequence, several phosphorothioate oligodeoxynucleotides (PS-ONs) have entered clinical trials (Dean et al., 1996a; Wagner and Flanagan, 1997; Webb et al., 1997). Because of its usefulness, this modification is also being used in the next generation of antisense drugs, in mixed-type compounds (Agrawal and Iyer, 1995).
The specific action of antisense oligonucleotides has been shown numerous times under well-controlled conditions in cell culture systems as well as in animal models (e.g., Chiang et al., 1991; Wagner et al., 1993; Dean and McKay, 1994; Dean et al., 1996b; Monia et al., 1996; Neurath et al., 1996; Wagner et al., 1996; Bennett et al., 1997). However, little is currently known about how and where within cells the antisense molecules bind to their target RNA and exert their activity. Only in a few cases has more than circumstantial evidence been provided for their mechanism of action: a RNase H-dependent degradation of the target RNA was most convincingly shown in Xenopus oocytes (Shuttleworth and Colman, 1988) and in a reversibly permeabilized cell system (Giles et al., 1995). In another case, a PS-ON led to cleavage of its endogenous target RNA and at the same time inhibited splicing of a particular intron (Condon and Bennett, 1996).
It is thought that PS-ONs enter the cell through endocytotic mechanisms (Beltinger et al., 1995; Tarrason et al., 1995, and references therein). It is of key importance for antisense oligonucleotides to then escape the endosomal/lysosomal compartments into the cytoplasm. In many reports antisense activity was dependent on the delivery of PS-ONs in complex with a transfection agent that promotes the escape from vesicular trapping (e.g., Bennett et al., 1992; Wagner et al., 1993). However, in other reports a transfection agent was not necessary to achieve specific antisense activity (e.g., Offensperger et al., 1993; Nestle et al., 1994; Neurath et al., 1996). These differences may be related to differences in cell type as well as experimental protocol but are at this time unresolved. In vivo, transfection agents also do not seem to be a prerequisite for efficient antisense activity (e.g, Offensperger et al., 1993; Dean and McKay, 1994; Monia et al., 1996; Neurath et al., 1996). In many cases, whenever significant antisense activity was displayed a significant, if not predominant, fraction of the PS-ONs localized to the nucleus. A nuclear localization was also predominant shortly after microinjection of PS-ONs into the cytoplasm (Chin et al., 1990; Leonetti et al., 1991; Fisher et al., 1993; Wagner et al., 1993). Thus, the nucleus has been especially implicated as an important site for the action of antisense PS-ONs. That antisense activity can indeed occur in the nucleus is clear from work in which treatment with an antisense PS-ON resulted in the appearance of an aberrant RNA species in the nucleus (Condon and Bennett, 1996) and from an example in which the target RNA sequences only existed in the nucleus (intron sequences; Wagner et al., 1993). On the other hand, antisense oligonucleotides certainly are also able to act in the cytoplasm, e.g., by inhibiting the formation of a translation initiation complex (Baker et al., 1997).
In the present report we characterized at high resolution the spatial and temporal distribution of PS-ONs in the nucleus under conditions in which they have the potential to display specific antisense activity. We find that the introduction of PS-ONs into cells leads to the formation of nuclear bodies.
MATERIALS AND METHODS
Cell Culture, Observation of Living Cells, and Induction of Intercellular Adhesion Molecule (ICAM)-1 Expression
HeLa cells were grown on glass coverslips at 10% CO2 and 37°C in high glucose, HEPES-buffered DMEM (Life Technologies, Gaithersburg, MD) supplemented with 10% FBS (Hyclone, Logan, UT) and 50 U/ml penicillin and 0.1 mg/ml streptomycin.
For observation of living cells, HeLa cells were grown on special coverslips fitting into a FCS2 live-cell microscopy chamber (Bioptechs, Butler, PA). After administration of the oligonucleotides as described below, the chamber was assembled and mounted on a Zeiss Axiovert 405 fluorescence microscope (Carl Zeiss, Thornwood, NY). The temperature was kept constant at 37°C, and fresh medium was pumped in every 2 h. Images were acquired with a Photometrics Nu200 cooled CCD camera (1,320 × 1,035 array, 6.7-μm pixel size, Photometrics, Tucson, AR).
To induce ICAM-1 expression, HeLa cells were treated with 10 ng/ml recombinant tumor necrosis factor-α (TNF-α, RD Systems, Minneapolis, MN) for 18 h after the delivery of oligonucleotides.
Oligonucleotides
All oligonucleotides employed in the present study are fully modified phosphorothioate oligodeoxynucleotides. 4298-F is a 18-mer of the sequence 5′-TGGGAGCCATAGCGAGGC-3′ and conjugated at its 5′-end to fluorescein. It is complementary to sequences −8 to +10 of the translational start region of human ICAM-1, and its parent molecule without fluorescent tag has been shown to have specific antisense activity (Chiang et al., 1991). 12182-T, a 5′-Texas Red–conjugated 20-mer of the sequence 5′-CTTCAGAGCGCTCCAGCTCT-3′ targets exon 5 of rat β-tropomyosin and has specific antisense activity in a microinjection assay (our unpublished results). 11068-F is a 20-mer of the sequence 5′-TGCATCCCCCAGGCCACCAT-3′, harbors a 5′-end fluorescein, and targets the 3′-untranslated region of murine ICAM-1 (Stepkowski et al., 1994). 11721-F is a 5′-fluorescein–conjugated 20-mer of the sequence 5′-TTCCCCAGATGCACCTGTTT-3′ and targets the 3′-untranslated region of human E-selectin. This PS-ON has antisense activity against E-selectin but shows no effect on ICAM-1 expression (Bennett et al., 1994) and therefore is employed as a control. Furthermore, E-selectin is not expressed in HeLa cells used for analyses. The PS-ONs were synthesized using conventional solid-phase triester chemistry (Beaucage and Iyer, 1992). 2′-Deoxy and 5′-amino–modified phosphoramidites were purchased from commercial suppliers (PerSeptive Biosystems, Framingham, MA; Glen Research, Sterling, VA). The fluorophores were conjugated postsynthetically using FITC or Texas Red Sulfonylchloride (Molecular Probes, Eugene, OR) according to the manufacturer’s protocols. Free fluorophores were separated from oligonucleotides by gel filtration using NAP-25 columns (Pharmacia, Piscataway, NJ).
Delivery of Oligonucleotides into Cells
PS-ONs were administered to cells by two methods. For lipofection (Chiang et al., 1991) 0.25 μM PS-ON was combined with 10 μg/ml Lipofectin (Life Technologies) in serum-free DMEM according to the manufacturer’s protocol. Cells were washed twice with serum-free DMEM and then incubated in the oligonucleotide/Lipofectin-containing medium for 4 h. After lipofection, cells were washed twice in complete medium and incubated for the indicated times, but at least for 1 h in DMEM, 10% FBS before they were fixed. When added to the medium without Lipofectin, the concentration of PS-ON was 5 μM.
Using microinjection, 30 μM oligonucleotide with 6 mg/ml dextran-Cascade Blue (Molecular Probes) in injection buffer (10 mM NaH2PO4, 10 mM K2HPO4, 80 mM KCl, 4 mM NaCl, pH 7.2) was directly delivered into the cytoplasm or nucleus of cells. We used an Eppendorff microinjector 5242 and micromanipulator 5170 setup mounted on a Zeiss Axiovert 10 inverted microscope and self-made microinjection needles. The needles were pulled on a Brown-Flaming pipette puller (model P80, Sutter Instruments, San Francisco, CA) from GC120TF-10 glass capillaries (Warner Instruments, Hamden, CT). The solutions to be injected were centrifuged for at least 30 min at 14,000 × g at 4°C to remove any particulate material. One can assume a 1:20 dilution of the concentrations in the needle upon injection into the cell. In some experiments the PS-ONs were added to permeabilized cells. Cells were permeabilized in CSK buffer containing 0.5% Triton X-100 as described below and then incubated in 0.2 μM PS-ON in CSK buffer lacking the detergent for 30 min at 37°C. Cells were rinsed twice in CSK without detergent and fixed.
Immunostaining and Fluorescence Microscopy
Cells were fixed for 15 min in PBS, 4% paraformaldehyde and then washed for 5 min with PBS three times. If antibody staining was involved, cells were permeabilized in PBS, 0.5% Triton X-100 for 5 min. After three washes for 5 min with PBS, cells were incubated for 30 min in primary antibody diluted in PBS, 1% normal goat serum. Bound primary antibodies were detected with secondary antibodies coupled to various fluorochromes (Jackson Immunoresearch, West Grove, PA), again diluted in PBS, 1% normal goat serum, and incubated for 30 min. Between the antibody incubations and after the secondary reagent, cells were washed for 10 min in PBS three times. Finally, the coverslips with the cells were mounted in 90% glycerol in 0.2 M Tris-base (pH 8) containing 1 mg/ml p-phenylenediamine. All incubations were carried out at room temperature. Human ICAM-1 (CD54) protein was visualized with monoclonal antibody 84H10 (Immunotech, Westbrook, ME). Primary antibodies for colocalization studies of PS-ONs with known nuclear domains/structures included mouse anti-SC35 (1:3000; Fu and Maniatis, 1990), mouse anti-U2-B" (1:3; Habets et al., 1989), mouse anti-m3G (1:5; Krainer, 1988), human anti-centromere (1:200; Moroi et al., 1980), human anti-SM (1:600; gift from Joe Kraft, Yale University), and mouse 5E10 anti-PML (1:3; Stuurman et al., 1992).
Images were acquired using a Nikon Microphot-FXA fluorescence microscope equipped with a Photometrics SenSys cooled CCD camera (1,320 × 1,035 array, 6.7-μm pixel size, Photometrics, Tucson, AZ) controlled by a Macintosh computer with Oncor Image (Oncor, Gaithersburg, MD) software. For colocalization analysis, a Zeiss confocal microscope LSM410 was used.
Nuclear Fractionation
HeLa cells grown on a glass coverslip in a 3.5-cm dish were lipofected with fluorescently tagged PS-ONs as described above. The coverslip was cut into four equal pieces. The first piece was fixed immediately after lipofection. With the other pieces, nuclear fractionation was carried out in three steps as described (Huang et al., 1994). First, cells were incubated on ice for 10 min in CSK buffer (10 mM PIPES, pH 6.8, 3 mM MgCl2, 100 mM NaCl, 300 mM sucrose, 0.5% Triton X-100, 10 mM leupeptin, 2 mM vanadyl adenosine). Then they were further extracted at 4°C for 5 min in extraction buffer (10 mM PIPES, pH 6.8, 3 mM MgCl2, 250 mM ammonium sulfate, 300 mM sucrose, 0.5% Triton X-100, 10 μM leupeptin, 2 mM vanadyl adenosine). Finally, DNA was removed by treatment for 1 h at 37°C with 100 U/ml RQ-DNase (Promega, Madison, WI) in CSK buffer containing 50 mM instead of 100 mM NaCl. After each step of the fractionation, samples were quickly rinsed once in the buffer used for the previous incubation, one coverslip piece removed, and fixed in 4% paraformaldehyde in PBS as described above. To check the deoxyribonuclease (DNase) digestion, cells were stained for 5 min with 0.5 μg/ml DAPI (Molecular Probes). RNase treatment was performed by substituting RQ-DNase with 0.4 mg/ml RNase A (Sigma Chemical, St. Louis, MO; boiled for 10 min to destroy residual DNase in the preparation) and omitting vanadyl adenosine in the buffer. The fractionated cells were incubated in RNase A solution or buffer alone for 30 min at 37°C, rinsed twice in CSK, 50 mM NaCl, and fixed.
Enzyme Immunoassay to Assess the Amount of Oligonucleotides within Cells
A cell-based enzyme immunoassay was employed for comparative measurements of PS-ONs in cells after the different steps of the nuclear fractionation procedure or after the RNase treatment. HeLa cells (5000) were seeded in each well of a 96-well plate; 2.5 d later, 0.25 μM fluorescein-conjugated PS-ON was introduced into cells by means of 10 μg/ml Lipofectin as described above. Then nuclear fractionation of the cells was performed in each well of the plate using the same procedure as for cells on coverslips. At different steps of the procedure a subset of wells was fixed with 4% paraformaldehyde in PBS. After fixation, all samples were washed with PBS. Cells directly fixed without fractionation were permeabilized with 0.5% Triton X-100 in PBS. All other samples were permeabilized by the nuclear fractionation protocol. Unspecific binding sites were blocked by incubation in PBS, 1% BSA, 0.02% Tween 20 (buffer PBT) overnight at 4°C. Wells were washed with buffer PBT and incubated with a monoclonal antibody against the fluorescein tag of the PS-ONs (Boehringer Mannheim, Indianapolis, IN), diluted 1:60 in buffer PBT, for 2 h at room temperature. Wells were washed three times, 10 min each, and then incubated with polyclonal sheep anti-mouse IgG antibody conjugated with Peroxidase (Cappel/ICN, Costa Mesa, CA) for 1 h at room temperature. After three 10-min washes, peroxidase substrate o-phenylendiamine (Sigma-Fast tablets, Sigma) was added. Finally, 30 min after substrate addition, the absorbance of each well at 450 nm was measured with an EIA reader (model 450; Bio-Rad, Hercules, CA). Control values of cells that were not lipofected with oligonucleotides were subtracted from the results. The means of six different wells per condition were calculated and referred to the mean of the directly fixed cells, which was set to 100%. In indirect immunofluorescence staining the anti-FITC antibody detected a similar intracellular distribution of PS-ONs, as observed with the fluorochrome-conjugated PS-ONs. There was some higher cytoplasmic diffuse staining, and the contrast between the PS foci and the diffuse nucleoplasmic molecules in the nucleus was less pronounced.
Correlative Electron Microscopy (EM)
To analyze the ultrastructural features of phosphorothioate bodies (PS bodies), we matched the pattern observed on the light microscopy level with the pattern of structures seen in the same cell on the EM level. To that end we microinjected 60 μM PS-ON 11068-F (conjugated to fluorescein) into the cytoplasm or nucleus of HeLa cells growing on gridded coverslips (Bellco, Vineland, NJ). One hour after injection, cells were fixed with 4% paraformaldehyde in PBS and quickly examined under the Nikon FXA fluorescence microscope. Particular cells were photographed using the cooled CCD camera (see above), and their position on the coverslip was registered by means of the grids. Then cells were fixed again in 3% glutaraldehyde in 0.2 M cacodylate buffer, pH 7.4, for 45 min at room temperature. The coverslips with the cells were washed in 0.2 M cacodylate buffer, pH 7.4, postfixed in 1% osmium tetroxide, 0.015% potassium ferro-cyanide for 1 h at room temperature, and dehydrated through incubation in a series of ascending concentrations of ethanol. Finally, the samples were embedded in Epon/Araldite, and the glass was etched off with hydrofluoric acid. Regions containing photographed cells were retrieved with the help of the grids of the coverslips, which had been imprinted into the Epon/Araldite plastic during embedding. Cells were serially sectioned (80-nm sections), and the sections were counterstained with 2% uranyl acetate and Reynold’s lead citrate. The sections were examined at 75 kV with a transmission electron microscope (H-7000, Hitachi Scientific Instruments, Mountain View, CA). The same cell photographed at the light microscopy level was located, and the section was identified and photographed that displayed a pattern of structures corresponding to the PS bodies.
RESULTS
Correlation of Intracellular Distribution and Antisense Activity
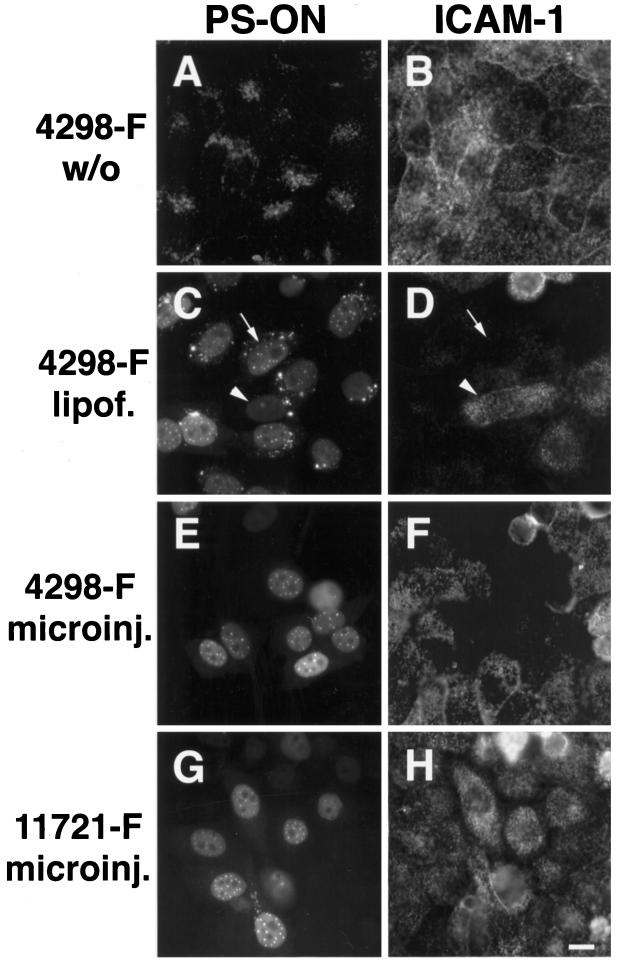
To simultaneously monitor antisense activity and PS-ON distribution in single cells, we delivered fluorescently tagged PS-ONs in the absence or presence of cationic lipids to HeLa cells. Alternatively, PS-ONs were microinjected into the cytoplasm or nucleus of the cells. After induction of endogenous ICAM-1 expression with TNF-α, single-cell analysis of antisense activity was performed by staining the treated cells with an antibody against ICAM-1 (Figure 1). Loss of the ICAM-1 signal indicated antisense activity. When added to the medium without cationic lipids, the antisense PS-ONs stayed trapped in vesicular structures in the cytoplasm and did not show antisense activity (Figure 1, A and B). In contrast, antisense PS-ONs administered to cells by lipofection predominantly localized to the nucleus and inhibited the expression of ICAM-1 protein (Figure 1, C and D). The concentration dependency of this antisense effect was evident by comparing cells with strong or weak oligonucleotide signals, i.e., high or low amounts of internalized PS-ONs: a high amount of oligonucleotide coincided with total loss of ICAM-1 protein staining in that cell (Figure 1, C and D, arrow), whereas a low amount of the antisense molecule still permitted the expression of ICAM-1 protein (Figure 1, C and D, arrowhead). When PS-ONs were microinjected into cells, the oligonucleotide-containing cells displayed bright nuclear fluorescence and strong repression of ICAM-1 protein expression in comparison with surrounding noninjected cells (Figure 1, E and F). Thus, in cells displaying antisense activity, PS-ONs localized predominantly to the cell nucleus. The nuclear distribution in most cells consisted of numerous, bright spherical structures (usually 20–30 per nucleus under the conditions used), hereafter termed PS bodies, which were set against a diffuse nucleoplasmic population (see Figure 2 for a higher magnification). The nucleoli were mostly free of PS-ON staining. This distribution reflected the situation 20 h after the delivery of the PS-ONs to the cells, but it was also representative of the distribution immediately after oligonucleotide delivery, e.g., after lipofection or nuclear microinjection, and coincided with the distribution in living cells (our unpublished results and Figure 8). A control PS-ON without antisense activity against ICAM-1 that reached the nuclear compartment showed a distribution indistinguishable from an active oligonucleotide (Figure 1, G and H). We never detected any differences, within the resolution of the light microscope, between the localization of active and control PS-ONs or in the presence or absence of the endogenous target RNA. We found the same pattern for all PS-ONs tested irrespective of their sequence or the label and means for detection (e.g., different fluorochromes, biotin-tag, or use of an antibody against the PS-ON itself; our unpublished results). Furthermore, the same distribution was seen in cell types other than HeLa, e.g., BHK, A549, IMR-90, Ref52, 3T3, or Rat-1 cells (our unpublished results). In addition, the same nuclear distribution pattern was obtained with antisense PS-ONs against rat β-tropomyosin upon coinjection of oligonucleotides and a plasmid-encoded target gene (Lorenz and Spector, unpublished). With respect to microinjection, there was no difference in the steady-state distribution when the PS-ONs were injected into the cytoplasm or the nucleus (our unpublished results). We also used polyethyleneimine as a transfection agent (Boussif et al., 1995) and found the same distribution as with PS-ONs administered by cationic lipids (our unpublished results). The mostly perinuclear vesicular staining in cells that received PS-ONs with the help of Lipofectin (or polyethyleneimine) was never seen upon microinjection and probably reflects the uptake pathway through endocytotic compartments and inefficient release of PS-ONs from endosomal and/or lysosomal vesicles.
Figure 1.
Simultaneous evaluation of PS-ON localization and antisense activity. Fluorescein-tagged antisense (4298-F) or control (11721-F) PS-ONs were delivered by different means to HeLa cells. They were added to the medium in the absence (A and B, 4298-F; concentration 5 μM) or presence of Lipofectin (C and D, 4298-F; at 0.25 μM), or were directly microinjected (E and F, 4298-F; G and H, 11721-F; concentration in the injection solution 30 μM). ICAM-1 expression was induced by TNF-α for 18 h after the PS-ON delivery. The distribution of the PS-ONs was monitored by means of their fluorescence label (A, C, E, and G), and ICAM-1 expression was detected by indirect immunofluorescence microscopy (B, D, F, and H). When trapped in the cytoplasm, 4298-F (A) did not show any effects on ICAM-1 expression (B). In contrast, a predominant nuclear distribution of 4298-F (C and E) coincided with strong antisense activity (D and F). The arrows or arrowheads point to particular cells with high or low PS-ON content and concomitant low or high ICAM-1 protein staining, respectively (C and D). The control PS-ON 11721, despite showing a similar nuclear distribution as the antisense molecule (G), did nevertheless not display antisense effects (H). Bar, 10 μm.
Figure 2.
Colocalization analysis by confocal microscopy of PS-ONs 11068-F (A) or 12182-T (D and G) microinjected into HeLa cells and three different nuclear structures, “speckles” enriched in splicing factor SC35 (B), PML nuclear bodies (E), and centromeres (H). The nuclear domains were labeled with specific antibodies by indirect immunofluorescence. In all cases the PS-ONs did not accumulate at the respective structures (see merge of the respective images in C, F, and I). Bar, 5 μm.
Figure 8.
PS-ON patterns were stable over a period of 6.5 h. HeLa cells received 0.25 μM PS-ON 11068-F complexed to Lipofectin. Six hours after lipofection time-lapse microscopy of living cells was started and fluorescence images were taken at the indicated times from the same two cells. The arrowheads point to three particular PS bodies in the nucleus that were held in focus throughout the experiment. Their location with respect to each other and to other nuclear structures like the nucleoli did not change noticeably. Note that apparent changes in the PS body pattern, especially in the upper cell, are solely due to focus changes caused by the movements of the cell, e.g. rotations of the nucleus. Bar, 5 μm.
Relationship of PS Bodies to Known Nuclear Structures and Antigens
To determine to which subnuclear structures the PS-ONs might preferentially bind and to see whether the PS bodies represented a known nuclear domain, we used confocal microscopy to compare the PS-ON pattern to various nuclear domains visualized by specific antibodies. Figure 2 compares the distribution of microinjected PS-ONs with the serine/ arginine protein splicing factor SC35, PML nuclear bodies, or centromeres, respectively, which were chosen because they also display foci in their distribution. We did not find any accumulation of PS-ONs at these sites nor were these antigens enriched in the PS bodies. Occasional touching or overlap of the PS bodies with SC35 was observed; however, we think that this association is random and attributed to the limited extrachromosomal space. The patterns of the endogenous nuclear structures looked similar in cells with or without oligonucleotides, suggesting that the PS-ONs did not perturb nuclear architecture. Other structures we found negative for PS-ON accumulation were small nuclear ribonucleoprotein particles (snRNPs), coiled bodies, DNA, DNA replication sites (visualized by bromodeoxyuridine incorporation as described in O’Keefe et al., 1992; our unpublished results), and transcription sites (detected by in situ transcription using bromouridine triphosphate as described in Huang and Spector, 1996; our unpublished results). We also tested colocalization of PS-ONs with heterogeneous nuclear ribonucleoproteins (hnRNPs) A1 and C, two diffusely localized nucleoplasmic proteins. Both are RNA-binding proteins, and A1 has been shown to bind to oligoribonucleotides in vitro (Burd and Dreyfuss, 1994; Abdul-Manan and Williams, 1996). The diffuse localization of the two hnRNPs made it difficult to assess accumulation in PS-ONs. However, at the sites of higher oligonucleotide concentration, i.e., the PS bodies, there was never a higher concentration of hnRNPs A1 and C (our unpublished results). In conclusion, PS-ONs were not specifically attracted by, nor were PS bodies enriched in, any known nuclear protein/structure, although some degree of colocalization may exist due to the diffuse nucleoplasmic distribution of the oligonucleotides.
Characterization of the Formation of PS Bodies
As we did not find a known nuclear structure to colocalize with the prominent PS bodies, the question arises whether these sites reflected binding to unknown nuclear structures or whether they were the result of aggregation of PS-ON–containing complexes. The formation of PS bodies was concentration dependent. When treating HeLa cells in the presence of Lipofectin with 10 nM PS-ON, only 14% of the cells containing the oligonucleotides displayed PS bodies. In contrast, addition of 50 nM or 250 nM PS-ON raised this percentage to 28% or 86%, respectively. At the same time, the number of PS bodies per cell increased. In cells containing very high amounts of PS-ONs, thread-like structures were observed (our unpublished results). The concentration dependency of formation can also be seen in Figure 1C where some cells with fainter fluorescence signals and thus lower oligonucleotide content (e.g., the cell marked by the arrowhead) do not display PS bodies. Since PS-ON signals in the nucleus could be observed in the absence of PS bodies, these structures do not represent sites of highest affinity for PS-ONs. PS bodies formed rapidly within minutes after microinjection (our unpublished results). Chilling of cells strongly reduced the formation of PS bodies by microinjected PS-ONs. Concentration of PS-ONs in discrete foci was rarely observed, and the typical PS body pattern was lacking even after prolonged incubation of the injected cell on ice for 4 h (Figure 3A). Furthermore, PS bodies did not form upon nuclear injection of PS-ONs into formaldehyde-fixed cells (Figure 3 B). Treatments of cells with α-amanitin or 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole, either before or after administration of PS-ONs, indicated that neither the formation nor the persistence of PS bodies was dependent on ongoing RNA polymerase II transcription (Figure 3C and our unpublished results). ATP depletion of cells by treatment with deoxyglucose and NaN3 did not abolish the formation of PS bodies (our unpublished results). These results argue that the formation of PS bodies is concentration dependent, temperature sensitive, but energy and transcription independent.
Figure 3.
Formation of PS bodies was restricted at 4°C, did not occur in fixed cells, and was independent of ongoing RNA-polymerase II transcription. (A) HeLa cells were chilled for 30 min on ice, injected quickly within about 2 min with the PS-ON, and incubated for another 4 h on ice until they were fixed. (B) HeLa cells were fixed and washed and then injected with the PS-ON. (C) HeLa cells were treated for 4 h with 50 μg/ml α-amanitin and then injected with the PS-ON. After another 30 min in the presence of the drug, cells were fixed and stained with antibody against SC-35 to check for the effects of transcription inhibition (SC-35 staining not shown). The noticeable changes of brighter PS-ON signals around the reorganized nucleoli are very likely the result of the nuclear reorganization induced by transcription inhibition (Haaf and Ward, 1996). In all cases, 30 μM PS-ON 11068-F was used for injection. Bar, 5 μm.
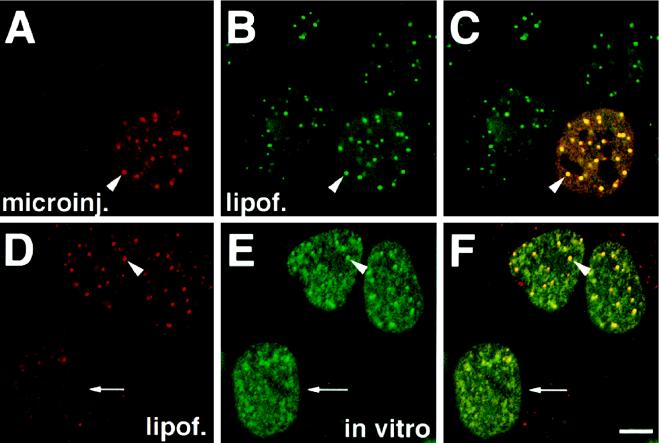
To determine whether two PS-ONs with different sequences localize to the same PS body or accumulate at distinct sites, we introduced the two molecules conjugated with fluorescein or Texas Red, respectively, independently of each other into HeLa cells. The first PS-ON was microinjected into the cytoplasm, and then the second PS-ON was added to the medium complexed with cationic lipids. When such cells were fixed and analyzed by confocal microscopy, the oligonucleotides completely colocalized at each PS body (Figure 4 A–C). No PS bodies were formed by the second PS-ON alone, suggesting that additional PS-ON molecules became attracted to the preexisting PS bodies. When the first PS-ON was delivered to living cells, e.g., using Lipofectin, and the second PS-ON was added to permeabilized cells, these PS-ONs bound to preexisting PS bodies as well (Figure 4, D–F). However, in cells without preexisting PS bodies, the PS-ONs added to the permeabilized cells were not able to form PS bodies alone. They only displayed granular signals (Figure 4, D–F, arrow). Similar results were obtained in cells permeabilized with digitonin, a procedure that retains more nuclear proteins than Triton X-100 permeabilization, and in other buffer systems (our unpublished results). These data are consistent with the notion that the formation of the PS bodies required activities and/or components of living cells and that PS-ONs are not simply binding to preexisting nuclear structures in cells.
Figure 4.
Two different PS-ONs introduced into cells independently of each other colocalize at the same nuclear structures. (A–C) Both PS-ONs were administered into living cells; 12182-T (A) was microinjected and then 11068-F (B) was delivered with the help of cationic lipids. All foci seen with 12182-T were also occupied by 11068-F. (D–F) 12182-T (D) was introduced into living cells by means of cationic lipids. Then cells were permeabilized and incubated with 11068-F at 37°C in vitro (E). In vitro administered 11068-F bound to PS bodies preformed by 12182-T but failed to display such structures in cells without such bodies preformed in the living state. Analysis by confocal microscopy with panels C and F representing merges of images A and B or D and E, respectively. The arrowheads point to particular PS bodies; the arrow marks the cell which has no PS bodies. Bar, 5 μm.
PS Bodies at the Ultrastructural Level
Electron microscopy provides the means to detect nuclear substructures at high resolution. We performed correlative fluorescence and electron microscopy on HeLa cells injected with fluorescently tagged PS-ONs and fixed to give optimal structural preservation. The PS-ON patterns of individual cells were visualized by fluorescence microscopy and photographed, after which the same cells were processed for electron microscopy. The PS body pattern seen at the light microscope level (Figure 5C) was recognizable in the electron microscope sections (Figure 5, A and B). The PS bodies were characterized as 150–300 nm in diameter, electron-dense, homogenous spherical nuclear bodies without obvious substructures. This appearance was clearly different from the ultrastructure of coiled bodies and PML bodies (Brasch and Ochs, 1992; Stuurman et al., 1992; Koken et al., 1994; Bohmann et al., 1995). Interestingly, nuclear bodies of the same size and similar morphology were also present in cells without PS-ONs, although their number was much lower (Figure 5, D and E). This raised the possibility that PS-ON induced the formation of a certain kind of nuclear body. However, there was no indication that the presence of the PS-ONs at the concentration used perturbed nuclear architecture.
Figure 5.
(A–C) On the ultrastructural level PS bodies were electron-dense, 150–300 nm in diameter, spherical nuclear bodies without apparent substructure. HeLa cells injected with 11068-F were fixed, photographed under epifluorescence to record the PS-ON signals, and processed for transmission electron microscopy under conditions that optimally preserve ultrastructure. Then thin sections were prepared from the injected cells. By matching patterns of PS bodies of the fluorescence image with patterns of structures seen on an EM section of the same cell, the PS bodies in the EM image were identified. The arrows point to four PS bodies displaying a characteristic pattern in the EM image of a 80-nm section (A), a higher magnification of the region of the same section (B), and the fluorescence photomicrograph of the same cell (C). (D and E) The ultrastructure of a control nucleus revealed spherical bodies (arrows) similar in appearance to PS bodies. Transmission electron micrograph showing a nucleus of a HeLa cell (D) and an enlargement of two nuclear bodies of the same cell (E), which had not been injected with PS-ONs but was grown on the same coverslip as the injected cells. Bars, 1 μm.
PS-ONs Associate with the Nuclear Matrix
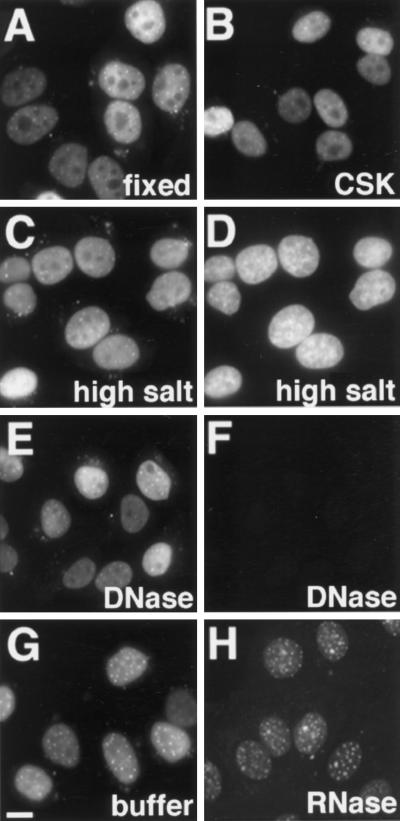
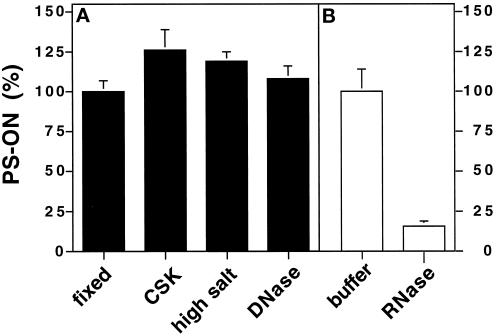
We analyzed how tightly the PS-ONs were bound to nuclear structures. To this end we performed in situ fractionation, which consisted of a series of treatments of unfixed cells that remove soluble fractions and DNA-attached components to obtain the nuclear matrix fraction (Fey et al., 1986). Fluorescently tagged PS-ONs were administered into HeLa cells by means of cationic lipids; the cells were subsequently treated with buffer containing Triton X-100, and then high salt buffer, and finally cells were incubated with DNase I to remove the DNA. At each step a sample was fixed, and images of the PS-ON signal were acquired by fluorescence microscopy (Figure 6, A, C, and E). Throughout the whole procedure the PS-ON distribution was not altered. There were no striking differences in the levels of fluorescence of the PS-ON in the nucleus during fractionation. To compare the amount of the remaining oligonucleotides in a more quantitative manner, we performed an indirect enzyme immunoassay (see MATERIALS AND METHODS) with antibodies against the fluorescein tag of the PS-ONs (Figure 7A). We measured an initial gain of signal between the directly fixed cells and the fractions, but within the fractions there were only small differences. The slight decreases in the signals were probably due to some cell loss during the procedure. The lower values of the directly fixed cells were likely caused by reduced accessibility to the oligonucleotides for the antibodies in comparison with extracted cells. When treated with RNase A instead of DNase, a significant amount of the PS-ONs in the nucleoplasm was removed (Figure 6, compare G with H). In contrast, the PS bodies appeared to be mostly inert against the action of RNase A. The loss of oligonucleotides was clearly evident when measured by enzyme immunoassay (Figure 7B). Our results imply that the majority of PS-ON molecules bind to components of the nuclear matrix and that PS bodies are resistant to both, DNase or RNase A.
Figure 6.
Nuclear fractionation suggested that the majority of intranuclear PS-ONs resided at the nuclear matrix as the nuclear binding was resistant to detergent, salt, and DNase but not to RNase treatment. HeLa cells lipofected with 0.25 μM PS-ON 11068-F were either directly fixed (A) or underwent consecutive treatments with CSK buffer containing 0.5% Triton X-100 (B), extraction buffer containing high salt concentration (C and D) and DNase (E and F) before they were fixed. DAPI staining verified the digestion of the DNA (D vs. F). Cells were also treated with buffer alone (G) or RNase A (H) instead of DNase. Epifluorescence images of the oligonucleotide signals (A–C, E, G, and H) and DNA signals (D and F) were taken from samples of the same lipofection and were acquired and processed in the same manner for panels A–C and E; panels D and F; panels G and H. Bar, 5 μm.
Figure 7.
Comparative measurements of cell-based PS-ONs by indirect enzyme immunoassay corroborated the binding of a major amount of the oligonucleotides to the nuclear matrix. (A) HeLa cells grown in 96-well plates were lipofected with 0.25 μM 11068-F, fixed directly (fixed) or fractionated consecutively with CSK buffer (CSK), extraction buffer (high salt), and DNase (DNase). There was little loss during fractionation. (B) Experiment in which the DNase step had been substituted with buffer alone (buffer) or RNase A (RNase). RNase treatment resulted in a strong displacement of PS-ONs from the nuclear remnants. The values represent the means ± SD of six wells of a representative experiment corrected for the means obtained for cells that had not been lipofected with oligonucleotide. They were normalized against the mean obtained for directly fixed cells (A) or buffer-treated cells (B), which was set at 100%. Similar results were obtained with a different PS-ON.
Dynamics of PS Bodies
To determine whether PS bodies are static or mobile structures, we observed their distribution in living cells over time. We observed the distribution of a fluorescently tagged PS-ON in a single cell by time lapse microscopy keeping particular PS bodies in focus throughout the experiment. Over 6.5 h, the PS body pattern was unchanged, the number of foci was the same, and individual PS bodies did not significantly change their location with respect to each other and, for example, nucleoli (Figure 8). In addition, there was no noticeable increase in cytoplasmic signal. This suggested that PS bodies were rather immobile, stable structures and that during the observed time interval there was neither disintegration nor de novo formation.
To follow the fate of PS-ONs during mitosis, PS-ONs were delivered into interphase cells, either by microinjection or by lipofection, and their distribution was analyzed 6–12 h thereafter. During mitosis, PS-ONs were distributed throughout the mitotic cytoplasm with exclusion of the chromosomes (Figure 9, B and D). In addition, in metaphase, anaphase, or telophase cells, bright spherical foci of accumulated molecules were observed (Figure 9, B and D, arrowheads). As these structures were fewer in number than the PS bodies in interphase cells (compare numbers of PS bodies per nucleus in Figure 9A with 9B and 9D), they were probably made up of several PS bodies fused to each other. Indeed, electron microscopy revealed that these large foci coincided with 500–600 nm in diameter, electron-dense bodies that were morphologically indistinguishable from interphase PS bodies (our unpublished results). However, we cannot exclude that some PS bodies might have disintegrated. We followed a single anaphase cell loaded with PS-ONs through completion of mitosis and entry into the next cell cycle and found that the oligonucleotides reentered the newly formed daughter nuclei (Figure 9, J and K). The reentry of the PS-ONs into the daughter nuclei was later than the reentry of the nucleolar protein fibrillarin, but earlier than the reentry of hnRNP-A1 (our unpublished results). The bright mitotic foci apparently stayed behind in the cytoplasm (Figure 9, F, J, and K, arrowheads) where they decreased in brightness over time until they dissappeared. This can either be explained by a late movement of oligonucleotide molecules from the foci into the daughter nuclei or degradation in the cytoplasm. PS bodies in the daughter nuclei were not evident until the cells were well into G1 (Figure 9, F and K, arrows). Thus, their formation apparently occurred de novo from the diffuse PS-ON pool.
Figure 9.
PS-ONs during mitosis. PS-ONs were excluded from the chromosomes but otherwise dispersed throughout the mitotic cytoplasm. Typical PS bodies (arrows) were not observed in mitotic cells, but fewer and larger foci (arrowheads) were present, which stayed behind the diffusely distributed material with respect to reentry into the daughter nuclei. PS bodies formed late when most of the PS-ON molecules were already nucleoplasmic and the cell was already well into G1 phase. Interphase HeLa cells were injected with 30 μM 11068-F and fixed 2 h later (A–G). To display all PS-ON foci in a cell, confocal z sections throughout the cell were taken and projected on top of each other. Shown are three interphase cells (A), a metaphase (B), a late anaphase (D), and G1 phase (F) cells, the last three identified by their respective Nomarski images (C, E, and G). Time-lapse epifluorescence microscopy of a HeLa cell lipofected with PS-ON 11068-F following it from anaphase through completion of mitosis and entry into G1 phase of the next cell cycle (H–K). Bars, 5 μm.
DISCUSSION
It is important to know the cellular pharmacokinetics of a compound intended for clinical use with respect to its specific and also its unspecific effects. Here we have demonstrated that, under conditions of specific antisense activity against ICAM-1, the predominant localization of PS-ONs was nuclear. This was unequivocally demonstrated by the simultaneous single cell analysis of PS-ON antisense activity and distribution. Since localization of a control PS-ON without antisense effect was indistinguishable from an active molecule, this distribution cannot be considered causally related to specific antisense activity. Nevertheless, the nuclear localization after administration of PS-ONs can serve as an indicator that the oligonucleotides escaped their trapping in vesicular structures in the cytoplasm, which has been suggested to be responsible for failure to detect significant antisense effects (Bennett et al., 1992; Wagner et al., 1993). Furthermore, the predominant nuclear localization of PS-ON molecules argues that the potential of the phosphorothioate modification as a drug for specific activity, but also side-effects, is greater in the nucleus. Even in cases where the antisense molecule acts in the cytoplasm, it might actually hybridize to its target in the nucleus and be exported complexed to RNA (Baker et al., 1997).
The PS-ON localization sites in the nucleus consisted of numerous bright spherical structures designated PS bodies and a diffuse nucleoplasmic population excluded from the nucleoli. Similar distributions of PS-ONs have been observed previously in studies that did not simultaneously measure antisense activity (Chin et al., 1990; Bennett et al., 1992; Fisher et al., 1993; Nestle et al., 1994; Shoeman et al., 1997). Under our experimental conditions, we have no evidence for significant damage to the organization of the nucleus on the light and electron microscopy level after introduction of PS-ONs. Also, DNA replication and in situ transcription using bromodeoxyuridine or bromouridine triphosphate, respectively, were similar to control cells (our unpublished results). The nuclear distribution was similar for all PS-ONs analyzed irrespective of their sequence or of the means used for their detection, e.g., fluorescence tag or indirect staining with an antibody. It was also similar immediately after nuclear injection or minutes after cytoplasmic injection and in living cells up to at least 6.5 h after administration, arguing that what we observed reflected mostly full-length oligonucleotides at their location within live cells. Consistent with that suggestion, stability measurements of PS-ONs delivered with liposomes into tissue culture cells have shown about 80% intact PS-ONs after 6 h and about 70% intact molecules 1 d after delivery (Thierry and Dritschilo, 1992).
PS Bodies, Nuclear Structures Formed by PS-ONs
The formation of the PS bodies, which represent the sites of highest PS-ON accumulation in the nucleus, do not coincide with known nuclear structures, and none of the investigated nuclear antigens were accumulated at these sites. We found the formation of PS bodies to be characteristic of the phosphorothioate modification. A phosphodiester oligonucleotide of the same sequence with 2′-O-propyl ribose moieties for nuclease stability never displayed such a structure (our unpublished results). Furthermore, oligonucleotides of other modifications that contained phosphorothioate internucleoside bridges (e.g., 2′-O-methoxyethyl-PS-ONs or 2′-F-PS-ONs) show PS bodies as well and colocalize with PS bodies formed by plain PS-ONs (Lorenz, Baker, Bennett, and Spector, unpublished). PS bodies have also been observed in previously published reports (Chin et al., 1990; Bennett et al., 1992; Fisher et al., 1993; Shoeman et al., 1997). In contrast to the present study, none of these reports provided a detailed characterization of these structures. The concentration dependency of the formation of PS bodies argues that they might result from aggregation of PS-ONs or PS-ON–containing complexes. However, several lines of evidence suggest that the formation of PS bodies is not simply a random self-aggregation process but requires the environment of a living cell for its induction: first, PS bodies were not observed upon delivery of PS-ONs to fixed or permeabilized cells. Second, even after prolonged incubation at 4°C, the typical PS body pattern was lacking. Third, on the ultrastructural level PS bodies resemble nuclear bodies that can be found, notably with less frequency, in control cells in the absence of PS-ONs. Fourth, the PS bodies show dynamic changes during mitosis and then form de novo in the newly assembled daughter nuclei. Thus, it is likely that PS bodies formed de novo once a certain concentration was exceeded and that this event takes place only in intact cells with all their components and activities.
Once formed, existing PS bodies appear to act as nucleation sites for additional PS-ONs, even molecules delivered to permeabilized cells. This also provides the explanation for the colocalization of two PS-ONs, delivered independently into living cells, at each PS body. Since the formation of the PS bodies occurred very quickly and in ATP-depleted cells, any cellular components involved are likely to already be present in the nucleus and more time-consuming processing steps or metabolic energy for the assembly of the PS bodies do not appear to be required.
The composition and function of nuclear bodies are poorly defined (Brasch and Ochs, 1992). Our results showed that PS bodies neither colocalize nor structurally coincide with coiled or PML bodies, two of the better characterized nuclear bodies. The PS bodies appear to fit most closely into the class of simple nuclear bodies (Chaly et al., 1983; Brasch and Ochs, 1992). Coiled and PML bodies have been shown to respond to cell cycle, metabolic status, transformation, or viral infection by changes in their composition, increased formation, or disintegration (Spector et al., 1992; Andrade et al., 1993; Ochs et al., 1995; Terris et al., 1995; Ishov and Maul, 1996; Ahn and Hayward, 1997). In contrast, we have no evidence for significant differences in the number and pattern of PS bodies in quiescent versus cycling or in primary versus transformed cells (our unpublished results). In addition, we did not find snRNPs or nucleolar antigens to be enriched in PS bodies as in the case of coiled bodies (Bohmann et al., 1995). PS bodies share some similarities in their behavior during mitosis with PML or coiled bodies: with respect to their presence in mitotic cells, PS bodies behave similarly to PML bodies (Stuurman et al., 1992). However, considering that PS bodies apparently assembled de novo in the daughter nuclei from the diffusely localized molecules, in that respect they behaved in a similar manner to coiled bodies that reassemble in G1 phase (Brasch and Ochs, 1992; Bohmann et al., 1995). Exclusion of larger particles from reentry into the daughter nuclei has also been observed for the splicing factor SC-35 (Spector et al., 1991), the meaning of which is unknown.
One possible explanation for the accumulation of PS-ONs in PS bodies is a reaction of a cell to sequester an overload of stable oligonucleotide to render it unable to exert toxic effects. The formation of nuclear bodies might also be coupled to the degradation of PS-ONs. The disappearance of the large mitotic PS-ON foci in the cytoplasm of G1 phase cells could reflect one such degradation pathway. It will be useful to identify host cell factors required for the formation of PS bodies. One might find at these sites oligonucleotide-metabolizing activities, such as nucleases, which are still ill-defined on the molecular level and with respect to their specific cellular location. Sequestration in PS bodies might limit the amount of free or loosely bound PS-ON available for hybridization to the target RNA or, on the contrary, might provide a depot of antisense molecules where PS-ONs are bound through relatively low-affinity interactions in comparison with the high affinities displayed toward their target sequences in the RNA. Such depot effects have been suggested to be one explanation for the unaffected potencies of C-5 propyne PS-ONs upon changes in the levels of expressed target RNA (Flanagan et al., 1996). Another possibility is that small cationic molecules, such as the polyamines spermine and spermidine, play a role in binding PS-ONs and forming PS bodies. Polyamines have been localized to the cell nucleus by light and electron microscopy and are implicated in the condensation of chromatin (Hougaard, 1992; Roch et al., 1997 and references therein). However, interactions between PS-ONs and polyamines have not been investigated so far. Coinjection of a 100-fold excess of spermine and spermidine over a PS-ON did not result in any noticeable redistribution of the oligonucleotides or change of the PS body structures (our unpublished results). In addition, coinjection of 20 mM of a potentially competing polyanion, heparin, did not abolish formation of PS bodies (our unpublished results). It will be interesting to determine whether PS bodies are formed in vivo in the tissues of an organism treated with PS-ONs as well.
PS-ONs Are Associated with the Nuclear Matrix
The nuclear pools of PS-ONs were found to bind to the nuclear matrix, which is operationally defined as the nuclear remnants persisting after removal of DNA and loosely bound nuclear components (Berezney, 1991). It consists of a peripheral component made up of the nuclear lamina and an internal matrix including residual nucleoli and an RNP network. As most PS-ON molecules were resistant to the removal of loosely attached material and DNA, associating proteins likely have to be sought in this fraction. The internal nuclear matrix is made up of proteins and RNA. The latter explains why RNase treatment extracts nuclear matrix material. RNase treatment also displaced most of the bound PS-ON with the noticeable exception of the PS body-bound molecules. Therefore, it is likely that PS-ONs are directly or indirectly bound to an RNP. In contrast, PS bodies appear to be tightly bound to the nuclear matrix independently of RNA, or RNase may not be accessible to these nuclear structures. Among the most abundant nuclear matrix proteins are hnRNPs (Mattern et al., 1996). Many proteins, however, have been found in the nuclear matrix fraction, and many nuclear functions such as transcription and DNA replication and splicing are thought to be linked to it (Berezney, 1991). Among the proteins/complexes we analyzed for colocalization with the PS-ONs were SC-35, snRNPs, hnRNP-A1, and hnRNP-C, all of them known to be associated with the nuclear matrix. The serine/arginine protein-splicing factor SC-35 and snRNPs are both found in “speckles” (Spector, 1993). In contrast, PS-ONs never showed a “speckled” pattern. It has been argued previously that there is some degree of overlap between snRNPs and PS-ONs (Chin et al., 1990; Shoeman et al., 1997), but when we used four different antibodies to antigens in “speckles,” we clearly saw no enrichment of PS-ON at these structures. In the other studies, higher concentrations of PS-ONs than used here were injected, which might influence the results. hnRNPs A1 and C are both diffusely localized in the nucleoplasm, and therefore their signals colocalize with the diffuse PS-ON signals. At the sites of higher PS-ON concentration, however, i.e., the PS bodies, we never detected higher concentrations of hnRNPs. Two other observations also speak against a stable association of hnRNP-A1 and PS-ONs. Upon transcription inhibition, the shuttling of hnRNP-A1 is inhibited and the A1 molecules become trapped in the cytoplasm (Piñol-Roma and Dreyfuss, 1992), a phenomenon we did not observe for PS-ONs (our unpublished results). Also, reentry into newly formed daughter nuclei occurred earlier for PS-ONs than for hnRNP-A1 (our unpublished results). However, there are many other possible proteins in nuclear matrix fractions that might have affinities for oligonucleotides. If a PS-ON–interacting molecule is diffusely localized or PS-ONs bind to a vast variety of different nuclear molecules, it would also prevent us from observing complexes between PS-ONs and particular antigens by fluorescence microscopy. A number of proteins have been cross-linked to PS-ONs that have been added to isolated nuclei (Leonetti et al., 1991), and two categories of high- and low- affinity binding sites totaling about 6 million per cell nucleus have been postulated (Clarenc et al., 1993). Others have found a high extent of complex formation between PS-ONs and nuclear proteins in gel shift assays (Brown et al., 1994). However, no protein has been identified, and the result might not reflect the in vivo situation. Many PS-ON–binding proteins thus far identified are cell surface molecules (reviewed by Stein, 1996). One exception is the predominantly nucleolar protein nucleolin, which binds PS-ONs in vitro (Weidner et al., 1995). However, we did not usually find PS-ONs within the nucleolus. Only when the nucleoplasm was saturated with high amounts of oligonucleotides did some overlap with nucleolar proteins occur at the nucleolar borders (our unpublished results).
Taken together, our results provide more insight into the spatial and temporal distribution of PS-ONs in the nucleus under conditions at which specific antisense activity can occur. We have characterized a nuclear structure that was formed after PS-ONs were introduced into the cell and that may be involved in sequestering oligonucleotides. Clearly, more has to be learned about the cell biology of antisense compounds to make this approach a true rational drug design. The questions where exactly antisense molecules hybridize to their target, what determines accessibility for this hybridization, what is the fate of the oligonucleotide/target RNA hybrid and where and through what mechanisms the antisense activity is actually exerted are all largely unanswered on the molecular level. Furthermore, many intracellular factors which interact with the phosphorothioate modification remain to be identified.
ACKNOWLEDGMENTS
We are particularly grateful to Tamara Howard for excellent electron microscopy. We thank Joe Kraft, Adrian Krainer, Eng Tan, and Roel Van Driel for antibodies; John Brugger, Sheri Manalili, Henri Sasmor and Mary Ann Zounes at ISIS Pharmaceuticals for synthesis of the oligonucleotides; the members of the Spector laboratory for helpful discussions and Tom Misteli for many valuable discussions and critical reading of the manuscript. P.L. was a recipient of a research fellowship from the Deutsche Forschungsgemeinschaft. This work was supported by grants from the National Insitutes of Health (NIGMS 42694) and ISIS Pharmaceuticals to D.L.S.
REFERENCES
- Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S. Antisense oligonucleotides: towards clinical trials. Trends Biotechnol. 1996;14:376–387. doi: 10.1016/0167-7799(96)10053-6. [DOI] [PubMed] [Google Scholar]
- Agrawal S, Iyer RP. Modified oligonucleotides as therapeutic and diagnostic agents. Curr Opin Biotechnol. 1995;6:12–19. doi: 10.1016/0958-1669(95)80003-4. [DOI] [PubMed] [Google Scholar]
- Ahn JH, Hayward GS. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J Biol Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- Beaucage SL, Iyer RP. Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron. 1992;48:2223–2311. [Google Scholar]
- Beltinger C, et al. Binding, uptake, and intracellular trafficking of phosphorothioate-modified oligodeoxynucleotides. J Clin Invest. 1995;95:1814–1823. doi: 10.1172/JCI117860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Chiang MY, Chan H, Shoemaker JE, Mirabelli CK. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- Bennett CF, Condon TP, Grimm S, Chan H, Chiang MY. Inhibition of endothelial cell adhesion molecule expression with antisense oligonucleotides. J Immunol. 1994;152:3530–3540. [PubMed] [Google Scholar]
- Bennett CF, et al. An ICAM-1 antisense oligonucleotide prevents and reverses dextran sulfate sodium-induced colitis in mice. J Pharmacol Exp Ther. 1997;280:988–1000. [PubMed] [Google Scholar]
- Berezney R. The nuclear matrix: a heuristic model for investigating genomic organization and function in the cell nucleus. J Cell Biochem. 1991;47:109–123. doi: 10.1002/jcb.240470204. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Santama N, Weis K, Lamond AI. Molecular analysis of the coiled body. J Cell Sci Suppl. 1995;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualcqh F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S, Xu X, Nerenberg MI. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaly N, Setterfield G, Kaplan JG, Brown DL. Nuclear bodies in mouse splenic lymphocytes: II. Cytochemistry and autoradiography during stimulation by concanavalin A. Biol Cell. 1983;49:35–43. doi: 10.1111/j.1768-322x.1984.tb00220.x. [DOI] [PubMed] [Google Scholar]
- Chiang MY, Chan H, Zounes MA, Freier SM, Lima WF, Bennett CF. Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J Biol Chem. 1991;266:18162–18171. [PubMed] [Google Scholar]
- Chin DJ, Green GA, Zon G, Szoka FC, Jr, Straubinger RM. Rapid nuclear accumulation of injected oligodeoxyribonucleotides. New Biol. 1990;2:1091–1100. [PubMed] [Google Scholar]
- Clarenc JP, Lebleu B, Leonetti JP. Characterization of the nuclear binding sites of oligodeoxyribonucleotides and their analogs. J Biol Chem. 1993;268:5600–5604. [PubMed] [Google Scholar]
- Coats S, Flanagan WM, Nourse J, Roberts JM. Requirement of p27Kip1 for restriction point control of the fibroblast cell cycle. Science. 1996;272:877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- Condon TP, Bennett CF. Altered mRNA splicing and inhibition of human E-selectin expression by an antisense oligonucleotide in human umbilical vein endothelial cells. J Biol Chem. 1996;271:30398–30403. doi: 10.1074/jbc.271.48.30398. [DOI] [PubMed] [Google Scholar]
- Crooke ST. Progress in antisense therapeutics. Med Res Rev. 1996;16:319–344. doi: 10.1002/(SICI)1098-1128(199607)16:4<319::AID-MED2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Crooke ST. Advances in understanding the pharmacological properties of antisense oligonucleotides. Adv Pharmacol. 1997;40:1–49. doi: 10.1016/s1054-3589(08)60136-2. [DOI] [PubMed] [Google Scholar]
- Crooke ST, Bennett CF. Progress in antisense oligonucleotide therapeutics. Annu Rev Pharmacol Toxicol. 1996;36:107–129. doi: 10.1146/annurev.pa.36.040196.000543. [DOI] [PubMed] [Google Scholar]
- Dean NM, McKay R. Inhibition of protein kinase C-alpha expression in mice after systemic administration of phosphorothioate antisense oligodeoxynucleotides. Proc Natl Acad Sci USA. 1994;91:11762–11766. doi: 10.1073/pnas.91.24.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean NM, McKay R, Miraglia L, Geiger T, Muller M, Fabbro D, Bennett CF. Antisense oligonucleotides as inhibitors of signal transduction: development from research tools to therapeutic agents. Biochem Soc Trans. 1996a;24:623–629. doi: 10.1042/bst0240623. [DOI] [PubMed] [Google Scholar]
- Dean N, et al. Inhibition of Growth of Human tumor cell lines in nude mice by an antisense of oligonucleotide inhibitor of protein kinase C-alpha expression. Cancer Res. 1996b;56:3499–3507. [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TL, Terhorst T, Cao X, Wagner RW. Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res. 1993;21:3857–3865. doi: 10.1093/nar/21.16.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan WM, Kothavale A, Wagner RW. Effects of oligonucleotide length, mismatches and mRNA levels on C-5 propyne-modified antisense potency. Nucleic Acids Res. 1996;24:2936–2941. doi: 10.1093/nar/24.15.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Giles RV, Spiller DG, Tidd DM. Detection of ribonuclease H-generated mRNA fragments in human leukemia cells following reversible membrane permeabilization in the presence of antisense oligodeoxynucleotides. Antisense Res Dev. 1995;5:23–31. doi: 10.1089/ard.1995.5.23. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp Cell Res. 1996;224:163–173. doi: 10.1006/excr.1996.0124. [DOI] [PubMed] [Google Scholar]
- Habets WJ, Hoet MH, De Jong BA, Van der Kemp A, Van Venrooij WJ. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- Hougaard DM. Polyamine cytochemistry: localization and possible functions of polyamines. Int Rev Cytol. 1992;138:51–88. doi: 10.1016/s0074-7696(08)61587-9. [DOI] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken MH, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16:9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc Natl Acad Sci USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger SM, Ratajczak J, Ratajczak MZ, Kuczynski WI, DiPaola RS, Ngo W, Clevenger CV, Gewirtz AM. A functional analysis of protooncogene Vav’s role in adult human hematopoiesis. Blood. 1996;87:1326–1334. [PubMed] [Google Scholar]
- Mattern KA, van Goethem RE, de Jong L, van Driel R. Major internal nuclear matrix proteins are common to different human cell types. J Cell Biochem. 1997;65:42–52. doi: 10.1002/(sici)1097-4644(199704)65:1<42::aid-jcb5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Monia BP, Johnston JF, Geiger T, Muller M, Fabbro D. Antitumor activity of a phosphorothioate antisense oligodeoxynucleotide targeted against C-raf kinase. Nat Med. 1996;2:668–675. doi: 10.1038/nm0696-668. [DOI] [PubMed] [Google Scholar]
- Moroi Y, Peebles C, Fritzler MJ, Steigerwald J, Tan EM. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Mitra RS, Bennett CF, Chan H, Nickoloff BJ. Cationic lipid is not required for uptake and selective inhibitory activity of ICAM-1 phosphorothioate antisense oligonucleotides in keratinocytes. J Invest Dermatol. 1994;103:569–575. doi: 10.1111/1523-1747.ep12396876. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Pettersson S, Meyer zum Buschenfelde KH, Strober W. Local administration of antisense phosphorothioate oligonucleotides to the p65 subunit of NF-kappa B abrogates established experimental colitis in mice. Nat Med. 1996;2:998–1004. doi: 10.1038/nm0996-998. [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Stein TW, Jr, Andrade LE, Gallo D, Chan EK, Tan EM, Brasch K. Formation of nuclear bodies in hepatocytes of estrogen-treated roosters. Mol Biol Cell. 1995;6:345–356. doi: 10.1091/mbc.6.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offensperger WB, Offensperger S, Walter E, Teubner K, Igloi G, Blum HE, Gerok W. In vivo inhibition of duck hepatitis B virus replication and gene expression by phosphorothioate modified antisense oligodeoxynucleotides. EMBO J. 1993;12:1257–1262. doi: 10.1002/j.1460-2075.1993.tb05767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Roch AM, Nicolas MT, Quash G. Ultrastructural immunolocalization of polyamines in HeLa cells subjected to fast-freezing fixation and freeze substitution. Histochem Cell Biol. 1997;107:303–312. doi: 10.1007/s004180050115. [DOI] [PubMed] [Google Scholar]
- Sharma HW, Narayanan R. The therapeutic potential of antisense oligonucleotides. Bioessays. 1995;17:1055–1063. doi: 10.1002/bies.950171210. [DOI] [PubMed] [Google Scholar]
- Shoeman RL, Hartig R, Huang Y, Grueb S, Traub P. Fluorescence microscopic comparison of the binding of phosphodiester and phosphorothioate (antisense) oligodeoxyribonucleotides to subcellular structures, including intermediate filaments, the endoplasmic reticulum, and the nuclear interior. Antisense Nucleic Acid Drug Dev. 1997;7:291–308. doi: 10.1089/oli.1.1997.7.291. [DOI] [PubMed] [Google Scholar]
- Shuttleworth J, Colman A. Antisense oligonucleotide-directed cleavage of mRNA in Xenopus oocytes and eggs. EMBO J. 1988;7:427–434. doi: 10.1002/j.1460-2075.1988.tb02830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell. 1992;3:555–569. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA. Phosphorothioate antisense oligodeoxynucleotides: questions of specificity. Trends Biotechnol. 1996;14:147–149. doi: 10.1016/0167-7799(96)20006-X. [DOI] [PubMed] [Google Scholar]
- Stepkowski SM, Tu Y, Condon TP, Bennett CF. Blocking of heart allograft rejection by intercellular adhesion molecule-1 antisense oligonucleotides alone or in combination with other immunosuppressive modalities. J Immunol. 1994;153:5336–5346. [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Tarrason G, Bellido D, Eritja R, Vilaro S, Piulats J. Digoxigenin-labeled phosphorothioate oligonucleotides: a new tool for the study of cellular uptake. Antisense Res Dev. 1995;5:193–201. doi: 10.1089/ard.1995.5.193. [DOI] [PubMed] [Google Scholar]
- Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- Thierry AR, Dritschilo A. Intracellular availability of unmodified, phosphorothioated and liposomally encapsulated oligodeoxynucleotides for antisense activity. Nucleic Acids Res. 1992;20:5691–5698. doi: 10.1093/nar/20.21.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RW, Flanagan WM. Antisense technology and prospects for therapy of viral infections and cancer. Mol Med Today. 1997;3:31–38. doi: 10.1016/S1357-4310(96)10053-8. [DOI] [PubMed] [Google Scholar]
- Wagner RW, Matteucci MD, Grant D, Huang T, Froehler BC. Potent and selective inhibition of gene expression by an antisense heptanucleotide. Nat Biotechnol. 1996;14:840–844. doi: 10.1038/nbt0796-840. [DOI] [PubMed] [Google Scholar]
- Wagner RW, Matteucci MD, Lewis JG, Gutierrez AJ, Moulds C, Froehler BC. Antisense gene inhibition by oligonucleotides containing C-5 propyne pyrimidines. Science. 1993;260:1510–1513. doi: 10.1126/science.7684856. [DOI] [PubMed] [Google Scholar]
- Webb A, Cunningham D, Cotter F, Clarke PA, di Stefano F, Ross P, Corbo M, Dziewanowska Z. BCL-2 antisense therapy in patients with non-Hodgkin lymphoma. Lancet. 1997;349:1137–1141. doi: 10.1016/s0140-6736(96)11103-x. [DOI] [PubMed] [Google Scholar]
- Weidner DA, Valdez BC, Henning D, Greenberg S, Busch H. Phosphorothioate oligonucleotides bind in a non sequence-specific manner to the nucleolar protein C23/nucleolin. FEBS Lett. 1995;366:146–150. doi: 10.1016/0014-5793(95)00517-d. [DOI] [PubMed] [Google Scholar]