Abstract
Mammals have circadian clocks in peripheral tissues, but there is no direct evidence of their physiological importance. Unlike the suprachiasmatic nucleus clock that is set by light and drives rest–activity and fasting–feeding cycles, peripheral clocks are set by daily feeding, suggesting that at least some contribute metabolic regulation. The liver plays a well known role in glucose homeostasis, and we report here that mice with a liver-specific deletion of Bmal1, an essential clock component, exhibited hypoglycemia restricted to the fasting phase of the daily feeding cycle, exaggerated glucose clearance, and loss of rhythmic expression of hepatic glucose regulatory genes. We conclude that the liver clock is important for buffering circulating glucose in a time-of-day-dependent manner. Our findings suggest that the liver clock contributes to homeostasis by driving a daily rhythm of hepatic glucose export that counterbalances the daily cycle of glucose ingestion resulting from the fasting–feeding cycle.
Keywords: glucose homeostasis, liver
Intrinsic daily rhythms of physiology and behavior are driven by circadian clocks, cell-autonomous oscillators built on a molecular feedback loop (1). In mammals, the circadian clock of the suprachiasmatic nucleus (SCN) acts as the central pacemaker driving circadian rhythms of behavior (2). Recently it has been recognized that circadian clocks are also found in additional brain regions (3), the retina (4), and in many peripheral tissues (5–7). Evidence for the importance of circadian clocks outside the SCN is only just emerging (8–10). In Drosophila, there is evidence that the antenna clock is sufficient to drive circadian rhythms of olfactory physiology (11).
Under conditions of continuous food availability, the SCN clock sets the phases of peripheral tissue clocks (6). However, when animals are placed on a daily feeding schedule that differs from the daily cycle of spontaneous feeding, peripheral tissue clocks, but not the SCN clock, are set to a new phase determined by the imposed feeding schedule (7, 12). The ability of the SCN to set the phases of peripheral tissue clocks thus appears to depend on its ability to drive daily rhythms of feeding behavior, resulting in daily fasting–feeding cycles (13). In principle, the entrainment of peripheral clocks by brain-driven fasting–feeding cycles allows peripheral tissues to anticipate daily fasting and daily food consumption, potentially optimizing processes required for food ingestion, metabolism, and energy storage and utilization.
Recent genetic studies strongly suggest an important role for circadian clocks in energy balance and glucose homeostasis (14–16). The mice in these studies had genetic alterations of circadian clock function in all tissues, including the SCN, and consequently had grossly defective regulation of sleeping, waking, locomotor activity, and feeding behavior. Thus it cannot be determined from these studies to what extent the observed metabolic abnormalities resulted from abnormal behavioral or physiological outputs of the SCN clock or from a loss of circadian clock function in one or more peripheral tissues involved in energy and glucose metabolism.
The liver plays a well known role in glucose homeostasis (17). When circulating glucose is abundant, the liver absorbs and stores glucose by converting it to glycogen, and when it is in demand, the liver produces glucose, derived from de novo synthesis (gluconeogenesis) or stored glycogen (glycogenolysis), and exports it into the circulation. A number of genes expressed in the liver that function in hepatic glucose metabolism exhibit robust circadian regulation (18–20), and mice with altered circadian clocks in all tissues have evidence of impaired gluconeogenesis (19), raising the possibility that the liver circadian clock plays a significant role in hepatic glucose metabolism. However, lesion studies have suggested that hepatic glucose metabolism is regulated at least in part by the SCN via autonomic projections (21), so it is possible that impaired hepatic gluconeogenesis in germ-line circadian clock mutant mice is secondary to impaired function of the SCN clock.
Results
To investigate the physiological functions of the liver clock, we generated mice with a liver-specific disruption of Bmal1 (Mop3), an essential component of the circadian clock (22). Our first step was to assess glucose homeostasis and energy balance in mice lacking Bmal1 function in all tissues (and therefore circadian clock function in all tissues) in the same genetic background as the planned liver-specific Bmal1 disruption (C57BL/6 × 129). Failure to identify metabolic abnormalities in this background would make it unlikely that a liver-specific disruption of Bmal1 would produce a relevant phenotype.
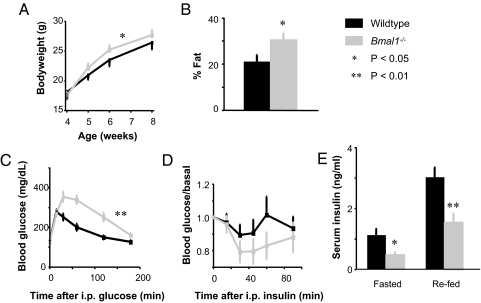
Because Bmal1−/− mice begin to develop a progressive arthropathy at 14 weeks of age (23), we used mice between 8 and 12 weeks old. As expected, Bmal1−/− mice in the hybrid C57BL/6 × 129 background had no detectable daily rhythms of locomotor activity or feeding behavior in constant darkness or under a light–dark cycle, instead manifesting characteristic ultradian behavior (data not shown). In a standard light–dark cycle, Bmal1−/− mice gained weight somewhat more rapidly than wild-type littermates between 4 and 8 weeks of age (Fig. 1A) (P < 0.03, genotype × time, ANOVA), but by young adulthood they had normal bodyweight (Fig. 1A). However, Bmal1−/− mice had increased total fat content (Fig. 1B) (P < 0.05, t test), perhaps reflecting systemic influences on fat, given a possible positive role of Bmal1 in adipocyte differentiation (24). Although Bmal1−/− mice had normal resting blood glucose, they showed intolerance to a bolus of glucose, responding with abnormally high blood glucose concentrations that were restored to normal values far more slowly than in wild-type littermates (Fig. 1C) (P < 0.01, genotype × time, ANOVA). Similar to a previous report (14), they exhibited a trend toward insulin hypersensitivity (Fig. 1D). Defects of glucose regulation typically exhibit increasing severity with age (25), so it is notable that Bmal1−/− mice at only 8–12 weeks exhibit glucose intolerance comparable to that of mice lacking AKT2, a major integrator of insulin signaling (26). After an overnight fast or after an overnight fast followed by refeeding, Bmal1−/− mice had significantly reduced serum insulin (Fig. 1E) (P < 0.02, P < 0.01, respectively, t test). Overall, we found a very similar pattern of metabolic defects in Per1−/−, Per2−/− double-mutant mice (129 background) [supporting information (SI) Fig. S1], in which circadian clock function is disrupted in a manner different from that of Bmal1−/− mice (27). These results indicate that Bmal1 function is important for the regulation of total body fat, glucose clearance, and insulin production and that these phenotypes very likely reflect the role of Bmal1 in the circadian clock mechanism, in the function of either the SCN clock or clocks at other sites (or both).
Fig. 1.
Glucose intolerance and abnormal energy balance in mice lacking Bmal1 function in all tissues. Shown are comparisons of Bmal1−/− mice and wild-type littermates (C57BL/6 × 129). (A) Bodyweight (ANOVA). (B) Total body fat content (t test). (C) Glucose tolerance (ANOVA); Zeitgeber time (ZT, in h) 4.5. (D) Insulin tolerance (ANOVA; the trend toward hypersensitivity was not significant); ZT 8.5 (in a 12-h light/12-h dark cycle). (E) Serum insulin concentration after overnight fast with or without refeeding (t test); ZT 2.5 and 4.5, respectively. Shown are mean and SEM of 7–9 mice of each genotype. *, P < 0.05; **, P < 0.01.
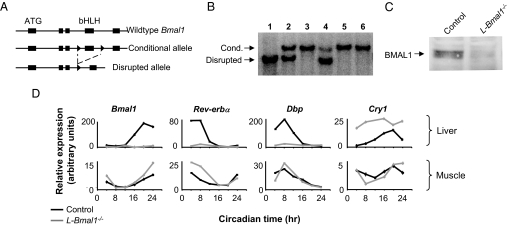
To generate mice with liver-specific loss of circadian clock function, we bred mice with a conditional Bmal1 allele (Fig. 2A) (10) with mice carrying a Cre recombinase transgene under control of the albumin promoter (albumin-Cre), which acts exclusively in hepatocytes (28). All experiments described below were performed with paired male littermate mice (C57BL/6 × 129) carrying a single copy of the albumin-Cre transgene and homozygous either for the Bmal1 conditional allele (L-Bmal1−/− mice) or for the wild-type Bmal1 allele (controls). This arrangement controls for any potential phenotype caused by the persistent expression of the Cre recombinase protein in hepatocytes.
Fig. 2.
Liver-specific loss of circadian clock function. (A) Conditional Bmal1 allele and disruption by Cre recombinase. Boxes, exons; ATG, translation start site; bHLH, basic helix–loop—helix domain; triangles, loxP sites. (B) Liver-specific disruption of Bmal1 conditional allele: genomic Southern blot showing fragments diagnostic of the conditional or disrupted Bmal1 alleles, as marked. Lanes 1–4, liver genomic DNA. Lane 1, homozygous Bmal1 conditional, ubiquitous Cre; lane 2, heterozygous for disrupted allele; lane 3, homozygous Bmal1 conditional, no Cre; and lane 4, homozygous Bmal1 conditional, albumin-Cre. Lanes 5 and 6, genomic DNA from skeletal muscle and kidney, respectively, from same mouse as lane 4. (C) Loss of BMAL1 protein in the liver: anti-BMAL1 Western blot of protein extracts from livers of L-Bmal1−/− and littermate control mice, as indicated. (D) Liver-specific loss of molecular circadian rhythms: quantitative reverse-transcriptase PCR (Q-PCR) showing temporal expression profiles of Bmal1 and other clock-associated genes, as indicated, in liver and muscle of L-Bmal1−/− mice and littermate controls. Shown are the mean and SEM of triplicate assays (most error bars are too small to be seen at this scale). Dbp, D-site albumin promoter binding protein; Cry1, Cryptochrome 1.
L-Bmal1−/− mice showed the predicted disruption of the Bmal1 gene in the liver but not in kidney or skeletal muscle (Fig. 2B). The residual nonrecombined allele in the liver (Fig. 2B, lane 4) was likely derived from nonhepatocyte cells, such as bile duct cells and vascular epithelial cells, in which Cre recombinase is not expressed. In addition, the mice showed the expected loss of BMAL1 protein from the liver (Fig. 2C), with trace residual protein also likely derived from nonhepatocyte cells. As expected, Bmal1 expression was severely reduced across the circadian cycle in the livers of L-Bmal1−/− mice but showed a normal rhythm of expression in skeletal muscle from the same animals (Fig. 2D). Bmal1-dependent genes Dbp and Rev-erbα (29) exhibited a nearly complete loss of expression in the liver but showed normal rhythmic expression in skeletal muscle, and Cry1, a core clock gene, showed the up-regulated expression expected in the absence of Bmal1 function (30) only in the liver (Fig. 2D). In contrast, Per2 transcript and protein showed persistent circadian regulation in the livers of L-Bmal1−/− mice (data not shown), confirming that Per2 rhythms in the liver can be driven by external signals in the absence of intrinsic clock function (31). Histopathological examination of livers from adult L-Bmal1−/− mice and littermate controls revealed no apparent differences in tissue architecture between genotypes (data not shown).
The circadian organization and overall quantity of both locomotor activity and feeding behavior of L-Bmal1−/− mice did not differ from controls (Fig. S2), demonstrating intact SCN function. In addition, serum concentrations of corticosterone and glucagon at two times of day did not significantly differ between L-Bmal1−/− mice and controls (Table S1). Together our results indicate that L-Bmal1−/− mice have a liver-specific loss of circadian clock function, allowing investigation of the physiological functions of the liver clock independently of other clocks and in a context of normal circadian regulation of feeding behavior and locomotor activity.
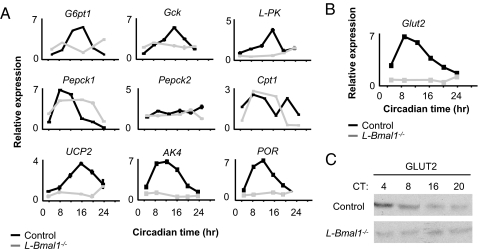
We next tested the expression of selected genes involved in hepatic physiology that were previously shown to have robust circadian expression profiles in the liver (18–20), including some important for hepatic glucose metabolism (Fig. 3A). Most, but not all (e.g., Pepck1), lost circadian regulation in L-Bmal1−/− mice (Fig. 3), consistent with the observation that many clock-regulated genes in the liver are under local circadian control, but a small minority can apparently be driven directly by rhythmic hormonal or neural signals (31). These results suggest that a loss of clock function in the liver is likely to have a significant impact on liver physiology.
Fig. 3.
Loss of rhythmic expression of clock-regulated metabolic genes in the livers of L-Bmal1−/− mice. (A) One-day temporal expression profiles of selected hepatic metabolic genes. Shown are the mean and SEM of Q-PCR triplicate assays (most error bars are too small to be seen at this scale). G6pt1, glucose-6-phosphate translocase 1; Gck, glucokinase; L-PK, liver pyruvate kinase; Pepck1, phosphoenolpyruvate carboxykinase 1; Pepck2, phosphoenolpyruvate carboxykinase 2; Cpt1, carnitine palmitoyltransferase 1; Ucp2, uncoupling protein 2; AK4, adenylate kinase 4; POR, P450 oxidoreductase. (B) One-day temporal expression profile of hepatic Glut2 (glucose transporter 2) transcript in the indicated genotypes. Shown are mean and SEM of Q-PCR triplicate assays (most error bars are too small to be seen at this scale). (C) One-day expression profile of GLUT2 protein. Shown is a Western blot of liver protein extracts obtained from mice of the indicated genotypes. CT, circadian time.
In addition, L-Bmal1−/− mice lost circadian expression of the glucose transporter 2 (Glut2) transcript and protein, both of which were detectable but appeared to be fixed at the approximate level of their normal daily minimum of expression (Fig. 3 B and C). Loss of function of the Glut2 gene in humans causes Fanconi–Bickel syndrome (32), a complex metabolic disorder in which one abnormality is fasting hypoglycemia, apparently caused by defective hepatic glucose export (33). In the livers of wild-type mice, Glut2 transcript and protein showed peak circadian expression during the subjective day (circadian time 0–12 h), corresponding in mice to the fasting phase of the circadian behavioral cycle, and trough expression during subjective night (circadian time 12–24 h), corresponding to the feeding phase of the cycle (Fig. 3 B and C). These observations suggest that the innate rhythm of feeding behavior entrains the liver circadian clock such that it drives maximal expression of GLUT2 during the fasting phase (i.e., at a time of little or no glucose ingestion), facilitating glucose export into the circulation, and minimal expression during the feeding phase (i.e., at a time of high glucose ingestion), inhibiting hepatic glucose export and thereby favoring storage and import. The observed phase of the GLUT2 protein rhythm in the liver thus suggested that the liver clock contributes to systemic glucose homeostasis by anticipating the innate fasting–feeding cycle and driving an autonomous rhythm of glucose export that matches the systemic requirements.
If the observed circadian rhythm of hepatic GLUT2 protein expression is physiologically meaningful, then the level of GLUT2 at its trough should be rate-limiting for hepatic glucose export. Even though L-Bmal1−/− mice differ from Fanconi–Bickel syndrome patients in that they make a constant low level of hepatic GLUT2 protein (Fig. 3C) rather than lack functional GLUT2 and that the defect is restricted to the liver, this hypothesis predicts that L-Bmal1−/− mice should have a similar phenotype—they should be hypoglycemic during the normal fasting phase of their daily behavioral cycle (when the hepatic GLUT2 level is considerably below that of controls) but not during the feeding phase of their daily cycle (when hepatic GLUT2 is comparably low in both L-Bmal1−/− and control mice).
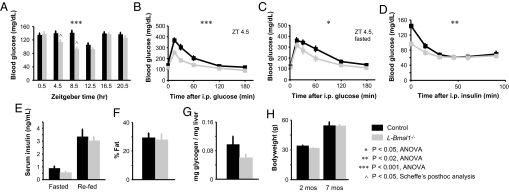
To test this prediction we compared glucose homeostasis in L-Bmal1−/− mice and littermate controls. Over a 24-h cycle, L-Bmal1−/− mice had a significantly lower overall profile of resting blood glucose than littermate controls (P < 0.001, genotype × time, ANOVA) (Fig. 4A). This difference was accounted for by hypoglycemia restricted to time points within the fasting phase of the daily cycle (P < 0.05 for each, Scheffé's post hoc analysis) (Fig. 4A). In sharp contrast to the glucose intolerance found in mice lacking circadian clocks in all tissues (Fig. 1 and Fig. S1), L-Bmal1−/− mice were more efficient than littermate controls at restoring basal blood glucose after a bolus of glucose (P < 0.001, genotype × time, ANOVA) (Fig. 4B). Enhanced glucose clearance was also observed after an overnight fast (P < 0.05, genotype × time, ANOVA) (Fig. 4C). L-Bmal1−/− mice also differed from mice lacking Bmal1 in all tissues (compare with Fig. 1) in that they had a normal or blunted sensitivity to insulin (Fig. 4D), normal insulin production (Fig. 4E), and normal total body fat content (Fig. 4F).
Fig. 4.
Hypoglycemia restricted to the fasting phase and exaggerated glucose clearance in mice with a liver-specific loss of circadian clock function. (A) Resting blood glucose of mice with the indicated genotypes measured across a 24-h cycle. The left half of the profile corresponds to the fasting phase of the daily behavioral cycle. The difference between genotypes over time is accounted for by the differences at fasting phase time-points ZT 4.5 and ZT 8.5. (B) Glucose tolerance; ZT 4.5. (C) Glucose tolerance after overnight fasting; ZT 4.5. (D) Insulin tolerance. The curves converge, with the significant difference between genotypes arising from the lower initial blood glucose in L-Bmal1−/− mice; ZT 8.5. (E) Serum insulin concentrations after mice were fasted overnight or fasted overnight and refed, as labeled. (F) Total body fat content. (G) Liver glycogen content. (H) Bodyweight at the indicated ages. For E–H, there were no significant differences between genotypes. Shown are the mean and SEM for 7–9 mice of each genotype. *, P < 0.05, ANOVA; **, P < 0.02, ANOVA; ***, P < 0.001, ANOVA; ∧, P < 0.05, Scheffé's post hoc analysis.
Thus L-Bmal1−/− mice have a distinct metabolic abnormality, characterized by hypoglycemia restricted to the fasting phase of the daily cycle and exaggerated glucose clearance in the presence of normal insulin production. This combination strongly points to a defect in hepatic glucose export into the circulation, consistent with the prediction from the circadian profile of hepatic GLUT2 expression and the phenotype of patients with Fanconi–Bickel syndrome. The livers of L-Bmal1−/− mice showed normal or possibly reduced glycogen content (Fig. 4G), revealing that the loss of hepatic Bmal1 function did not simply favor glycogen storage over glucose production. Taken together, the results are consistent with defective liver gluconeogenesis accompanied by a compensatory glycogenolysis or with an impairment of both liver glucose storage and production. The phenotype presumably results from the combined effects of a loss of daily BMAL1-driven increase in the expression of Glut2 and at least several other genes involved in hepatic glucose storage, transport, and export (Fig. 3A). Mice lacking Bmal1 function in all tissues likely have a similar defect in hepatic glucose metabolism (14), but expression of the resulting hypoglycemia is presumably masked by the hyperglycemia caused by an additional impairment of insulin production (Fig. 1E), likely reflecting a loss of Bmal1 function in the pancreas, brain, or both.
Discussion
Our results indicate that Bmal1 function in the liver is required for circadian regulation of hepatic glucose regulatory genes and for regulation of systemic glucose homeostasis in a time-of-day-dependent manner. Unlike metabolic phenotypes reported to date for germ-line circadian clock mutations, the defect in glucose homeostasis in L-Bmal1−/− mice occurred in a context of normal feeding behavior and locomotor activity, indicating a primary defect in metabolic responses. We cannot exclude a hypothetical noncircadian function of Bmal1 in the liver as the cause of the phenotype, but several considerations argue strongly for a defective liver circadian clock as the primary factor. The sheer number and variety of genes (18–20, 31) and proteins (34) with circadian expression profiles in the liver strongly suggests a fundamental role of the clock in liver physiology. The circadian phases of expression of hepatic glucose regulatory genes are consistent with their known functions, with those involved in glucose export (Glut2) showing peak expression during the fasting phase of the cycle, whereas those involved in glucose import and storage (Gck, L-PK) show peak expression during the feeding phase of the cycle, ≈8 h later (Fig. 3). The hypoglycemic phenotype is expressed in a time-of-day-dependent manner, strongly implying an underlying circadian process (Fig. 4).
Thus our findings strongly suggest that, in addition to well described acute hepatic responses to circulating glucose levels (17), the liver circadian clock drives a daily rhythm of hepatic glucose export timed so as to counterbalance the brain-driven fasting–feeding cycle, thereby buffering blood glucose concentrations over the course of the daily behavioral cycle (Fig. S3). The liver clock is likely important for additional aspects of liver physiology, such as clearance of xenobiotics, impaired in mice lacking the clock-regulated transcription factors DBP, TEF, and HLF (35). The work presented here provides direct evidence that a circadian clock in a peripheral tissue has a significant physiological function in vivo.
The presence of circadian rest–activity cycles in animals from insects to mammals (36) suggests that daily rhythmic organization of behavior has provided a strong selective advantage, but the resultant daily rhythms of food ingestion and energy expenditure likely result in major challenges for systemic homeostasis. It is possible that peripheral tissue circadian clocks have been selected over evolutionary time at least in part to drive rhythms of physiological processes that counteract undesirable physiological consequences of daily behavioral rhythms.
Materials and Methods
Mice and Behavioral Assays.
Mice were weighed weekly and entrained to a 12-h light/12-h dark cycle for 2 weeks before experiments. Genotyping was as described: Bmal1−/− (22); conditional Bmal1 and L-Bmal1−/− (10); Per1−/−; Per2−/− (27). Primers for Cre were forward 5′-gcg gtc tgg cag taa aaa cta tc and reverse 5′-gtg aaa cag cat tgc tgt cac tt. Running-wheel activity and feeding activity (feeding monitor; Mini Mitter) were recorded (Clocklab; Actimetrics) for a period of 2–6 weeks under the indicated conditions.
Insulin Tolerance Tests.
Mice were placed in clean cages (without food) at Zeitgeber time (ZT) 6.5 and injected i.p. 2 h later with 0.5–1.0 unit/kg of body weight of Novolin-R in 0.9% NaCl. Blood glucose was measured by using a One Touch Basic glucometer (Lifescan) before injection of insulin and at 15, 30, 45, 60, and 90 min after insulin injection.
Glucose Tolerance Tests.
Mice were placed in clean cages (without food) 2 h before the experiment (unless otherwise noted) and were injected with glucose (2 mg/g of body weight; 20% glucose in 0.9% NaCl). Blood glucose levels were measured before the injection of glucose and at 15, 30, 60, 120, and 180 min after injection.
Glucose and Insulin Determination.
Mice were placed in clean cages (without food) at ZT 0. Sixteen hours later (ZT 2 of the next day), blood glucose was measured, and blood (∼50 μl) was collected on ice for serum insulin measurement. The mice were then provided with chow, and 2 h later blood was collected for glucose and insulin measurement. Blood collected for serum insulin measurement was allowed to clot for 20 min at room temperature and cleared by centrifugation for 5 min at 3,000 × g. Insulin was measured by using the UltraSensitive Rat Insulin ELISA kit (CrystalChem).
Body Composition Analysis.
Mice were killed by CO2 inhalation (ZT 7–9), and the stomach and intestines were removed. The carcasses were weighed and then dried at 60°C. Dried carcasses were saponified in a solution of two parts ethanol to one part 30% potassium hydroxide (KOH) at 60°C, which converts all triglycerides to glycerol by de-esterification, and the resulting samples were analyzed for glycerol content by using the Free Glycerol Reagent (Sigma) according to the manufacturer's protocol.
Liver Glycogen Determination.
Mice were killed (ZT 7–9), and liver pieces were collected and frozen in liquid nitrogen. Pieces weighing <30 mg were boiled in 700 μl of 2 M hydrochloric acid for 3 h with constant agitation, which converts glycogen to glucose or glucose 6-phosphate. The solution was neutralized by addition of 700 μl of 2 M NaOH and 15 ml of 1 M Tris (pH 7.4), and glucose was measured with the glucose–hexokinase reagent (Amresco).
Quantitative Reverse-Transcriptase PCR.
Tissues were collected and immediately frozen in liquid nitrogen. RNA was prepared by using TRIzol (Invitrogen), RNeasy Plus Mini Kit (Qiagen), or RNeasy Fibrous Tissue Mini Kit (for skeletal muscle samples; Qiagen). Samples were adjusted to RNA concentrations of 1 μg/7 μl, and cDNA was prepared from 1 μg of RNA by using SuperScriptIII reverse transcriptase (Invitrogen). cDNA samples were then diluted 4-fold in water, and 2 μl was used per reaction for real-time PCR using iQ SYBR mix (Bio-Rad) on a thermocycler with Opticon Monitor 2 software (MJ Research). All amplified products were normalized to Hprt. Primer sequences are available upon request.
Western Blots.
Analysis was performed with rabbit anti-BMAL1 antiserum (a gift from U. Schibler) (1:500) or with anti-GLUT2 antibody (a gift from B. Thorens) (1:2,000).
Supplementary Material
Acknowledgments.
We thank Christopher Bradfield (University of Wisconsin, Madison, WI) for Bmal1−/− (Mop3−/−) mice, David Weaver and Steven Reppert (University of Massachusetts Medical School, Worchester, MA) for Per1−/− and Per2−/− mice, Ueli Schibler (University of Geneva, Geneva, Switzerland) for anti-BMAL1 antiserum, Bernard Thorens (University of Lausanne, Lausanne, Switzerland) for anti-GLUT2 antiserum, Ming Liu for expert technical assistance, and Daniel Konrad and Andrew Greenberg for helpful discussions. This work was supported by National Institutes of Health grant NS43491 (to C.J.W.). K.A.L. is a Merck Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 14753.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806717105/DCSupplemental.
References
- 1.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 2.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 3.Abe M, et al. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 5.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in cultured Rat-1 fibroblasts. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 7.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durgan DJ, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–24269. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- 9.McDearmon EL, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storch KF, et al. Intrinsic circadian clock of the mammalian retina: Importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 13.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: Time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 14.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oishi K, et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 17.Klover PJ, Mooney RA. Hepatocytes: Critical for glucose homeostasis. Int J Biochem Cell Biol. 2004;36:753–758. doi: 10.1016/j.biocel.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 19.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 20.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 21.Cailotto C, et al. The suprachiasmatic nucleus controls the daily variation of plasma glucose via the autonomic output to the liver: Are the clock genes involved? Eur J Neurosci. 2005;22:2531–2540. doi: 10.1111/j.1460-9568.2005.04439.x. [DOI] [PubMed] [Google Scholar]
- 22.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunger MK, et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 24.Shimba S, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neubauer N, Kulkarni RN. Molecular approaches to study control of glucose homeostasis. ILAR J. 2006;47:199–211. doi: 10.1093/ilar.47.3.199. [DOI] [PubMed] [Google Scholar]
- 26.Cho H, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 27.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 28.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 29.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 30.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- 31.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santer R, et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat Genet. 1997;17:324–326. doi: 10.1038/ng1197-324. [DOI] [PubMed] [Google Scholar]
- 33.Santer R, Steinmann B, Schaub J. Fanconi-Bickel syndrome—a congenital defect of facilitative glucose transport. Curr Mol Med. 2002;2:213–227. doi: 10.2174/1566524024605743. [DOI] [PubMed] [Google Scholar]
- 34.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 36.Hendricks JC, Sehgal A. Why a fly? Using Drosophila to understand the genetics of circadian rhythms and sleep. Sleep. 2004;27:334–342. doi: 10.1093/sleep/27.2.334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.