Abstract
MicroRNAs (miRNAs) are short noncoding RNAs that down-regulate gene expression by silencing specific target mRNAs. While many miRNAs are transcribed from their own genes, nearly half map within introns of ‘host’ genes, the significance of which remains unclear. We report that transcriptional activation of apoptosis-associated tyrosine kinase (AATK), essential for neuronal differentiation, also generates miR-338 from an AATK gene intron that silences a family of mRNAs whose protein products are negative regulators of neuronal differentiation. We conclude that an intronic miRNA, transcribed together with the host gene mRNA, may serve the interest of its host gene by silencing a cohort of genes that are functionally antagonistic to the host gene itself.
INTRODUCTION
RNA interference (RNAi) is a physiological pathway in which short double-stranded RNA (dsRNA) silences specific target RNA. In the normal cell, RNAi is triggered by microRNAs (miRNAs) that are ∼22-nt long noncoding RNAs found in diverse organisms from plants to metazoan animals (1–7). In the predominant mechanism, the miRNAs bind to imperfectly matched miRNA response elements (MREs) within the 3′-UTR of the target mRNAs. This leads to the recruitment of key proteins at the site leading to the assembly of the RNAi-induced silencing complex (RISC) that blocks translation initiation or elongation, or cleaves the target RNA (7–11). There are a few hundred experimentally sequenced and predicted human miRNAs (12,13) that together hold the potential to regulate thousands of genes impacting a large variety of biological processes.
The majority of miRNAs are produced from their own independent transcription units in intergenic regions; the primary transcript of these genes, pri-miRNA, is first trimmed by the nuclear RNase III-like enzyme, Drosha (7,14,15). The product, pre-miRNA, is further processed in the cytoplasm by the RNase III-like enzyme, Dicer, to generate the final miRNA. In contrast, ∼40% of mammalian miRNA sequences map within the introns of protein-coding ‘host’ genes (16–22), many of which appear to be conserved across species. The regulation of such ‘intron-derived’ (or ‘intronic’) miRNAs and their relationship with their host genes remain a mystery. Recent evidence that many of them are generated through unique processing steps bypassing Drosha hints that potentially novel features of this class of miRNAs still await discovery (23–25).
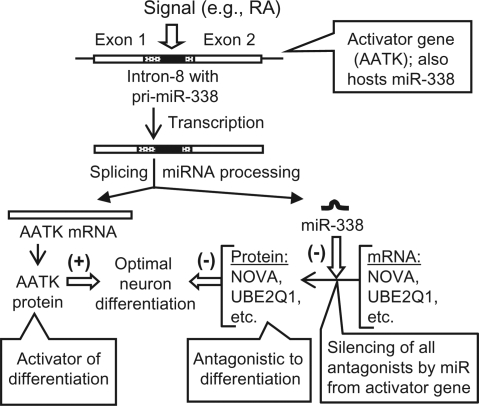
To investigate the significance of physiologically regulated intronic miRNAs, we took advantage of the fact that the precursor sequence for miR-338 is located within the eighth intron (intron-8) of the apoptosis-associated tyrosine kinase (AATK) gene (19) and that the AATK kinase activity plays an essential role in promoting neurite extension in developing neurons (26–28). This differentiation process can be induced in cultured neurons by all-trans retinoic acid (RA), 12-O-Tetradecanoyl phorbol 13-acetate (TPA) and insulin-like growth factor (IGF), all of which activated de novo AATK gene expression (26). Although continued overexpression of recombinant AATK from cloned cDNA also triggered this process, additional induction of the chromosomal AATK gene by RA, TPA or IGF promoted faster and more robust neurite growth (26,28). We hypothesized that optimal neurite growth requires not only the enzymatic activity of AATK but also the intronic miR-338 generated from the chromosomal AATK gene. We further hypothesized that miR-338 suppresses the translation of a select group of cellular mRNAs whose protein products are negative regulators of neurite growth. In the rest of this article, we provide proof of this novel positive feed-back miRNA circuit (See Model in Figure 6).
Figure 6.
Model for silencing of antagonistic genes by intronic miRNA. A pathway (e.g. differentiation) is activated by the product of the gene (e.g. AATK) that also hosts the miRNA (e.g. miR-338) that silences antagonistic genes, providing optimal stimulation of the pathway. Note that many variations of the theme are possible. For example, the mechanism would also apply to examples of repression if the first gene were a repressor of the pathway; the miRNA would then silence a family of antagonistic activators of the pathway to promote optimal repression.
MATERIALS AND METHODS
Neurite growth
Human neuroblastoma SH-SY5Y and M17 cells were grown on collagen-coated dishes, and induced and measured essentially as described (26). In brief, growth medium was RPMI1640 supplemented with 10% heat-inactivated FBS, penicillin (100 U/ml) and streptomycin (100 U/ml). Cells were photographed in a Nikon TE2000-E2 imaging station. Neurite differentiation was routinely induced by RA (10 μM), although IGF (10 nM) and TPA (16 nM) (all from Sigma-Aldrich, St. Louis, MO, USA) were also tested initially and showed similar effects. Differentiated cells were quantified by counting about 400 cells in random fields at 20× magnification. Cells were considered differentiated if they had at least one process (neurite) longer than the cell body. The lengths of the neurites were measured in at least three independent experiments and the results expressed as mean ± standard error.
Cloning and generation of stable cell lines
Stable Tet-responsive SH-SY5Y (or M17) cell lines were created using the double-stable Tet-ON system (Clontech, Mountain View, CA, USA). The cells were first stably transfected with the pTET-On-Advanced plasmid that produces the Tet regulator and clones resistant to G418 (150 μg/ml) were selected. Clones with low background but high induction of luciferase by doxycycline (Dox, a tetracycline derivative; 100 ng/ml) from the test plasmid pTRE-Tight-Luc were then transfected with AATK or ubiquitin-conjugating enzyme E2Q1 (UBE2Q1) expression plasmids as follows. To construct the AATK clones the 4206-nt long human AATK-coding sequence (NM_001080395.1) was amplified by RT–PCR using total RNA from 3-day RA-induced SH-SY5Y cells as template and was cloned into the EcoRI-XbaI sites of pTRE-Tight. To insert the intron-8 into the AATK cDNA, SphI and NdeI restriction sites were first created in the left- and right-flanking exons, respectively, by site-specific mutagenesis of appropriate sequences to synonymous codons (e.g. GCCTGT was changed to GCATGC, to become an SphI site but coding for the same amino acids, Ala and Cys) using the QuikChange™ kit (Stratagene, La Jolla, CA, USA). The 1412-nt long intron-8 of AATK, along with portions of the flanking exons, was then amplified with primers containing SphI and NdeI sequences using genomic DNA as template, restricted with these two enzymes and cloned into same two sites of the modified cDNA clone described above. The Tet-regulator-expressing SH-SY5Y cell was transfected with the AATK cDNA clones (with and without intron-8), and stable cell lines were obtained by hygromycin (150 μg/ml) selection. The AATK genes in these clones are tightly regulated and inducible by Dox (Figure 4A).
Figure 4.
Dox-inducible coexpression of recombinant AATK and its intronic miR-338 is sufficient to promote optimal neurite growth. Note that there is no RA treatment in this panel of experiments, and therefore, the endogenous AATK gene is silent. (A) Measurement of neurite growth. SH-SY5Y cells-containing stable AATK transgene with or without intron-8 were induced with Dox where indicated for 4 days (+) or 6 days (++), or uninduced (−). Synthetic miR-338 mimic was transfected where indicated (+50 nM; ++100 nM). Control miR-139 (100 nM) was transfected in lane c. Transfection of siRNA (50 nM) against MAP1A, NOVA1 or UBE2Q1 was done in lanes M, N and U, respectively. Neurite length was measured and standard error bars from three experiments are shown. AATK and control profilin protein levels in the cell extracts were determined by immunoblot. In the miR-338 panel, intron-8-generated miR-338 was detected as described in Materials and methods section. ND, not done. (B) Representative photomicrographs of cells corresponding to data 1, 2, 4 and 6 of panel A.
The 1269-nt long UBE2Q1-coding sequence plus 599 nt of 3′-UTR (total 1868 nt) containing all three predicted MREs was amplified by RT–PCR using total cellular RNA as template and cloned within EcoRI and XbaI sites of pTRE-Tight. The following primers were used with the restriction sites underlined and the extra sequences in lowercase added for efficient restriction: agatacGAATTCATGCAGCAGCCGCAGCCGCAG and agatacTCTAGAGTTTCTCTTGCTTTAGCAGCAAAATG. The reverse transcription was performed at 65°C for 3 h using Tth DNA polymerase (Roche), followed by PCR using a Taq:Pfu polymerase mix (20 : 1) and the following parameters: initial denaturation at 94°C, 4 min; 40 cycles (94°C, 40 s; 65°C, 30 s; 72°C, 4 min); final extension at 72°C, 8 min; hold at 4°C. The product was gel-purified, restricted and cloned. The three MRE sites (Figure 2A) were sequentially mutated (GCUG → CGAC) using the QuikChange™ kit. The C351A mutant (TGC → GCC) of this clone was then constructed using the same kit.
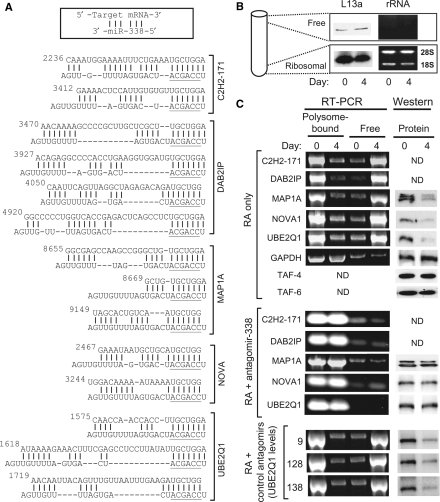
Figure 2.
Putative miR-338 response sites and translational status of target mRNAs. (A) Multiple predicted MRE-miR-338 hybrids for the indicated target genes are depicted in the pattern shown above, i.e. for each hybrid, the top sequence is MRE and bottom is miR-338. The seed sequence of miR-338 is underlined. For each target, the number on the left is the position of this nucleotide in the NCBI entry (whose accession numbers are provided in the Results section). (B) Validity of ribosomal fractionation. The ribosome-free fraction from the top and the ribosomal/polyribosomal fraction from the bottom of the gradient were analyzed for the ribosomal protein L13a in western blot (left) and for the presence of rRNA by agarose gel electrophoresis (right). Note that similar fractions were analyzed in (C) as well. Day 0 and 4 mean no RA treatment and 4 days of RA treatment, respectively. (C) Polysomal association of target mRNAs. SH-SY5Y cell were treated as shown on left (i.e. RA treatment with or without transfection with antagomirs against miR-9, -128, -138). Before (day 0) and after 4 days (day 4) of RA treatment, the polysomal and free RNAs were quantified by qRT–PCR and representative samples analyzed on 2% agarose–EtBr gels and photographed under UV light (left panels). Immunoblot (western) of total extract of the corresponding cells shows the different protein levels (right panels). ND, not done.
Transfection
Plasmids were prepared using the Midi kit (Qiagen, Valencia, CA, USA). Transfection of cultured cells with plasmids, synthetic siRNA (Ambion, Austin, TX, USA), miRIDIAN miRNA Mimics (Dharmacon, Lafayette, CO, USA) and 2′-O-methyl antagomirs (Ambion) was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as described (18). Three siRNAs each were designed against each of the targets [UBE2Q1, microtubule-associated proteins 1A (MAP1A), neuro-oncological ventral antigen 1 (NOVA1)] and were all effective within a range of 65–80% knockdown, showing specificity. However, we have provided results with the most effective siRNA for brevity (Table 1). Control luciferase siRNA has been described (29).
Table 1.
siRNA and primer sequences
| siRNA | Sequence |
| AATK | 5′ GGGCCCUGAAGCACAGCAAdTdT 3′ |
| 3′ dTdTCCCGGGACUUCGUGUCGUU 5′ | |
| MAP1A | 5′ GUGGGCAGGCAAUAGUAAAdTd 3′ |
| 3′ dTdTCACCCGUCCGUUAUCAUUU 5′ | |
| NOVA1 | 5′ GCACAGCAGGUCUGAUAAUdTdT 3′ |
| 3′ dTdTCGUGUCGUCCAGACUAUUA 5′ | |
| UBE2Q1 | 5′ GGGCAAGAAAUCUGAAGAUdTdT 3′ |
| 3′ dTdTCCCGUUCUUUAGACUUCUA 5′ | |
| qRT–PCR primers | Sequence (5′ → 3′) |
| AATK | AGCAGTGAGGATGAGGACAC (sense) |
| GCCTCCTTGAGCACAAACTC (antisense) | |
| C2H2-171 | GGACCAGCTGGACAAAAGAG (sense) |
| TGCTAGCACGTCTTCAATGG (antisense | |
| DAB2IP | GCAGCCTTTCCGAGAAGAG (sense) |
| GCTCTTGGTGCGCTTGATG (antisense) | |
| MAP1A | GTTTAGAGCCTGGGGGAGAC (sense) |
| TCCCAGGGTCTCTATTGCTG (antisense) | |
| NOVA1 | AAACTGGAGCCACCATCAAG (sense) |
| CACATTTTGGGGCATTTCTC (antisense) | |
| UBE2Q1 | CACTGCAACATCACGGAGTC (sense) |
| TTATGTCCACCAGCCTCTCC (antisense) | |
| GAPDH | CAATGACCCCTTCATTGACC (sense) |
| GACAAGCTTCCCGTTCTCAG (antisense) |
Assays
Quantitative real-time reverse transcription PCR (qRT–PCR) with total cellular RNA was performed as described (30) using mRNA-specific primers (Table 1). Genomic amplification was ruled out by three reasons: (i) the RNA isolation procedure involved RNase-free DNaseI treatment; (ii) no product was obtained in control PCR without reverse transcription; (iii) the two primers of a pair were designed against two exons spanning an intron, and a single product was generated in qRT–PCR, corresponding in size to the intronless processed mRNA. We have shown product bands from a representative cycle (usually between 25 and 30) when amplification was still exponential.
Immunoblots (western blot) were performed with 40 μg total cell lysate loaded per SDS–PAGE lane. The primary antibodies were: rabbit polyclonal antibody against human ribosomal L13a (31), rabbit anti-AATK (kindly provided by Dr Stacey Baker and Dr E. P. Reddy); rabbit anti-MAP1A (Santa Cruz Biotechnology, Santa Cruz, CA, USA); goat anti-NOVA1 (Santa Cruz Biotechnology); polyclonal mouse anti-UBE2Q2 (Novus Biologicals, Littleton, CO, USA), which was found to cross-react with UBE2Q1 due to substantial homology (not shown); TAF-4, TAF-6 (Sigma-Aldrich); and monoclonal mouse anti-GAPDH (Ambion). HRP-conjugated secondary antibodies (Sigma-Aldrich) were developed with the ECL method using the SuperSignal West Dura substrate (Pierce/Thermo Scientific, Rockford, IL, USA). The ECL luminescence and EtBr fluorescence of DNA in agarose gels were captured in Fujifilm LAS-3000.
The miRNA levels were measured using the mirVana kit (Ambion) and the manufacturer's procedure, based on solution hybridization with labeled antisense RNA, followed by RNase digestion, gel electrophoresis and autoradiography. For northern quantitation of AATK and β-actin mRNAs, about 300-bp long cDNA segments, amplified and 32P-labeled by PCR, was used as probe.
To analyze the ribosomal profile, SH-SY5Y cells were treated with RA alone or transfected with the indicated antagomirs as well as treated with RA. Cells were lyzed 4 days later and the polysomal mRNA was isolated exactly as described (31). Control RA-untreated cells were processed similarly. The preparation was analyzed by centrifugation through 20% sucrose. The ribosome-bound (polysomal) pellet from the bottom and the free RNA from the top of the tube were collected, deproteinized and subjected to qRT–PCR using mRNA-specific primers (Table 1). Portions of the isolated RNA were tested for the presence of 18S and 28S rRNA. Where mentioned, polysomal fractions were incubated with puromycin (90 µg/ml) for 1 h at 4°C, followed by centrifugation through sucrose as before, and the dissociation of GAPDH mRNA from the polysomal to the ‘free’ fraction (top of gradient) was tested by RT–PCR of the fractions.
Statistical analysis
Changes were analyzed by one-way ANOVA and by Student's t-test with Bonferroni correction. All numerical data were collected from at least 3 separate experiments. Results were expressed as mean ± SEM (error bars in graphs). Differences were considered to be significant at P < 0.05.
RESULTS
Neurite growth in culture parallels activation of the AATK gene and generation of miR-338
We first tested if induction of the chromosomal AATK gene indeed generated miR-338. We induced differentiation in two kinds of neuroblastoma cells, M17 and SH-SY5Y, by treating with RA, and confirmed the development of the neurites (Figure 1A), with robust growth observed by day 4. In both cells, AATK mRNA and protein levels rose to optimal levels by days 2 and 3, respectively (Figure 1B). When we measured miR-338 levels, we detected a strong and sustained increase, whose kinetics matched that of the AATK mRNA (Figure 1B), as one would expect if both are processed from the same primary transcript. We note that maximal neurite length was previously reported on day 7 and 8 after induction of the AATK gene (26); however, as our main focus is on the effect of miR-338, which is synthesized before AATK protein is translated, we chose the 4-day time point for routine assays to strike an optimal balance between the effects of AATK kinase activity and the effects of miR-338. Control β-actin mRNA and other miRNAs (miR-139, miR-143) were unaffected at all times.
Figure 1.
Neuronal differentiation parallels AATK mRNA and miR-338 levels. (A) Photomicrograph of M17 and SH-SY5Y cells after 4 days of RA treatment or no treatment. (B) Northern measurement of mRNAs and miRNAs and immunoblot of the proteins in two different cells after the indicated days of RA treatment. In the lowest panels, the M17 cells were transfected with AATK siRNA (Table 1) 18 h before the addition of RA.
To directly demonstrate an essential role of AATK in neurite growth, we performed a control experiment in which AATK expression was knocked down by pretransfection with specific siRNA. As shown (Figure 1B, lowest panel), this resulted in degradation of AATK mRNA and loss of AATK protein, causing little or no neurite growth (Figure 1A, lowest panel). In contrast, the miR-338 levels remained largely unaffected, which supports the view that intronic miRNA is produced in the nucleus before the processed mRNA is exported to the cytoplasm, where RISC-mediated degradation of mRNA occurs. Together, these results show a strong correlation between miR-338 biogenesis and AATK gene induction and also demonstrate an essential role of the native AATK enzyme in RA-induced differentiation.
Each pre-miRNA precursor can potentially generate two processed species, viz., the miRNA from one strand that enters the RISC and the miRNA* that generally acts as the carrier strand but nonetheless may potentially function as miRNA as well (32). However, we could not detect any miR-338* by northern under the same conditions that revealed the induction of miR-338 (data not shown). We presume that miR-338* is rapidly degraded upon biogenesis and thus not relevant for our studies.
Translational silencing of miR-338 targets occurs during neurite growth
First, in order to identify target mRNAs regulated by miR-338, we used multiple search programs, namely, PicTar (33), miRANDA (12), TargetScan (34,35) and DIANA (36), and then visually verified the top scorers from each program by the comprehensive set of miR-target base-pairing rules such as multiple MRE and by conservation across species (8,34–36). Based on these, we selected five candidates for further study (Figure 2A): zinc-finger protein 238 (ZNF238, also called C2H2-171; NM_205768), DAB2-interacting protein (DAB2IP; NM_032552), MAP1A (NM_002373), NOVA1 (NM_002515) and UBE2Q1 (NM_017582). All the predicted MREs in these targets perfectly paired with the seed region nucleotides 2–7 at the 5′-end of miR-338, considered essential for miRNA function (Figure 2A). Translational arrest by miRNA involves a variety of mechanism that may operate at either initiation or elongation stage of translation and may lead to dissociation of the target mRNA from the polysomal or a polysome-like fraction with or without destabilization of the mRNA (9,10,11,37). To investigate the status of our candidate miR-338 target mRNA, we isolated the polysomal and free (nonpolysomal) mRNA fractions from RA-treated and untreated cells. We first confirmed the identity of the fractions by the presence of ribosomal proteins and rRNAs specifically in the ribosomal/polysomal fraction at the bottom and their absence in the ‘free’ fraction (Figure 2B). We then determined the specific mRNA levels in these fractions by qRT–PCR. Note that we have presented results of SH-SY5Y cells only to conserve space, but the M17 cells produced essentially similar results that are not shown. Our results (Figure 2C) show that upon RA treatment, large portions of all five mRNAs were released free from the polysomal fraction. A concomitant reduction in the protein levels was evidenced by immunoblot using antibodies available against three target proteins. These changes did not occur in the RA-untreated cells or to the control GAPDH mRNA. Incubation of the polysomal fraction with puromycin (see Materials and methods section) led to release of the GAPDH mRNA to the top of the gradient, confirming the validity of the fractionation procedure (data not shown). TAF-4 and TAF-6, two subunits of the general transcription factor TFIID, also acted as negative controls since their protein levels remained unaffected by RA; their polysomal association was, therefore, not studied. Overall, we conclude that translational silencing of the miR-338 target genes correlated with neurite growth.
Antagomirs are short synthetic RNAs that are antisense to specific miRNAs and inhibit the function of the miRNAs against which they are designed (38). To obtain direct evidence that the observed silencing was indeed due to miR-338, we transfected the cells with antagomir-338, which restored polysomal association of the target mRNAs as well as protein levels (Figure 2C). Control antagomirs against three irrelevant miRs (miR-9, -128, -138) had no effect on the polysomal association or protein levels of the targets, as shown for UBE2Q1 as representative (Figure 2C), even though these three miRs are naturally abundant in neuronal tissues including the CNS (16,39). Together, these results validate selected miR-338 targets predicted by bioinformatics and demonstrate that they are indeed silenced during neuronal differentiation.
Optimal neurite growth requires AATK activity as well as miR-338
Having shown that miR-338, originating from an AATK intron, indeed translationally silences its predicted targets, we sought to determine the relative effects of miR-338 and AATK on neuron differentiation. We first knocked down AATK protein expression by specific synthetic siRNA, which almost completely abrogated RA-mediated differentiation (Figure 3, lane 3), confirming the indispensable role of AATK (26–28). Neutralization of miR-338 by antagomir-338 also strongly reduced neurite length by about 50% without affecting AATK protein levels (Figure 3, lanes 5 and 6), suggesting a stimulatory role of miR-338 in differentiation. Joint transfection with anti-AATK siRNA and antagomir-338 was as inhibitory as anti-AATK siRNA alone (Figure 3, lane 7). Control siRNA (against luciferase) and control antagomirs affected neither AATK levels nor differentiation (Figure 3, lanes 8–10). Clearly, AATK kinase is absolutely essential for differentiation whereas miR-338 serves as an accessory to promote optimal neurite growth. This is consistent with the generally accepted notion that miRNAs are not major developmental regulators but rather support developmental transcriptional programs (40).
Figure 3.
Induced endogenous intronic miR-338 promotes neurite growth. Standard SH-SY5Y monolayer was treated with RA and/or transfected with siRNA or antagomirs in the following combination, and at 4 days thereafter, neurite lengths were measured and then cell extracts were made to determine AATK and control profilin protein levels by immunoblot. 1, no treatment; 2, RA alone; 3, RA + anti-AATK siRNA (50 nM); 4, RA + anti-Luc siRNA (100 nM); 5, RA + antagomir-338 (50 nM); 6, RA + antagomir-338 (100 nM); 7, RA + anti-AATK siRNA (50 nM) + antagomir-338 (50 nM); 8, RA + control antagomir-9 (100 nM); 9, RA + control antagomir-128 (100 nM); and 10, RA + control antagomir-138 (100 nM).
Recombinant AATK and miR-338 can promote optimal neuronal differentiation without RA
To rule out any indirect effect of RA on the neurons and clearly dissociate the role of miR-338 from AATK activity, we created neural cell lines containing Tet-inducible recombinant AATK gene with or without intron-8, to permit the study of neuronal differentiation without activating the endogenous AATK gene by RA. Basically, two types of AATK plasmids were constructed using the pTRE-Tight Tet-ON vector, one containing the AATK protein-coding sequence only (without introns) (26) and the second, containing coding sequence plus intron-8 in its natural position. The plasmids were transfected into SH-SY5Y cells constitutively expressing the Tet trans-activator, and stable cell lines were obtained by hygromycin selection. These cells were first confirmed to express AATK (Figure 4A); both intronless and intron-containing AATK transgenes produced AATK protein following addition of Dox without RA (Figure 4A), as expected. Biogenesis of miR-338 from the Dox-induced intron-containing AATK (lane 2) but not from the intronless gene (lane 4) was confirmed. Both cell types also underwent differentiation (Figure 4B, panels 2 and 4). Most significantly, however, the average length of neurites expressing intronless AATK (Figure 4A, lanes 4 and 5) was considerably shorter than those expressing intron-8-containing AATK (Figure 4A, lane 2). The length of the former could be restored by transfection of synthetic miR-338 (Figure 4A, lanes 6 and 7), proving that the role of intron-8 was to provide miR-338. Control miR-139 had no effect (Figure 4A, lane 8). These results lend additional support to our hypothesis that miR-338 from intron-8 of AATK is essential for optimal neurite length.
If the role of miR-338 is indeed to silence key mRNA targets, we reasoned that silencing the same targets with an alternative approach would also have a similar effect, i.e. help neurite growth. To test this, we knocked down MAP1A, NOVA1 and UBE2Q1 with synthetic siRNA. These three were chosen because, as shown in Figure 1, their antibodies were available to monitor silencing at the protein level. The siRNAs were transfected into SH-SY5Y cells expressing intronless AATK mRNA induced by Dox. Loss of the specific target protein was confirmed by immunoblot that revealed 70–80% reduction whereas control luciferase siRNA had no effect (data not shown). All three siRNAs increased neurite lengths to various extents in the presence of AATK activity but without miR-338 (Figure 4A, lanes 9–11). We conclude that all three miR-338 targets have various degrees of neurite growth suppressive functions. Note that miR-338, when expressed, promoted even longer neurites (lanes 6, 7), which is expected if miR-338 should silence other antagonistic genes as well.
Overexpression of an unsilenced miR-338 target inhibits neuronal differentiation
In a reciprocal approach, we tested if overexpression of a miR-338 target is in fact detrimental to neurite growth. To accomplish this, we chose UBE2Q1, which contained three prospective MREs (Figure 2), and constructed Tet-inducible clones of the full-length UBE2Q1-coding sequence engineered to contain an N-terminal 6xHis-tag, followed by either a wild type 3′-UTR or a mutated 3′-UTR in which the GCUG sequence of all three MREs were mutated to CGAC such that they should no longer bind miR-338. We selected stable transfectants of SH-SY5Y cells containing these expression cassettes, induced the UBE2Q1 transgene with Dox and also activated the differentiation of the cells with RA. We monitored the expression of the recombinant His-tagged and endogenous UBE2Q1 proteins and measured the average neurite length. The results (Figure 5) showed that Dox indeed induced His-tagged UBE2Q1 that reacted with both anti-UBQ and anti-His antibodies (lanes 1 and 4). Control uninduced cells (lane C) showed only the endogenous UBQ band and no His-UBQ, also demonstrating tight repression by the Tet repressor. RA silenced the His-UBE2Q1 protein levels only when the transgene had functional MRE sites (lane 2) and not when the MREs were mutated (lane 5). The endogenous UBE2Q1 was always silenced by RA, as it contained the natural MRE sites. Direct evidence that the mutant MREs in the transgene were indeed mir-338 resistant was obtained by transfection of synthetic miR-338 that silenced the MRE-wild-type transgene (lane 3) but not the MRE-mutant (lane 6). When we measured the differentiation phenotype of these cells, RA-induced neurites of cells with low UBE2Q1 levels (lane 2) were about 60% longer than those with high UBE2Q1 levels (lane 5). Knockdown of the mir-338-resistant UBE2Q1 transgene with transfected siRNA also restored optimal neurite length (lane 7). Finally, members of the ubiquitin-conjugating enzyme E2 family contain a C-terminal catalytic domain, a key residue of which is a conserved Cys that forms thiol–ester linkage with the C-terminus of ubiquitin protein ligase enzymes, E3, an essential intermediate in ubiquitin-mediated protein degradation pathway (41). To obtain evidence that the neurite suppressive role of UBE2Q1 may be due to its conjugating function, we mutated the corresponding Cys351 of UBE2Q1 to Ala in the miR-338-resistant clone and expressed the recombinant C351A mutant in the same Dox-inducible manner. This mutant protein failed to inhibit neurite growth (lane 8; compare with lane 5). Together, these results prove that high ubiquitin-conjugating activity of UBE2Q1 is indeed detrimental to neurite growth, offering a persuasive reason for the cell to silence it by miR-338, generated during normal neuronal differentiation.
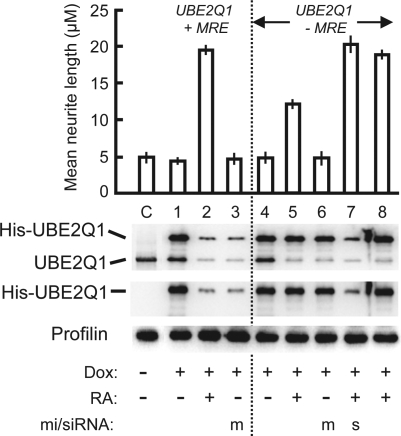
Figure 5.
Overexpression of recombinant UBE2Q1 protein impedes neurite growth in SH-SY5Y cells. Recombinant UBE2Q1 transgene with (lanes C and 1–3) or without (lanes 4–8) functional MREs was induced with Dox (+) or not induced (−). Cells were treated with RA (+) (to activate endogenous AATK and miR-338 synthesis) or left untreated (−). Lane 8 expressed the C351A mutant recombinant UBE2Q1 protein. Transfection with exogenous miR-338 mimic (m) and anti-UBE2Q1 siRNA (s) were performed where indicated. In the three immunoblot (western) panels, cell lysates were probed with (from top to bottom) anti-UBE2Q1, anti-His and antiprofilin antibodies. Note that the UBE2Q1 antibody reacts with both the His-tagged recombinant and the endogenous protein, whereas the His antibody reacts with the recombinant only, confirming the identity of the two bands. To achieve a substantial separation between the His-tagged and endogenous protein bands (that differ by ∼2000 Mr), the SDS–PAGE was performed in a 30-cm tall gel apparatus (normally used for DNA sequencing runs).
DISCUSSION
In this communication, we demonstrate that an intronic miRNA can silence genes that are functionally antagonistic to its host gene product, thus creating a positive feed-back loop that assists the physiological role of the host gene. Such an intronic miRNA can be viewed as a ‘slave’ serving the interests of its ‘master’ host gene on demand. The proposed mechanism is summarized in Figure 6.
The role of miR-338 in neuronal growth, as reported here, is consistent with previous studies in which a number of miRNAs, including miR-338, were detected in primary mammalian neurons (16,42). Although we do not know exactly how the various miR-338 targets may inhibit neurite growth, they clearly utilize diverse mechanisms working in parallel. Ubiquitin-mediated protein degradation, for example, regulates remodeling and degradation of neuronal processes, including dendritic pruning (43). Dysregulation of E2 activity is central to Wallerian axonal degeneration (43), characteristic of conditions such as multiple sclerosis. Conceivably, UBE2Q1 may be involved in the destabilization of key proteins essential in neurite growth. The high molecular mass MAP1A and MAP1B are expressed predominantly in cells of the nervous system (44). Expression of MAP1B is high during early stages of neuronal development and is down-regulated in the adult (45). Interestingly, MAP1A exhibits a reciprocal pattern of expression; it is all but absent in developing axonal fibers but reaches its peak in the adult brain, when neuronal differentiation is over (45). This is consistent with our finding that MAP1A levels may be low in developing neurons. Moreover, MAP1B mRNA does not contain a recognizable MRE for miR-338 (data not shown). Thus, although the exact function of the MAPs in neuronal differentiation is currently unclear, it is obvious that the use of miR-338 offers a way to preferentially silence MAP1A over MAP1B. NOVA1 is recognized as a regulator of brain-specific splicing (46) and we presume that it promotes the generation of splice variants of specific transcripts whose products suppress differentiation in the resting neuron. The DAB2IP gene is a tumor suppressor and its promoter is epigenetically silenced in various cancers, consistent with its general growth inhibitory role (47,48). It appears that the nerve cells have evolved the miRNA-based alternative and rapid mechanism to silence DAB2IP translation rather than transcription. C2H2-171, a novel POZ-domain zinc-finger protein (also known as ZNF238 or RP58, repressor protein 58 kDa) is a sequence-specific transcriptional repressor in brain, and other members of this family also function as repressors and/or developmental regulators (49). It is, therefore, reasonable to speculate that C2H2-171 represses specific developmental genes in the resting neurons and hence, must be silenced in order to activate such genes.
For the risks of toxicity and saturation of the RISC machinery, we did not attempt to silence all these miR-338 targets simultaneously by siRNA. It is also possible that there are other miR-338 targets in the differentiating neuron that we have not studied and that their combined silencing by miR-338 will boost neuron length most optimally. Nonetheless, in continuing our qRT–PCR-based screen of the polysomal fractions with and without antagomir-338 (similar to Figure 2C), we found that two other potential targets, highly scored by MIRANDA, are also regulated in the same manner (data not shown). These are: cerebellar degeneration-related protein 1 (CDR1) and 26S proteasome non-ATPase regulatory subunit 7 (PSMD7). There is in fact evidence that both are involved in neuronal degeneration and/or remodeling (50,51), and conceivably, need to be silenced for optimal neuron growth. Confidence in our study is further bolstered by the lack of this regulation in some other predicted targets, e.g. TAF-4 and TAF-6 (Figure 2C), which are TATA-binding protein-associated factors that function as subunits of the general transcription initiation factor TFIID and thus have no obvious biochemical basis to be detrimental to cell growth.
Although our model (Figure 6) is presented in terms of miR-338, the universality of this mechanism for other regulated intronic miRs is justifiable. In a recent study (52), miR-208, originating from an intron in the α-myosin heavy chain (αMHC) gene of the heart, was shown to silence the thyroid hormone receptor-associated protein 1 (THRAP1, also known as TRAP240), which in turn regulated the transcription of the βMHC gene. Thus, the αMHC gene, in addition to encoding a major cardiac contractile protein, also regulated βMHC in response to stress and hypothyroidism. Even though the full regulatory circuitry of miR-208 remains to be elucidated, we speculate that it regulates a family of targets in the heart to further αMHC function. In another potential example, miR-151 resides within intron-21 of the gene for focal adhesion kinase (FAK) (19), transcription of which is up-regulated in proliferative diseases such as cancer (53). It, therefore, appears logical that a number of bioinformatically predicted targets of miR-151 are regulators of cell cycle, whose expression is either expected or demonstrated to be reduced in various forms of cancer; such targets include SCC-112 (54), chromosome-associated kinesin KIF4A and growth arrest-specific protein GAS-2 (55).
Several unique features and advantages of our mechanism deserve mention: (i) It is an example of a regulatory noncoding RNA within a coding RNA. Coupled synthesis of the miRNA and its host transcript ensures that the miRNA will be available to assist the host gene whenever the latter is activated, but at no other time. This may also be a reason the miRNA evolved to be located within the host gene. (ii) As this regulation works in the form of RNA, it is independent of the biochemical nature of the protein product of either the host gene or the target genes. For example, it would have worked just as well if AATK were say, a neural transcription factor rather than a kinase. (iii) A given miRNA from one host gene can silence multiple antagonistic genes simultaneously. (iv) The concentration of the intronic miRNA may discriminate between high- and low-affinity MREs and thereby fine-tune their silencing. (v) This concentration will be regulated by the same signals and pathways that regulate the host gene promoter (e.g. RA, TPA, IGF for AATK). (vi) Though the mechanism of processing of the intronic miRNA is still not fully clear, it may be strategically regulated by the alternate splicing decisions of the intron. (vii) The inducible intronic miRNA offers a reversible silencing mechanism, i.e. when the host gene transcription will be shut off upon cessation of the activating signal, the target protein levels will also be restored. (viii) Although in our example, AATK is a ‘positive’ regulator of differentiation and the target genes are ‘negative’, the mechanism would work the same way if it were the reverse, i.e. if the host gene were a repressor of a pathway and the targets were activators of the same pathway. In fact, one can readily imagine more elaborate variations of this basic theme. For example, as mentioned before, miR-208 seems to help the function of its host gene (αMHC) by silencing the transcription regulator of the target βMHC gene without directly affecting βMHC mRNA (52). Indeed, the functional antagonism of the target genes toward the host gene is the fundamental aspect of the mechanism. Clearly, the coevolution of a miRNA-hosting intron and its multiple target gene MREs would be an interesting paradigm to study.
Funding
National Institutes of Health, USA (AI059267). Funding to pay the Open Access publication charges for this article was provided by the Department of Biochemistry and Molecular Biology, University of South Alabama.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
As a member of Lions-USA Eye Research group, the author is indebted to the Lions Club International Fund (LCIF) and the local Lions clubs for the gift of the Nikon TE2000E2 imaging station. Thanks are also due to Alla Musiyenko for expert technical assistance, Dr Stacey Baker and Dr E. P. Reddy (Fels Institute, Philadelphia, PA) for the AATK antibody, Dr Barsanjit Mazumder (Cleveland State University, OH) for the L13a antibody and constructive criticisms, Dr Chris Basler (Mount Sinai School of Medicine, NY) for various vectors and Dr Jonathan Scammell (Department of Comparative Medicine) for the SH-SY5Y and M17 cells.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates C. elegans developmental timing. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 12.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. Erratum in: PLoS Biol., (2005) 3 e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SL, Miller JD, Ying SY. Intronic microRNA (miRNA) J. Biomed. Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J. Mol. Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin SL, Chang D, Wu DY, Ying SY. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochem. Biophys. Res. Commun. 2003;310:754–760. doi: 10.1016/j.bbrc.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 22.Ying SY, Lin SL. Intron-derived microRNAs − fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 23.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 25.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raghunath M, Patti R, Bannerman P, Lee CM, Baker S, Sutton LN, Phillips PC, Damodar Reddy C. A novel kinase, AATYK induces and promotes neuronal differentiation in a human neuroblastoma (SH-SY5Y) cell line. Brain Res. Mol. Brain Res. 2000;77:151–162. doi: 10.1016/s0169-328x(00)00048-6. [DOI] [PubMed] [Google Scholar]
- 27.Tomomura M, Fernandez-Gonzales A, Yano R, Yuzaki M. Characterization of the apoptosis-associated tyrosine kinase (AATYK) expressed in the CNS. Oncogene. 2001;20:1022–1032. doi: 10.1038/sj.onc.1204210. [DOI] [PubMed] [Google Scholar]
- 28.Tomomura M, Hasegawa Y, Hashikawa T, Tomomura A, Yuzaki M, Furuichi T, Yano R. Differential expression and function of apoptosis-associated tyrosine kinase (AATYK) in the developing mouse brain. Brain Res. Mol. Brain Res. 2003;112:103–112. doi: 10.1016/s0169-328x(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 29.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2004;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 30.Bitko V, Garmon NE, Cao T, Estrada B, Oakes JE, Lausch RN, Barik S. Activation of cytokines and NF-kappa B in corneal epithelial cells infected by respiratory syncytial virus: potential relevance in ocular inflammation and respiratory infection. BMC Microbiol. 2004;4:28. doi: 10.1186/1471-2180-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri S, Vyas K, Kapasi P, Komar AA, Dinman JD, Barik S, Mazumder B. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA. 2007;13:2224–2237. doi: 10.1261/rna.694007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 36.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 38.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nature. 2005;438:685–689. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat. Genet. 2006;38(Suppl.):S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 41.Worthylake DK, Prakash S, Prakash L, Hill CP. Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 A resolution. J. Biol. Chem. 1998;273:6271–6276. doi: 10.1074/jbc.273.11.6271. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl Acad. Sci. USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 44.Noiges R, Eichinger R, Kutschera W, Fischer I, Nemeth Z, Wiche G, Propst F. Microtubule-associated protein 1A (MAP1A) and MAP1B: light chains determine distinct functional properties. J. Neurosci. 2002;22:2106–2114. doi: 10.1523/JNEUROSCI.22-06-02106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garner CC, Garner A, Huber G, Kozak C, Matus A. Molecular cloning of microtubule-associated protein 1 (MAP1A) and microtubule-associated protein 5 (MAP1B): identification of distinct genes and their differential expression in developing brain. J. Neurochem. 1990;55:146–154. doi: 10.1111/j.1471-4159.1990.tb08832.x. [DOI] [PubMed] [Google Scholar]
- 46.Jelen N, Ule J, Zivin M, Darnell RB. Evolution of Nova-dependent splicing regulation in the brain. PLoS Genet. 2007;3:1838–1847. doi: 10.1371/journal.pgen.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Toyooka S, Gazdar AF, Hsieh JT. Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J. Biol. Chem. 2003;278:3121–3130. doi: 10.1074/jbc.M208230200. [DOI] [PubMed] [Google Scholar]
- 48.Dote H, Toyooka S, Tsukuda K, Yano M, Ota T. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in gastrointestinal tumour. Br. J. Cancer. 2005;92:1117–1125. doi: 10.1038/sj.bjc.6602458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M. RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J. Biol. Chem. 1998;273:26698–26704. doi: 10.1074/jbc.273.41.26698. [DOI] [PubMed] [Google Scholar]
- 50.Fathallah-Shaykh H, Wolf S, Wong E, Posner JB, Furneaux HM. Cloning of a leucine-zipper protein recognized by the sera of patients with antibody-associated paraneoplastic cerebellar degeneration. Proc. Natl Acad. Sci. USA. 1991;88:3451–3454. doi: 10.1073/pnas.88.8.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen H, Korutla L, Champtiaux N, Toda S, LaLumiere R, Vallone J, Klugmann M, Blendy JA, Mackler SA, Kalivas PW. NAC1 regulates the recruitment of the proteasome complex into dendritic spines. J. Neurosci. 2007;27:8903–8913. doi: 10.1523/JNEUROSCI.1571-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 53.Han EK, McGonigal T. Role of focal adhesion kinase in human cancer: a potential target for drug discovery. Anticancer Agents Med. Chem. 2007;7:681–684. doi: 10.2174/187152007784111296. [DOI] [PubMed] [Google Scholar]
- 54.Kumar D, Sakabe I, Patel S, Zhang Y, Ahmad I, Gehan EA, Whiteside TL, Kasid U. SCC-112, a novel cell cycle-regulated molecule, exhibits reduced expression in human renal carcinomas. Gene. 2004;328:187–196. doi: 10.1016/j.gene.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 55.Brancolini C, Bottega S, Schneider C. Gas2, a growth arrest-specific protein, is a component of the microfilament network system. J. Cell Biol. 1992;117:1251–1261. doi: 10.1083/jcb.117.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]