Abstract
MicroRNAs (miRNAs) are a class of small regulatory RNAs that are thought to be involved in diverse biological processes by regulating gene expression. Numerous miRNAs have been identified in various species, and many more miRNAs remain to be detected. Generally, hundreds of mRNAs have been predicted to be potential targets of one miRNA, so it is a great challenge to identify the genuine miRNA targets. Here, we generated the cell lines depleted of Drosha protein and screened dozens of transcripts (including Cyclin D1) regulated potentially by miRNA-mediated RNA silencing pathway. On the basis of miRNA expressing library, we established a miRNA targets reverse screening method by using luciferase reporter assay. By this method, we found that the expression of Cyclin D1 (CCND1) was regulated by miR-16 family directly, and miR-16 induced G1 arrest in A549 cells partially by CCND1. Furthermore, several other cell cycle genes were revealed to be regulated by miR-16 family, including Cyclin D3 (CCND3), Cyclin E1 (CCNE1) and CDK6. Taken together, our data suggests that miR-16 family triggers an accumulation of cells in G0/G1 by silencing multiple cell cycle genes simultaneously, rather than the individual target.
INTRODUCTION
MicroRNAs (miRNAs) are an extensive class of small noncoding RNAs (18–25 nt), with profound impact on many biological processes in development, differentiation, growth and metabolism by regulating gene expression. miRNAs are initially transcribed as part of much longer primary transcripts (termed pri-miRNAs) (1). These primary transcripts are first trimmed into approximate 70-nt stem-loop forms (called pre-miRNAs) by the RNase III type endonucleases, Drosha, in the nucleus (2). Following this initial processing, pre-miRNAs are exported from the nucleus to the cytoplasm by Exportin5 (Exp5) and processed to generate approximate 22-nt mature miRNAs by Dicer, another RNase type III (3,4). Finally, the mature single-stranded miRNA binds to proteins of the Argonaute family to form the RNA–protein complex known as RNA-induced silencing complex (RISC) (5,6).
miRNAs can regulate gene expression essentially by two modes depending on the degree of complementarity with the mRNA targets. In plants, miRNAs are often fully complementary to their targets and elicit mRNA decay. In contrast, most animal miRNAs are only partially complementary to their targets and inhibit synthesis and function of proteins without affecting transcript levels (7–9). However, recent findings indicate that miRNAs can induce substantial mRNA degradation even in the absence of extensive base pairing to their targets (10,11). By regulating the expression of target genes, some miRNAs play a crucial role in the initiation and progression of human cancer. miR-15a and miR-16-1 are located at chromosome 13q14, a region deleted in more than half of B-cell chronic lymphocytic leukemia (CLL). Detailed deletion and expression analysis showed that miR-15a and miR-16-1 were located within a 30-kb region of loss in CLL, and that both genes were deleted or downregulated in ∼68% of CLL cases (12). Bottoni et al. (13) reported that miR-15a and miR-16-1 were expressed at lower levels in pituitary adenomas as compared to normal pituitary tissue. Their expression was inversely correlated with tumor diameter and with arginyl-tRNA synthetase expression, but was directly correlated with p43 secretion, suggesting that these miRNAs might influence tumor growth. Cimmino et al. (14) then demonstrated that miR-15a and miR-16-1 expressions were inversely correlated to Bcl2 expression in CLL and negatively regulated Bcl2 at a posttranscriptional level. Bcl2 repression by miR-15a and miR-16-1 induced apoptosis in a leukemic cell line model. Therefore, miR-15 and miR-16 were natural antisense Bcl2 interactors that could be used for therapy of Bcl2-overexpressing tumors. In addition, miR-15 and miR-16 can regulate Nodal/activin signaling by targeting the Nodal type II receptor Acvr2a, and restrict the size of Spemann's organizer in early Xenopus embryos (15).
Numerous miRNAs have been identified in diverse animals and plants. The miRNA database (16,17) currently contains 533 human miRNAs (release 10.0), and many more miRNAs may be still uncovered. In contrast to the big number of validated miRNA sequences, only a handful of comprehensive studies on miRNA function have been completed. One of the biggest obstacles to miRNAs function validation is the identification of their targets. Several computational algorithms have been established to predict target genes of miRNAs. According to these, a miRNA is generally thought to possibly have hundreds of or even to thousands of predicted target genes (18), therefore, it is difficult to determine which one or ones is/are true targets regulated by the miRNA to educe concrete biological functions. This leads to great trouble on miRNAs function analysis. Drosha is a key gene controlling miRNAs processing, and its downregulation can inhibit the expression levels of mature miRNAs and thus result in the upregulation of their target mRNAs (via miRNAs mediating RNAi mechanism). Therefore, the transcripts upregulated in Drosha-depleted cells are probably the targets of miRNAs. Likewise, Rehwinkel et al. (19) reported that depletion of Drosha triggered upregulation of numerous transcripts in Drosophila S2 cells, and many of these transcripts were genuine targets of the miRNA pathway. For those upregulated transcripts, computational algorithms can be used to predict their potential controlling miRNAs. Next, we can screen the miRNAs genuinely regulating these transcripts by using luciferase reporter assay.
Here, we established Drosha knockdown cell line and identified a series of transcripts regulated potentially by miRNA mediating RNAi mechanism. Among these candidate miRNA target transcripts, Cyclin D1 (CCND1) is a cell cycle-related gene and plays a key role in controlling G1/S transition. Moreover, it is a potential target for tumor gene therapies. On the basis of miRNAs expression library established (20), we developed a miRNA targets reverse screening method using luciferase reporter assay. By this method, CCND1 was found to be regulated by miR-16 family. Furthermore, our research demonstrated that miR-16 family regulated the expressions of several other cell cycle genes, including CCND3, CCNE1 and CDK6. All these data suggest that miR-16 family induces G1 arrest by regulating multiple downstream effectors simultaneously.
MATERIALS AND METHODS
Plasmid construction
Drosha shRNA expressing plasmid was generated by cloning annealed synthetic 63-mer oligonucleotides (See Supplementary Material) (21) into BamHI and HindIII sites within pSilencer hygro U6 vector (Ambion, Austin, TX, USA).
Eukaryotic expression plasmids
DNA fragments coding CCND1, CCND3, CCNE1 and CDK6 protein were amplified by PCR from A549 cell cDNA, and cloned into pIERS2-EGFP expression vector digested by XhoI and BamHI or EcoRI.
Firefly luciferase reporter constructs
Wild-type 3′ untranslated regions (UTRs) containing predicted miRNA target sites were amplified by PCR from HepG2 cell genomic DNA. Mutant 3′-UTRs were generated by overlap-extension PCR method. Both wild-type and mutant 3′-UTRs were cloned downstream of firefly luciferase coding region between the XbaI and NdeI sites of a modified pGL3-control plasmid (Promega, Madison, WI), as described before (20).
Cell culture, transfection and preparation of stable cell lines
HeLa, HepG2 and A549 cell lines were grown in DMEM containing 10% FBS and 100 μg/ml penicillin/streptomycin. DNA transfection was performed with FuGene® HD transfection reagent (Roche, Indianapolis, IN,USA) according to the manufacturer's protocol. To generate Drosha knockdown stable cell lines, HepG2 cells were transfected with pSilecer-U6-shDrosha and control plasmid. Cells were grown in the presence of 400 ng/ml hygromycin according to the manufacturer's protocol to select stably transfected clones. Single clone was selected to generate monoclonal cell lines, and total RNAs were extracted to test the knockdown effects of Drosha mRNA and miRNAs.
miRNAs, siRNAs and transfection
The miRNAs and siRNAs were designed and synthesized by GenePharma (Shanghai, China) (See Supplementary Material). siRNAs and miRNAs transfection were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA,USA). In brief, cells were plated in 6-well plate to 40% confluence. For each well, 5 μl siRNA or miRNA (20 μM) were added into 250 μl Opti-MEM medium, 5 μl of Lipofectamine 2000 into 250 μl Opti-MEM medium and then mixed siRNA or miRNA with Lipofectamine 2000. The mixture was added to cells and incubated for 6 h before replacing the medium. Total RNA and protein were prepared 48 h after transfection and were used for qRT–PCR or western blot analysis.
RNA extraction and qRT–PCR
Total RNA was extracted from the cultured cells using Trizol Reagent (Invitrogen) according to the manufacturer's protocol. qRT–PCR was used to confirm the expression levels of mRNAs and miRNAs. For mRNAs detection, reverse transcription was performed according to the protocol of Improm-II™ Reverse Transcriptase System (Promega); qPCR was performed as described in the method of SYBR premix Ex Taq (TaKaRa, Dalian, China) with Mx3000p (Stratagen, La Jolla, CA, USA) supplied with analytical software. GAPDH mRNA levels were used for normalization. For miRNAs detection (22), total RNA was polyadenylated by poly(A) polymerase (Ambion) first. The 50 µl polyadenylation reaction was set up with 10 µg total RNA and 1 µl (2 U) poly(A) polymerase according to the manufacturer's protocol. The reaction was incubated at 37°C for 60 min. After incubation, poly(A)-tailed total RNA was recovered by phenol/chloroform extraction and ethanol precipitation. Reverse transcription was performed using 1 µg poly(A)-tailed total RNA and 1 µg RT primer(GCG AGC ACA GAA TTA ATA CGA CTC ACT ATA GG(T)18VN) with 1 µl Improm-II™ Reverse Transcriptase according to the manufacturer's protocol; qPCR was performed as described in the method of Quantitect SYBR Green PCR Kit (Qiagen, Hilden, Germany) with Mx3000p (Stratagene) supplied with analytical software. One primer of miRNAs amplification is miRNA specific, and the other is a universal primer (GCG AGC ACA GAA TTA ATA CGA C). U6 snRNA levels were used for normalization.
mRNA microarray analysis
Stably transfected cells and normal HepG2 cells were harvested, and total RNA was extracted with Trizol reagent. Total RNA (5 μg) was reversely transcribed with the cDNA Synthesis Kit (TaKaRa, Dalian, China) and cRNA was produced by in vitro transcription (IVT) by T7 RNA polymerase using T7 RiboMAX Express Large Scale RNA Production System (Promega). cRNA (2 μg) was reversely transcribed by SuperscriptII reverse transcriptase and 9-nt random primer. The products of reverse transcription were labeled with klenow polymerase and 9-nt random primer. Labeled cDNA was hybridized to 35k human Genome Array Genechips (CapitalBio, Beijing, China). GenePix Pro 4.0 was used to analyze the image of microarray, and the data were normalized by the method of Lowess.
Western blot analysis
Protein extracts were prepared by a modified RIPA buffer with 0.5% sodium dodecyl sulfate (SDS) in the presence of proteinase inhibitor cocktail (Complete mini, Roche, Indianapolis, IN, USA). Polyacrylamide gel electrophoresis, tank-based transfer to Immobilon Hybond-C membranes (Amersham Biosciences) and immunodetection were performed with standard techniques. Antibodies against Drosha (sc-31159, Santa Cruz Biotechnology, Santa Cruz, CA, USA), CCND1 (K0062-3, MBL), CCND3 (K0013-3, MBL, Nagoya, Japan), CCNE1 (K0172-3, MBL), CDK6 (K0006-3, MBL), pRb (11131, Signalway antibody, Pearland, TX, USA) and β-actin (sc-1616-R, Santa Cruz) were used in western analysis in accordance with the manufacturer's instruction. Signals were visualized with SuperSignal® West Pico chemoluminescent substrate (Pierce, Rockford, Ill, USA) by exposure to films.
Luciferase assays
HepG2 cells were transfected in 24-well plates using FuGene® HD transfection reagent. The transfection mixtures contained 100 ng of firefly luciferase reporter plasmid and 400 ng of plasmids expressing miRNA primary transcripts. These plasmids expressing miRNAs come from a miRNA expression library (20). pRL-TK (Promega) was also transfected as a normalization control. Cells were collected 48 h after transfection, and luciferase activity was measured using a dual-luciferase reporter assay system (Promega).
Flow cytometry
Cells were transfected with miRNAs or siRNAs. Nocodazole (100 ng/ml; Sigma-Aldrich, St. Louis, MO, USA) was added 24 h after transfection, and cells were further incubated for 20 h. Floating and adherent cells were harvested, combined, washed once in phosphate-buffer saline (PBS), and fixed in 70% ethanol overnight. Staining for DNA content was performed with 50 μg/ml propidium iodide and 1 mg/ml RNase A for 30 min. Analysis was performed on a FACScalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) with Cell Quest Pro software. Cell-cycle modeling was performed with Modfit 3.0 software (Verity Software House, Topsham, ME, USA).
Production of recombinant adenovirus and cell infection
DNA fragment containing miR-195 was amplified from pcDNA-miR-195 (20), and cloned into pAdTrack-CMV (23) digested with SalI and XbaI, generating pAdTrack-CMV-miR-195. pAdTrack-CMV-miR-195 and pAdEasy-1 were homologously recombinated in bacteria BJ5183. The newly recombinated plasmid, pAd-miR-195, was tested by restriction endonuclease digestions. Ad-miR-195 was propagated in HEK293 cells; viruses were collected from these cells, and stored at −70°C.
A549 cells were grown in DMEM containing 10% FBS and 100 μg/ml penicillin/streptomycin. For 24 h prior to infection, 3 × 105 A549 cells were plated in each well of 6-well plates, then infected with recombinant viruses (Ad-miR-195 or Ad-empty) at a multiplicity of infection (MOI) of 100 for 1 h at 37°C, followed by the addition of fresh growth medium. Three days later, cells were harvested and cell cycles were analyzed by flow cytometry.
RESULTS
Microarray analysis of transcripts regulated by miRNA pathways
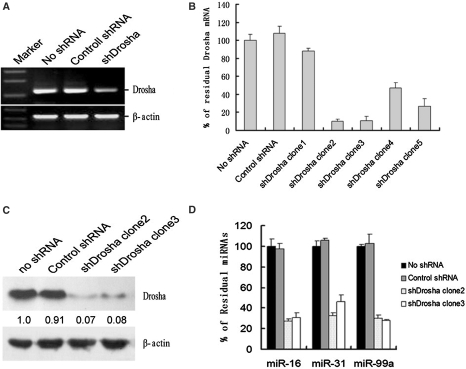
To screen the transcripts regulated by miRNA-mediated RNA silencing pathway in genome wide, we constructed the plasmid expressing shRNA targeting to Drosha first. To verify the knockdown efficiency, the plasmids were transfected into HeLa cells. Results of semiquantitative RT–PCR revealed that the level of Drosha mRNAs was strongly reduced (Figure 1A). Then, plasmids expressing the shRNA were transfected into HepG2 cells, and the stably transfected cell clones were selected using hygromycin. Quantitative RT–PCR was used to screen the stable cell lines in which Drosha was strongly knocked down (Figure 1B). qRT–PCR results indicated that Drosha mRNA was reduced in all selected stable cell clones, and the knockdown effect was also confirmed by western blot (Figure 1C).
Figure 1.
Generation and characterization of Drosha knockdown stable cell lines. (A) The effectiveness of Drosha depletion was analyzed by semi-quantitative RT–PCR. Normal HeLa cells and control shRNA transfected HeLa cells are in lanes 1 and 2. The β-actin mRNA served as an internal control. (B) The depletion effectiveness in Drosha knockdown stable HepG2 cell lines was analyzed by qRT–PCR. Drosha mRNA was reduced in all selected stable cell clones, five of which were shown respectively in bars 3–7 (bar 1: normal HepG2 cells; bar 2: control stably transfected HepG2 cells). shDrosha clone 2 in bar 4 demonstrated a strong downregulation to 95% of Drosha mRNA. (C) Drosha protein was detected by western blot. (D) miRNA expressions were detected by qRT–PCR. The expression levels of three miRNAs selected randomly, including miR-16, miR-31 and miR-99a decreased moderately in shDrosha clone 2 and clone 3 compared with normal HepG2 cells and control stably transfected HepG2 cells.
Drosha plays an important role in the processing of miRNAs, so the maturation of miRNAs should be repressed when Drosha was depleted (2,24). To analyze the effect of Drosha knockdown on miRNA expressions, we detected the expression levels of three randomly selected miRNAs by quantitative RT–PCR. As shown in Figure 1D, the expression levels of miR-16, miR-31 and miR-99a in clone 2 and clone 3 were reduced moderately compared with normal HepG2 cells and control plasmid transfected cells. The results above indicated that these cell lines could be used to screen the transcripts regulated by miRNA-mediated RNA silencing pathway globally. To identify transcripts whose levels are regulated by the miRNA pathway in HepG2 cells, we analyzed Drosha knockdown stable cell line, control plasmid stably transfected cell line and normal HepG2 cells using 35k human Genome Array (CapitalBio Corporation, Beijing, China). Table 1 summarizes analysis of transcripts changing expression levels upon knockdown of Drosha. We observed that there were 781 transcripts at least 1.5-fold upregulated in Drosha-depleted cells compared with normal HepG2 cells, and 558 transcripts compared with control plasmid stably transfected HepG2 cells. However, there were only 281 or 153 transcripts 2-fold upregulated compared with normal HepG2 cells or control stably transfected cells. The bulk of upregulated transcripts changed slightly (<2-fold) at the mRNA level, suggesting that miRNAs generally have a tuning role in regulating gene expression.
Table 1.
Distinct filters identify various numbers of differentially expressed (upregulated and downregulated) probe sets in individual samples
| shDrosha Clone2/no shRNA | shDrosha Clone2/control shRNA | Overlap | |
|---|---|---|---|
| All probes changed (upregulated and downregulated probes) | |||
| Changed 1.5-fold | 1497 | 1054 | 394 |
| Changed 2.0-fold | 491 | 282 | 90 |
| Upregulated probes | |||
| Upregulated 1.5-fold | 781 | 558 | 221 |
| Upregulated 2.0-fold | 281 | 153 | 53 |
Since inhibition of the miRNA pathway correlates with increased abundance of mRNAs targeted by miRNAs, the transcripts upregulated in Drosha-depleted cells may represent putative miRNA targets. Compared with normal HepG2 cells or control stably transfected cells, there were respectively 281 and 153 transcripts upregulated significantly in Drosha-depleted cells, in which 53 transcripts overlapped. We defined these 53 mRNAs as core transcripts, whose levels were potentially regulated by the miRNA pathway, that is, these mRNAs were potential miRNA targets. Besides, these transcripts represented 45 genes (Table 2), and they did not exhibit any apparent functional relationship.
Table 2.
mRNAs upregulated in Drosha depleted HepG2 cells compared with control transfected cells and normal HepG2 cells
| Gene name | Ratio of shDrosha /no shRNA | Ratio of shDrosha /control shRNA | Function |
|---|---|---|---|
| SPINK5L3 | 36.15 | 41.79 | |
| SERPINE1 | 11.18 | 13.26 | Blood coagulation, fibrinolysis, regulation of angiogenesis |
| CCND1 | 8.6 | 2.78 | Cell cycle, cell division, fat cell differentiation |
| F3 | 8.04 | 5.53 | Blood coagulation, immune response |
| GADD45A | 7.56 | 3.4 | DNA repair, apoptosis, cell cycle |
| DUSP5 | 6.09 | 3.58 | Protein amino acid dephosphorylation |
| FN1 | 5.74 | 2.43 | Acute-phase response, cell adhesion, cell migration, metabolic process, response to wounding |
| STS-1 | 5.53 | 3.63 | |
| MFAP5 | 5.27 | 12.71 | |
| SAT | 4.91 | 3.92 | Metabolic process |
| SLC19A2 | 4.69 | 3.02 | Sensory perception of sound, thiamin transport |
| ODC1 | 4.69 | 3.12 | Kidney development, polyamine biosynthetic process, cell proliferation |
| ANGPTL4 | 4.43 | 3.64 | Angiogenesis, cell differentiation, development, apoptosis, lipoprotein lipase activity, response to hypoxia |
| MATN2 | 4.35 | 8.4 | Biological process |
| ADM | 4.1 | 4.32 | Cell–cell signaling, circulation, female pregnancy, heart development, cell proliferation |
| NRG1 | 4.09 | 7.04 | Blood pressure regulation, cell differentiation, development, glucose transport, myelination, of transcription |
| CYR61 | 4.03 | 5.53 | Anatomical structure morphogenesis, cell adhesion, cell proliferation, chemotaxis, |
| DKK1 | 3.81 | 2.93 | Wnt receptor signaling pathway, embryonic limb morphogenesis, multicellular organismal development |
| ETS1 | 3.71 | 2.12 | Immune response, cell proliferation, erythrocyte differentiation, regulation of transcription |
| CTPS | 3.69 | 2.93 | Biosynthetic process, metabolic process, response to drug |
| RAPGEF2 | 3.69 | 5.12 | MAPKKK cascade, cAMP-mediated signaling, regulation of small GTPase-mediated signal transduction |
| SLC7A11 | 3.65 | 4.04 | Transport, protein complex assembly |
| CPA4 | 3.47 | 3.71 | Histone acetylation, proteolysis |
| ABHD5 | 3.28 | 3.44 | Lipid metabolic process, proteolysis |
| ENO2 | 3.14 | 6.48 | Glycolysis |
| RND3 | 3.09 | 3.32 | Actin cytoskeleton organization and biogenesis, cell adhesion, small GTPase-mediated signal transduction |
| BMP6 | 3.02 | 4.31 | BMP signaling pathway, development, inflammatory response, osteoblast differentiation, aldosterone biosynthetic process |
| CD44 | 3 | 2.1 | Cell adhesion |
| PPP1R3F | 2.99 | 4.41 | |
| Q9P2H4 | 2.81 | 2.63 | |
| SLPI | 2.75 | 3.45 | Serine-type endopeptidase inhibitor activity |
| GAD1 | 2.69 | 3.26 | Carboxylic acid metabolic process, neurotransmitter biosynthetic process, protein-pyridoxal-5-phosphate linkage, synaptic transmission |
| IGFBP3 | 2.61 | 18.36 | Signal transduction, apoptosis, myoblast differentiation, protein amino acid phosphorylation, cell growth |
| Q7Z2U1 | 2.61 | 2.39 | |
| HDHD1A | 2.51 | 2.13 | Metabolic process |
| DNAJC1 | 2.44 | 2.24 | Proteolysis, protein folding, protein secretion |
| OSMR | 2.36 | 2.4 | Cell proliferation, cell surface receptor linked signal transduction |
| TMEM50A | 2.3 | 3.07 | |
| C21orf34 | 2.26 | 3.29 | |
| ATF3 | 2.21 | 2.08 | Regulation of transcription, DNA-dependent |
| KRT17 | 2.13 | 3.37 | Biological process, epidermis development |
| ABCD3 | 2.12 | 2.78 | Peroxisome organization and biogenesis, transport |
| LIMA1 | 2.11 | 3.33 | Actin filament bundle formation, actin filament depolymerization, ruffle organization and biogenesis |
| C1orf181 | 2.09 | 2.29 | |
| CDKN1A | 2.09 | 5.87 | Cell cycle arrest, apoptosis, cell proliferation, response to DNA damage stimulus, response to UV |
As shown in Table 2, CCND1 was upregulated significantly in Drosha-depleted cells, which suggested CCND1 was a potential miRNA target. Cyclin D1, the product of CCND1 gene, is an important regulator of G1 to S phase progression in many different cell types. Cyclin D1, together with its binding partners cyclin-dependent kinase 4 and 6 (CDK4 and CDK6), forms active complexes that promote cell cycle progression by phosphorylating and inactivating the retinoblastoma protein (Rb) (25). In many human cancers, including parathyroid adenoma, breast cancer, colon cancer, lymphoma, melanoma and prostate cancer, CCND1 is often overexpressed and plays important roles in cancer development (26). Recent study validated that repression of CCND1 was a useful strategy to inhibit the growth of tumors, suggesting CCND1 was a potential anticancer drug target (27).
Reverse screening the miRNAs regulating CCND1 by Luciferase assay
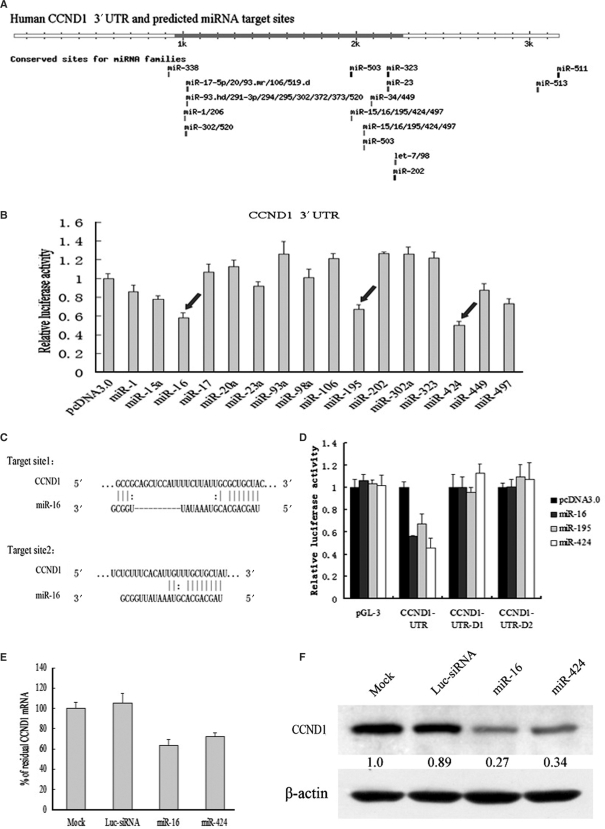
To identify the miRNAs controlling CCND1 expression, Targetscan software (www.targetscan.org) (28,29) was used to analyze potential miRNA target sites in 3′-UTR of CCND1. As shown in Figure 2A, the 3′-UTR of CCND1 mRNA contains many elements complementary to various miRNA seed regions, and these elements carry the identical sequences in human, mouse, rat, dog and chicken mRNA orthologues, which suggest that CCND1 could be regulated by diverse miRNAs.
Figure 2.
miR-16 family downregulated CCND1 protein and mRNA by targeting a putative binding site. (A) Human CCND1 3′-UTR and its possible miRNA target sites predicted by targetscan software. Many different miRNA seed regions are complementary to the elements of CCND1 3′-UTR, among which miR-16 family has two conserved sites. (B) Luciferase assays indicated that miR16 family could downregulate the expression of CCND1. pcDNA3.0 control plasmid and 16 different miRNA expressing plasmids were cotransfected with a modified pGL-3 control vector containing CCND1 3′-UTR. Luciferase assays were performed 48 h posttransfection. Histogram shows normalized mean values of relative luciferase activity form three independent experiments. Normalized luciferase activity in the absence of miRNAs expression vector was set to 100%. Arrows indicate a significant reduction of luciferase activity by miR-16/195/424. (C) Sequence inspection by bioinformatics reveals that target site 1(above) and target site 2 (below) are two conserved elements complementary to the seed region of miR-16 (nucleotides 2–8). (D) Luciferase assays indicated that miR-16 family downregulated the expression of CCND1 by targeting putative target site 2. pcDNA3.0 control plasmid, miR-16 expressing plasmid and miR-424 expressing plasmid were cotransfected with a modified pGL-3 control vector containing wild-type CCND1 3′-UTR or two deletants of CCND1 3′-UTR, respectively. The effectiveness of synthetic miR-16/424 on CCND1 mRNA and protein was respectively analyzed by qRT–PCR (E) and western blot (F).
To screen the miRNAs which can genuinely regulate the expression of CCND1 mRNA, the CCND1 3′-UTR was cloned into a modified pGL-3 control vector, placing the 3′-UTR with most of potential miRNA binding sites downstream of coding sequence of luciferase. The construct was cotransfected into HepG2 cells with control plasmid or plasmids expressing miRNAs regulating CCND1 potentially, which come from a constructed miRNA expression library (20). The results of luciferase assays revealed that overexpression of miR-16, miR-195 and miR-424 could reduce the luciferase activity from the reporter construct containing the CCND1 3′-UTR significantly, and that miR-15a and miR-497 could also reduce the luciferase activity moderately, whereas other miRNA expression plasmids and control plasmid had no effect on the luciferase activity (Figure 2B). These results of reverse screening suggested that miR-16, miR-195 and miR-424 could regulate the expression of CCND1 by targeting the complementary sites. And the seed hexamer of miR-16, miR-195 and miR-424 is identical, which suggests that they belong to the same miRNA family and regulate gene expression by targeting the same binding sites. Additionally, sequence inspection revealed the presence of two conserved elements complementary to the seed region of miR-16 family (nucleotides 2–8) (Figure 2C). To identify which putative target site was regulated by miR-16 family, two diverse deletants of CCND1 3′-UTR were cloned into the modified pGL3 control vector, of which the first contains putative target site 1 and the second contains no putative miR-16 target site at all. Luciferase assays indicated that overexpression of miR-16, miR-195 and miR-424 could reduce the luciferase activity from the reporter construct containing wild-type CCND1 3′-UTR (containing both putative binding sites), whereas had no effectiveness on any deletant of CCND1 3′-UTR (Figure 2D). These results reveal that miR-16, miR-195 and miR-424 may regulate the expression of CCND1 by targeting putative binding site 2.
Having established the ability of miR-16 and miR-424 to mediate repression of luciferase via the CCND1 3′-UTR, we next assayed their capacity to regulate the endogenous CCND1. Synthetic miR-16, miR-424 duplex and control Luc-siRNA were transfected to HepG2 cells. Western blot and qRT–PCR were used to detect the expression level of CCND1 protein and mRNA, respectively. qRT–PCR analysis showed a moderate reduction of CCND1 mRNA in HepG2 cells overexpressing miR-16 or miR-424 (Figure 2E). miR-16, miR-195 and miR-424 were downregulated in HepG2 cells depleted of Drosha (Figure 1D and Figure S1), this could explain the upregulation of CCND1 mRNA in clone 2 (Table 2). Western blot analysis revealed that miR-16 and miR-424 markedly reduced CCND1 protein levels compared to Luc-siRNA and mock transfected cells (Figure 2F). All the results above support the notion that miR-16 and miR-424 can regulate the expression of CCND1 directly in vivo.
miR-16 family induced cell cycle arrest partially by regulating CCND1
CCND1, in association with cyclin-dependent kinase (CDK4 and CDK6), phosphorylates the Rb, blocking its growth inhibitory activity, promoting cells to pass through the R point and thus driving them from G1 phase to S phase (30). Repression of CCND1 prevents cells from entering the S phase, causing an accumulation of cells in G0/G1. However, we detected no G0/G1 accumulation phenotype in HepG2 cells transfected with miR-16, miR-424 or siRNA against CCND1, although CCND1 protein levels reduced markedly (data not shown). It is possible due to insensitivity of HepG2 cells to miR-16 and the depletion of CCND1.
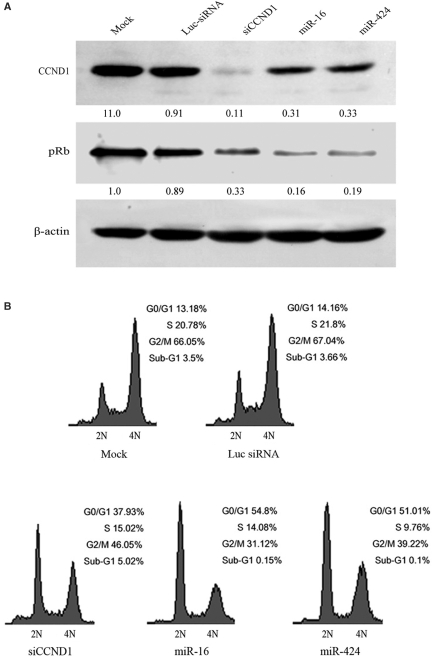
miR-16 family downregulates many transcripts and negatively regulates cell cycle progression by inducing the G0/G1 accumulation in A549 cells and some other cell lines (31). However, no genuine targets regulated by miR-16 family directly have been identified. To verify whether miR-16 family triggered G1 arrest by regulating the expression of CCND1, these miRNA and siRNA against CCND1 were transfected into A549 cells. The results revealed that miR-16, miR-424 and siRNA against CCND1 could depress the expression of CCND1 in A549 cells as well (Figure 3A). Furthermore, the levels of phosphorylated pRb decreased at the same time. Cyclin D1 levels were consistently found to be low in S phase, and high in G1 and G2 phases (32), but qRT–PCR analysis suggested that the expression levels of miR-16, miR-195 and miR-424 had no obvious change during the G0/G1, S and G2/M phases of the cell cycle (Figure S2). Compared with miR-16 and miR-424, siRNA against CCND1 had stronger inhibition effectiveness on CCND1; however, its inhibition effect on pRb was weaker than these miRNAs (Figure 3A). The results of cell cycle analysis confirmed that all of miR-16, miR-424 and siRNA against CCND1 could lead to G1 arrest in A549 cells (Figure 3B). But, siRNA against CCND1 (37.93%) triggered less G0/G1-cell accumulation than miR-16 (54.80%) or miR-424 (51.01%), despite its silencing effect on target gene was stronger than these miRNAs. All results above suggest that CCND1 is just one of the targets regulated by miR-16 family to control cell cycle progression, and some other targets may be involved in G0/G1 transition, too.
Figure 3.
miR-16 and miR-424 triggered G1 arrest partially by regulating CCND1. (A) The effectiveness of miR-16, miR-424 and siRNA against CCND1 on CCND1 and pRb was analyzed by western blot, with mock and control Luc-siRNA transfected A549 cells as controls. Antibody against β-actin was used as a loading control. (B) miR-16 and miR-424 triggered the accumulation of cells at G0/G1 stage. A549 cells were treated with nocodazole 24 h posttransfection and cell cycle distributions were detctected 20 h later. 2N, cells having diploid DNA content; 4N, cells having tetraploid DNA content.
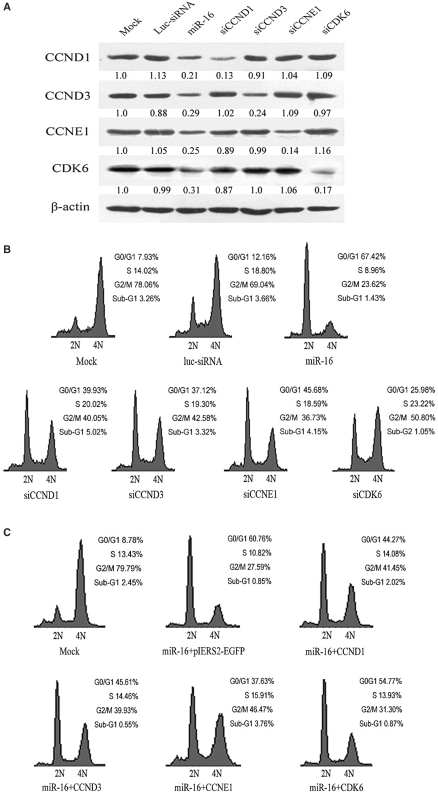
miR-16 family regulated the expressions of CCND3, CCNE1 and CDK6
The passage of cells through the cell division cycle is regulated by a family of kinases, the cyclin-dependent kinases (CDKs) and their activating partners, the cyclins. Different cyclin/CDK complexes regulate each of the cell cycle transitions. The G1/S phase transition is regulated primarily by D-type cyclins (D1, D2 or D3) in complex with CDK4/CDK6, and E-type cyclins (E1 or E2) in complex with CDK2. These complexes cooperate in phosphorylating and preventing Rb binding to E2F, thus activating E2F-mediated transcription and driving cells from G1 into S phase (27). The results of computational prediction (www.targetscan.org) revealed that the bulk of genes regulating G1/S transition directly, including CCND1, CCND2, CCND3, CCNE1, CDK6 and cdc25A, had the elements complementary to the seed hexamer of miR-16 (Figure S3), which suggested these genes were putative targets of miR-16 family.
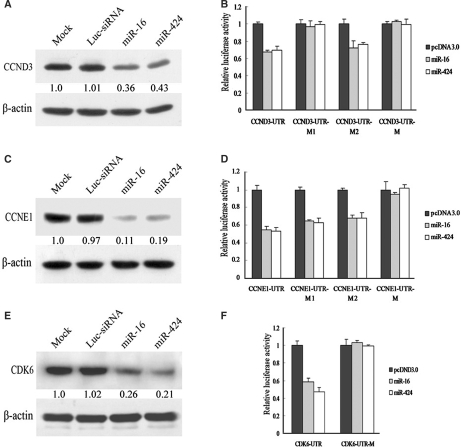
To analyze the effects of miR-16 family on these putative targets, miR-16 and miR-424 were transfected into A549 cells, and western blot was used to detect the protein expression levels of putative targets. As evident from Figure 4, miR-16 and miR-424 markedly reduced CCND3, CCNE1 and CDK6 protein levels compared with mock transfected cells (Figure 4A, C and E). In contrast, we observed no decrease in cdc25A protein (data not shown). The results above indicated that miR-16 family regulated the expressions of CCND3, CCNE1 and CDK6.
Figure 4.
miR-16 and miR-424 regulated a set of cell cycle genes. Western blot analysis of CCND3 protein(A), CCNE1(C) and CDK6(E) in A549 cells transfected with dsRNA indicated above the lanes. Antibody against β-actin was used as a loading control. Luciferase assays indicated that miR16 family downregulated the expression of CCND3(B), CCNE1(D) and CDK6(F) directly. pcDNA3.0 control plasmid, miR-16 expressing plasmid and miR-424 expressing plasmid were cotransfected with luciferase reporter vector containing wild-type 3′-UTR of CCND3/CCNE1/CDK6 and their own different mutants of 3′-UTR, respectively. For all histograms, mean values are shown and error bars represent SD from three independent experiments.
To test whether regulation was direct, 3′-UTRs of these selected targets were cloned into the modified pGL3-control vector, placing the 3′-UTRs with putative miR-16 binding sites downstream of the coding sequence of luciferase. Co-transfection with pcDNA-miR-16 and pcDNA-miR-424 but not pcDNA3.0 specifically decreased luciferase levels from each reporter (Figure 4B, D and F). Mutation in seed complementary site fully rescued repression for CDK6. For CCNE1, mutation of the single conserved seed complementary site had only a partial effect (Figure 4D), and mutations of both seed complementary sites fully rescued repression for CCNE1, indicating a combination of both sites effects of miR-16. For CCND3, only mutation of the first putative binding site rescued repression, which suggested that miR-16 regulated CCND3 by targeting the first seed complementary site.
miR-16 family triggered G1 arrest by regulating a set of cell cycle-related genes
To investigate the biological significance of these targets regulated by miR-16 family, we synthesized the siRNAs against these target mRNAs and transfected them into A549 cells. The results of western blot displayed that miR-16 repressed the expression of CCND1, CCND3, CCNE1 and CDK6 simultaneously, and each siRNA inhibited its target, respectively (Figure 5A). We then analyzed transfected cells for their cell cycle distributions. As shown in Figure 5B, silencing of these selected miR-16 targets by using siRNAs led to a substantial arrest in G1, partly phenocopy activation of miR-16. Linsley et al. (31) reported that siRNA pool targeting CARD10 induced the accumulation of cells in G0/G1 to the same extent as miR-16, but our luciferase assay results suggested that CARD10 was not the genuine target regulated by miR-16 directly (data not shown).
Figure 5.
miR-16-induced G1 arrest in A549 cells by regulating multiple cell cycle genes. (A) The downregulation effectiveness of miR-16 and siRNAs against different genes (lanes 3–7) was analyzed by western blot with antibodies indicated on the left. In lanes 1 and 2, mock and control Luc-siRNA transfected A549 cells served as controls. Antibody against β-actin was used as a loading control. (B) miR-16 and siRNAs induced G1 arrest in A549 cells. miR-16 triggered more accumulation of cells at G0/G1 stage than siRNAs against CCND1, CCND3, CCNE1 and CDK6, respectively. A549 cells were treated with nocodazole 24 h posttransfection and cell cycle distribution detected 20 h later. 2N, cells having diploid DNA content; 4N, cells having tetraploid DNA content. (C) miR-16 and cell cycle gene expression vectors were cotransfected into A549 cells, and cell cycle distributions were detctected by flow cytometry.
To finalize the function of miR-16-targeting these genes in cell cycle, rescue experiments were carried out. Utilizing pIERS2-EGFP expression vector, CCND1, CCND3, CCNE1 and CDK6 constructs without wild 3′-UTR were generated. These expression vectors were transiently cotransfected with miR-16. To remove the influence of transient low-transfection efficiency, the GFP-positive A549 cells were selected to perform cell cycle analysis. As shown in Figure 5C, when miR-16 was cotransfected with any of these cell cycle genes expression vectors, G1 arrest effectiveness of miR-16 was partially rescued, and CCNE1 had the strongest inhibiting effect on the activity of miR-16.
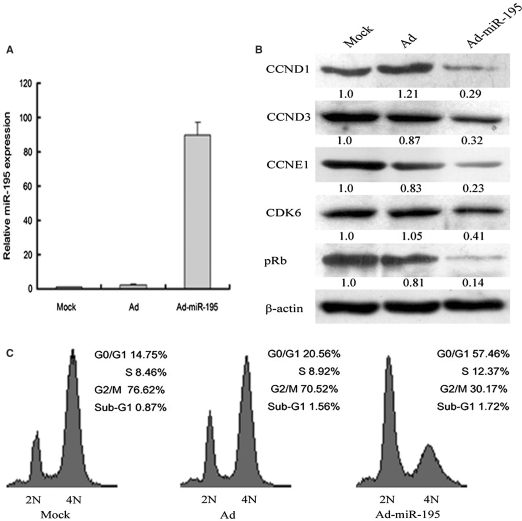
To confirm the cellular phenotype induced by miR-16 family is not artificial, we determined whether CCND1 downregulation and G0/G1 cell accumulation phenotype could be induced by miR-16 family expressed from genomic fragment. A549 cells were transfected with the plasmids expressing primary forms of miR-16 or miR-424. Flow cytometry analysis revealed that these plasmids could just induce slight G1 arrest in A549 cells (data not shown). It happened probably as a result of low-transient transfection efficiency. In addition, recombinant adenovirus was produced to express miR-195. When A549 cells were infected with Ad-miR-195, the expression level of miR-195 increased significantly (up to 90-fold) (Figure 6A). Western blot analysis revealed that the levels of CCND1, CCND3, CCNE1, CDK6 and pRb decreased in A549 cells infected with Ad-miR-195 (Figure 6B). Moreover, the results of cell cycle analysis confirmed that natural form of miR-195 could lead to G1 arrest in A549 cells (Figure 6C). Taken together, results above suggest that miR-16 family triggers an accumulation of cells in G0/G1 by regulating multiple downstream effectors including CCND1, CCND3, CCNE1 and CDK6.
Figure 6.
miR-195 repressed the expression of multiple cell cycle genes and induces cell cycle arrest. (A) The expression of miR-195 was detected by qRT–PCR, A549 cells were infected by Ad and Ad-miR-195. (B) The effectiveness of miR-195 on CCND1, CCND3, CCNE1, CDK6 and pRb was analyzed by western blot. (C) miR-195 induced G1 arrest in A549 cells. A549 cells were treated with nocodazole 48 h postinfection and cell cycle distributions were detected 20 h later.
DISCUSSION
To date, a large number of miRNAs have been identified in several organisms, including vertebrates and plants. In human, 533 miRNAs were validated (16,17), and some computational studies estimated that the number of human miRNAs is as many as 2- to 4-fold higher (33). However, only a handful of comprehensive studies on miRNA function have been completed, whereas the function of most miRNAs remains unknown. One of the main challenges in identification of miRNA function is to identify the genuine miRNA target gene (or genes) among many predicted targets.
In this study, we generated the cell lines depleted of Drosha protein, and indicated that 53 transcripts were upregulated remarkably in HepG2 cells depleted of Drosha. Given the roles of Drosha in the miRNA pathway (2,21,34), these upregulated transcripts may be regulated by miRNAs. Rehwinkel et al. (19) profiled the expression of mRNAs in Drosophila S2 cells depleted of Drosha, the results revealed that Drosha knockdown triggered upregulation of many transcripts. Furthermore, they concluded that the majority of those transcripts were genuine miRNA targets. Among these transcripts upregulated in HepG2 cells depleted of Drosha, CCND1 is a key cell cycle-related gene, and regulates G1/S transition. On the basis of miRNA expressing library (20), we established a miRNA targets reverse screening method by using luciferase reporter assay. By this method, multiple miRNAs including miR-16, miR-195 and miR-424 were identified to regulate the expression of CCND1. Furthermore, these miRNAs triggered G1 cell cycle arrest partially by repressing CCND1 in A549 cells (Figure 3). As all know, not all miRNA targets are downregulated at mRNA levels, thus, not all of them can be identified by the strategy analyzing mRNA expression in Drosha knockdown cells. Actually, for any genes we are interested in, this miRNA target reverse screening method can be used to analyze whether they are regulated by miRNAs, and screen the miRNAs regulating these mRNA genes. Using the reverse screening method, we found that several genes could be regulated by certain miRNAs, even some target genes could be regulated by multiple miRNAs simultaneously (data not shown). All these data suggest that this miRNA targets reverse screening method is very useful for identifying miRNA targets, and may play an important role in the characterization of miRNA function.
Although the biological roles of only a small fraction of identified miRNAs have been elucidated to date, the available evidence has shown that miRNA mutations or mis-expression correlate with various human cancers and that miRNAs can function as tumor suppressors and oncogenes to regulate key pathways involved in cellular growth control (35). Some miRNAs are involved in tumor genesis by modulating cell cycle progression (36). For example, aberrant expression of miR-221/222 was observed in diverse tumors, such as papillary thyroid carcinoma (37,38), pancreatic adenocarcinoma and glioblastoma (39). Further study revealed that ectopic overexpression of miR-221/222 repressed the expression of p27(Kip1) by targeting the binding sites in the 3′-UTR of p27 mRNA and strongly affected cellular growth potentially by inducing a G1 to S shift in the cell cycle. These results suggest that miR-221/222 can be regarded as a new family of oncogenes, directly targeting the tumor suppressor p27(Kip1), and that their overexpression might be one of the factors contributing to the oncogenesis and progression of prostate carcinoma through p27(Kip1) downregulation (40,41). Recently, miR-34 family members have been shown to be directly regulated by the tumor suppressor, TP53 (42–44). Overexpression of miR-34 causes cell cycle arrest at G1 phase by downregulation of a significantly great number of genes promoting cell cycle progression. Several downregulated cell cycle genes (including CCNE2, CDK4 and MET) have been validated as direct targets by showing miR-34 regulation of reporters engineered to contain the 3′-UTRs of the respective targets (44). Recently, Schultz et al. (45) have shown that let-7b regulates CCND1 and induces G1 arrest in malignant melanoma cells. Our studies have revealed that miR-34a (46) and miR-16 family can regulate CCND1 and induce G1 arrest in A549 cells too. To compare their effects on cell cycle, these miRNAs were synthesized and transfected into A549 cells. As shown in Figure S4, all of let-7b, miR-16 and miR-34a reduced CCND1 protein level and induced G1 arrest compared with Luc-siRNA. miR-16 and miR-34a had stronger inhibition effectiveness on CCND1 than let-7b, and miR-16 had the best effect on triggering accumulation of cells in the G0/G1 phase. Furthermore, compared with siRNA against CCND1, all these miRNAs had weaker inhibition effectiveness on CCND1, but triggered more prominent G0/G1-cell accumulation. These results suggest that all these miRNAs induce G1 cell cycle arrest by regulating multiple target genes, rather than a single key target.
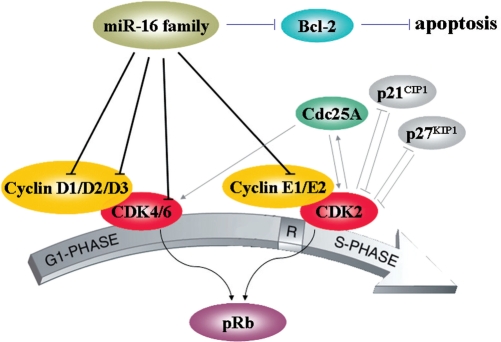
miR-16 family negatively regulates cellular growth and cell cycle progression. They trigger the G0/G1 accumulation phenotype in diverse cell lines, including HCT116, DLD-1, A549, MCF7 and Tov21G cells (31,36). Here, we found that miR-16 and miR-424 could regulate the expression of CCND1 by targeting putative target site (Figure 2). Further study revealed that several other cell cycle genes (such as CCND3, CCNE1 and CDK6) could also be regulated by miR-16 family (Figure 4), and all these cell cycle genes were involved in G1/S transition. Flow cytometry assay revealed that depletion of these targets led to a substantial arrest in G1, partly phenocopy activation of miR-16 (Figure 5B). All results above indicated that simultaneous silencing of these genes was more effective on blocking cell cycle progression than deletion of an individual gene. Taken together, both our results and findings of Linsley et al. (31) argue that miR-16 targets act in concert, rather than individually, to regulate G1/S transition (Figure 7). In other words, multiple targets regulated by an individual miRNA can act coordinately to regulate the same biological process (36).
Figure 7.
miR-16 family modulates G1/S transition by regulating Cdk4/6-cyclin D complexes and Cdk2-cyclin E complexes. The G1/S phase transition is regulated primarily by D-type Cyclins (D1, D2 or D3) in complex with CDK4/CDK6, and E-type Cyclins (E1 or E2) in complex with CDK2. These complexes cooperate in phosphorylating and preventing Rb binding to E2F, thus activating E2F-mediated transcription and driving cells from G1 into S phase. miR-16 family control the expression levels of Cyclin D1, Cyclin D3, Cyclin E1 as well as CDK6, inactivate Cdk2/4/6 kinases, and thus prevent phosphorylation of Rb proteins. Moreover, miR-16 can induce apoptosis by downregulation of antiapoptosis protein Bcl-2.
Just like siRNA against CCND1, miR-16 family repressed the expression of CCND1, but led to no G0/G1 accumulation phenotype in HepG2 cells. Likewise, miR-16 could not trigger cell cycle arrest in megakaryocytic cell line MEG-01 (data not shown). Cimmino et al. (14) reported that miR-16 induced apoptosis by targeting Bcl-2 in MEG-01 cells. However, we detected no significant apoptosis following transfection with miR-16 in A549 cells. These data suggest that the effect of miR-16 family on cell cycle arrest and apoptosis is cell type dependent, that is to say, miR-16 leads to cell cycle arrest in some types of cells, but induces apoptosis in others. It is possibly because some types of cells are sensitive to the downregulation of the targets regulated by miRNA, whereas others are not. Or the deletion, mutation or editing of binding sites in target mRNAs can also result in the inactivation of miRNA (47).
All data above argue that miR-16 family may participate in cell growth control and play tumor-suppressor roles by regulating cell proliferation and/or apoptosis pathway in various tumor cells. Both CCND1 (25) and Bcl-2 (48) play important roles in tumorigenesis, and serve as specific tumor therapeutic targets. And siRNAs against them can inhibit tumor cell growth significantly. Now that miR-16 can regulate multiple target genes promoting cell growth, including Bcl-2, CCND1, CCND3, CCNE1 and CDK6, and have stronger inhibitory effectiveness on cell growth than siRNAs against an individual gene, exogenous expression of miR-16 may be more effective for tumor therapy than depletion of single oncogene.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese State Key Projects for Basic Research (2006CB0D0807); National 863 plans projects (2006AA02Z127); Chinese National Natural Science Foundation project (30500299); Beijing Natural Science Foundation project (5072039) partially. Funding for open access charge: Chinese State Key Projects for Basic Research (2006CB0D0807).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 4.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 6.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Olsen RC, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking Lin-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 9.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterchronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J. Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 14.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Saini HK, Dongen SV, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krützfeldt J, Poy MN, Stoffel M. Strategies to determine the biological function of microRNAs. Nat. Genet. 2006;38:S14–S19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- 19.Rehwinkel J, Natalin P, Stark A, Brennecke J, Cohen SM, Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui J, Fu H, Feng J, Zhu J, Tie Y, Xing R, Zheng X. The construction of miRNA expression library for human. Prog. Biochem. Biophys. 2007;34:389–394. [Google Scholar]
- 21.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 22.Fu HJ, Zhu J, Yang M, Zhang ZY, Tie Y, Jiang H, Sun ZX, Zheng XF. A novel method to monitor the expression of microRNAs. Mol. Biotechnol. 2006;32:197–204. doi: 10.1385/MB:32:3:197. [DOI] [PubMed] [Google Scholar]
- 23.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 25.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol. Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 27.Grillo M, Bott MJ, Khandke N, McGinnis JP, Miranda M, Meyyappan M, Rosfjord EC, Rabindran SK. Validation of cyclin D1/CDK4 as an anticancer drug target in MCF-7 breast cancer cells: effect of regulated overexpression of cyclin D1 and siRNA-mediated inhibition of endogenous cyclin D1 and CDK4 expression. Breast Cancer Res. Treat. 2006;95:185–194. doi: 10.1007/s10549-005-9066-y. [DOI] [PubMed] [Google Scholar]
- 28.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing,often flanked by adenosines,indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim. Biophys. Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 31.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 33.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 34.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 35.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 36.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 37.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 39.Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 40.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LinksGalardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 42.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53–mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 46.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 47.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J. Biol. Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 48.Ghobrial IM, Witzig TE, Adjei AA. Targeting apoptosis pathways in cancer therapy. CA Cancer J. Clin. 2005;55:178–194. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.