Abstract
The construction of metagenomic libraries has permitted the study of microorganisms resistant to isolation and the analysis of 16S rDNA sequences has been used for over two decades to examine bacterial biodiversity. Here, we show that the analysis of random sequence reads (RSRs) instead of 16S is a suitable shortcut to estimate the biodiversity of a bacterial community from metagenomic libraries. We generated 10 010 RSRs from a metagenomic library of microorganisms found in human faecal samples. Then searched them using the program BLASTN against a prokaryotic sequence database to assign a taxon to each RSR. The results were compared with those obtained by screening and analysing the clones containing 16S rDNA sequences in the whole library. We found that the biodiversity observed by RSR analysis is consistent with that obtained by 16S rDNA. We also show that RSRs are suitable to compare the biodiversity between different metagenomic libraries. RSRs can thus provide a good estimate of the biodiversity of a metagenomic library and, as an alternative to 16S, this approach is both faster and cheaper.
INTRODUCTION
We live in a world dominated by microorganisms (1). However, very little is known about the role they play in our environment. One of the main questions that remains to be answered is how these microorganisms compete and communicate between themselves to get nutrients and produce energy in an ecosystem. To address this question, one has to overcome the limitations associated with the ‘uncultivability’ of at least 99% of the microorganisms in nature (2). The development of culture-independent methods applied to environmental samples was a turning point for the field. In 1985, Pace and colleagues (3) were the first to propose direct analysis of 5S and 16S rRNA gene sequences to describe the microbial diversity in an environmental sample without culturing. The 16S rRNA gene is highly conserved among all microorganisms, is of suitable length (about 1500 bp) for bioinformatic analysis and is an excellent molecule for discerning evolutionary relationships among prokaryotic organisms (4). For all these reasons, this molecule has given rise to a huge public database (RDPII: http://rdp.cme.msu.edu/containing 481 650 16S rRNAs, 13 February 2008) (5). Finally, defining phylotype (or species) on the basis of 16S rDNA sequences has been and remains the accepted standard for studies of uncultured microorganism diversity (6–10).
These molecular tools have revealed a wider microbial diversity than expected in several ecosystems (11,12). The functions, however, of the different groups of microorganisms are largely unknown. Pace proposed the first cloning of genomic DNA directly from environmental samples using a phage vector (13). Later, this approach, called metagenomics, inspired other groups to penetrate the microbial world from all sources including human faeces, whale falls, soil, marine and other aquatic ecosystems (14–18). Metagenomics, conducted on a massive scale, has provided dramatic insights into the structure and metabolic potential of microbiota (also used for microbial population) (19,20). Functional screening of metagenomic libraries has led to the assignment of functions to numerous ‘hypothetical proteins’, so far demonstrating the power of functional metagenomics (21). Metagenomics is a newly emerging technology, and has generated more than 100 projects in the GOLD Web site, Genomes OnLine Database (February 2008, http://www.genomesonline.org/gold.cgi), 31 of which have already been completed.
One of the approaches allowing the classification of metagenomic fragments is the sequence-composition-based method. It relies on the analyses of oligonucleotide frequencies that vary significantly among genomes, permitting discrimination of different species (22,23). This approach, which needs a training process in using genomic sequences available in databases, has been the method of choice for some analyses of microbial communities in recent years (24–26) and has been used in different software such as TETRA or PhyloPythia (27,28). However, it encounters limitations not only in the availability of genomic sequences in databases for their learning process, but also in the size of the analysed metagenomic fragments. As discussed by the authors themselves, the sequence-composition-based approach needs complementary methods to analyse short metagenomic fragments (<1 kb) such as single-read end-sequences.
Another approach to study microbial diversity is a large-scale screening for clones or contigs containing a phylogenetic gene marker such as 16S rRNA gene. To that end, clones harbouring 16S can be screened by several methods. The first consists of the extraction of the recombinant vectors to remove the genome of the organism in which the cloning has been performed, then selection of the 16S rRNA gene by DNA–DNA hybridization on a macroarray (18). The second method involves the massive sequencing of the whole-metagenome and subsequent in silico identification of the 16S rDNA sequences (16). PCR-based 16S rRNA gene sequence-based analysis, an approach that has been widely used in the literature to analyse the diversity of microbial communities could have been applied directly on the environmental samples. However, due to PCR bias and the fact that different DNA extraction methods have been used in both approaches (PCR and metagenomic libraries), we would not recover the same diversity.
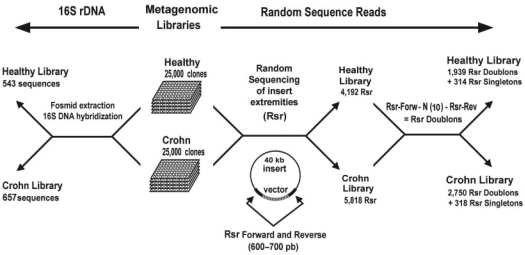
In a previous study (18), we built two metagenomic libraries to analyse the intestinal microbiota of healthy volunteers and patients with Crohn's disease. We then used the first method to analyse the diversity of our libraries, performing a high throughput extraction of recombinant vector to avoid the Escherichia coli genome and DNA–DNA hybridization on nylon membrane to target the 16S (Figure 1). This technique allowed us to identify 1200 clones containing 16S rDNA sequences from both libraries. The diversity analysis was based on a multiple alignment using ClustalW and the taxonomic assignment browser of the RDPII Release 9. This 16S analysis approach that has been habitually used to analyse environmental microbial diversity, is, however, very expensive in terms of both time and money required (in our case: 6 months, three persons and more than $70 000 of materials and equipment without taking into account the sequencing step that is much cheaper nowadays). An additional disadvantage associated with relying on 16S for estimates of species diversity and abundance is the varying number of copies of rRNA genes between taxa (a difference of more than an order of magnitude among prokaryotes) (29,30) leading to an overestimation of microorganisms containing a high copy number and an underestimation of those harbouring a low copy number.
Figure 1.
16S rDNA sequences and random sequence reads (RSRs). We have previously built two human faecal metagenomic libraries (healthy and Crohn) containing 25 000 clones each (18). Each clone contains an insert of 40 kb of prokaryotic genomic fragment. About 1200 16S rDNA sequences have been screened by DNA hybridization from the two libraries in the previous work (18). In the present study, about 4500 clones have been randomly chosen to perform a one read sequencing on the two extremities of the insert (Forward and Reverse). To further simplify the parsing, each RSR-Forward is then attached by 10 nt to the RSR-Reverse for each clone to form a RSR doublon.
The objective of this study is to demonstrate that analysis of random sequence reads can serve as a faster and cheaper alternative to 16S rDNA sequencing. Thus, we present metagenomic diversity analyses comparing the use of the 16S phylogenetic marker and random sequence reads (RSRs). To rule out the differential effects of more sophisticated taxonomic assignment protocols on the two set of sequences, we applied the same simple computational pipeline, TAP (Taxonomic Assignment Pipeline) based on the search against a prokaryotic sequence database using BLASTN (31) to both of them. We show, however, that when applied to the 16S sequences, this pipeline produces assignments comparable to those obtained when using a more sophisticated assignment method based on the detection of conserved regions across the 16S sequence alignments. We then used TAP to obtain taxonomic assignments on RSRs in a metagenomic library from the intestinal microbiota of healthy human volunteers, and we found no significant differences in the resulting assignments. Furthermore, we show that by using RSRs it is possible to identify taxonomic differences between the intestinal microbiota of healthy and Crohn-affected individuals, which cannot be detected using 16S rDNA sequences. Finally, we applied RSR-TAP to Sargasso Sea samples and showed that the diversity pattern was similar in broad outline to the previous 16S analysis made by Venter et al. (16).
METHODS
Random sequence reads (RSRs)
We have previously built two human faecal metagenomic libraries (healthy and Crohn) containing 25 000 clones each (18) (Figure 1). Each clone contains an insert of 40 kb of prokaryotic genomic fragment. In this study, about 4500 clones have been randomly chosen to perform a one read sequencing on the two extremities of the insert (5′ and 3′). All sequences were determined by Genoscope (Evry, France) on an ABI 3730 DNA sequencer. The sequencing provided 10 010 high-quality sequences (4192 for healthy and 5818 for Crohn library) with an average size of 615 bp ranging from 43 to 880 bp. To further simplify the parsing, each RSR-5′ was then computationally attached with 10 nt to the RSR-3′ for each clone to form a RSR doublon. In this way, we obtained 1939 RSR doublons and 314 RSR singletons (for which the counterpart sequencing has failed) for the healthy library and 2750 RSR doublons and 318 RSR singletons for the Crohn library. All our sequences have been deposited in GenBank (EU057993-EU068001), except one (LM0ACA7ZB01RM1) from the Crohn library that was too short (<50 bp).
16S rDNA sequences
To validate our computational approach, we used the 1200 16S sequences obtained from a previous work (Figure 1). These sequences ranged in size from 990 to 1330 bp. In Manichanh et al. (18), taxonomic analysis was based on a multiple alignment (ClustalW). Poorly aligned positions and divergent regions of the DNA alignment were removed with Gblocks (32) using parameters optimised for rRNA (33) and so, no manual refinement of the alignments was necessary. Distance matrices were computed with DNADIST v3.6 (34); and trees were constructed with NEIGHBOR v3.6, which implements the Neighbor-Joining method of Nei and Saitou, and the UPGMA method of clustering (35). We defined an operational taxonomic unit (OTU) as a cluster of 16S rDNA sequences sharing at least 98% similarity. We then taxonomically assigned each OTU using the online Seqmatch program of the RDPII webpage (http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp). One reference sequence for each OTU has been submitted to Genbank (healthy library: AY850400 to AY850487, Crohn library: AY850488 to AY850541). For the present analysis, we did not apply Gblocks and used the whole length of the 16S sequences.
Shotgun sequences from the Sargasso Sea
To analyse metagenomic libraries other than those from human gut, we downloaded 1 982 807 sequences from a shotgun dataset (cloning of random segments of 2–6 kb) of the Sargasso Sea (16). These sequences had an average length of 600 bp ranging from 100 to 1177 bp. To analyse a comparable number of RSRs than for the human gut metagenome, we selected randomly 5000 sequences (0.25%) from the downloaded dataset and used them as RSR singletons.
Statistical analyses
To compare the phylotype frequencies across different datasets and assignment procedures we computed a chi-square of homogeneity—which essentially tests whether the frequencies observed in the analysed samples are consistent with the hypothesis of the samples being randomly drawn from the same population. P-values smaller than 0.05 were considered enough to reject this hypothesis, and to denote therefore significant differences between the compared groups.
RESULTS
Metagenomic sequence datasets
Figure 1 illustrates the human gut datasets used in the present study. In Manichanh et al. (18), we built two metagenomic libraries containing 50 000 genomic fragments (inserts of 40-kb each) of human faecal microbiota collected from samples of six healthy volunteers and six patients with Crohn's disease (CD). From these libraries, two different sets of sequences were generated. The first set contains 16S sequences obtained from a high throughput screening using a DNA–DNA hybridization technique on both libraries. The screening and sequencing revealed 1200 clones containing 16S sequences of about 1200-bp each. These sequences permitted the phylogenetic analysis and comparison of the two healthy and Crohn libraries based on a computational method appropriate for 16S sequences (see ‘Methods’ section). The results indicated a reduced complexity of the Firmicutes bacterial phylum as a signature of the faecal microbiota in patients with CD. The second set, which has been specifically generated for this study, is a randomly generated collection of 5818 and 4192 high-quality sequence reads (from Crohn and healthy libraries, respectively). The total reads consisted of 6.3 Mbp. They were obtained by sequencing the two extremities (forward and reverse sequences) of each insert. Thus, our datasets consisted of RSR doublons from both human gut libraries (1939 from ‘healthy’ and 2750 from ‘Crohn’) and 4192 RSR singletons from ‘healthy’ human gut. We also added 5000 RSR singletons from the Sargasso Sea libraries.
The taxonomy assignment pipeline (TAP)
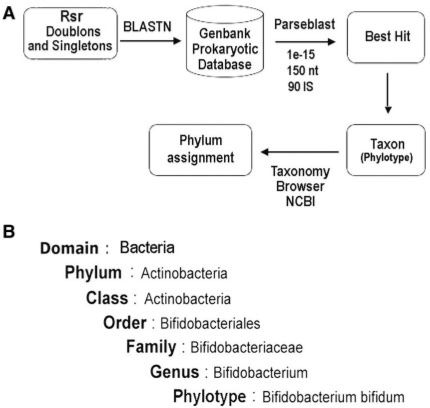
Figure 2A shows our processing pipeline. The sequences were compared against a database of GenBank prokaryotic sequences (GenBank_prok) using BLASTN. We applied this local alignment program to find the closest relative for each of the RSRs against the prokaryotic DNA sequence database of GenBank that contained 516 770 entries. HSPs which met specific cut-off parameters [expect value, HSP length and identity score (IS)], were further considered. For each of these parameters, we tested different values. We decided to keep these criteria (expect value <e−15, HSP length >150 nt and IS >90%) because the resulting microbial diversity was the most similar in terms of number of phylotypes (P = 0.14) found in each phylum to that obtained from the 16S phylogenetic analysis (18). We then took the top blast hit, identified the species to which the sequence belongs to, and used this to perform the taxonomic assignment. Finally, the phylum of each taxon was recovered from the NCBI Taxonomy browser (http://www.ncbi.nlm.nih.gov/entrez/batchentrez.cgi?db=Taxonomy). Bacterial nomenclature used in this study is based on the Bergey's Manual of Systematic Bacteriology (Figure 2B).
Figure 2.
Taxonomic assignment pipeline (TAP) to analyse the biodiversity using RSRs and BLASTN. (A) We applied BLASTN to find the closest relative for each of the RSRs against GenBank_prok HSPs which met the following cut-off parameters: expect value <1e−15, HSP length >150 nt and IS >90%, were further considered. We then took the top blast hit, identified the species to which the sequence belongs to, and used this to perform the taxonomic assignment. Finally, the phylum of each taxon was recovered from the NCBI Taxonomy browser (http://www.ncbi.nlm.nih.gov/entrez/batchentrez.cgi?db = Taxonomy). (B) Bacterial nomenclature used in this study is based on the Bergey's Manual of Systematic Bacteriology.
Biodiversity with 16S rDNA sequences for healthy and Crohn human gut libraries
The 1200 16S sequences, obtained from a high throughput DNA hybridization screening from two metagenomic libraries (healthy and Crohn), were previously analysed phylogenetically using a computational method developed specifically for 16S (see ‘Methods’ section). They represented 125 different phylotypes (or species), and each phylotype was defined as a cluster of sequences showing at least 98% similarity. Although a single sequence of each phylotype was deposited in GenBank, they were tagged as environmental sequences, and were therefore absent from the database subsequently used in TAP.
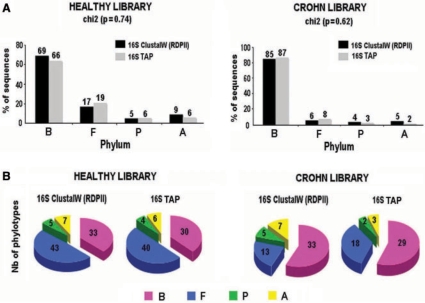
In the present work, we applied TAP to these 16S sequences. Of the 1200 sequences, 2% had no hits. We obtained, as expected, the four dominant bacterial phyla (Bacteroidetes, Firmicutes, Proteobacteria and Actinobacteria) found in human gut. Figure 2B shows the taxonomic nomenclature used in this study. Figure 3 shows a comparison of the TAP results and those obtained previously (18) using an approach for phylogenetic markers (see ‘Methods’ section). Tables S1 and S2 show this comparison at the genus and phylotype levels, respectively. The study in (18) was performed 2–3 years ago, and since then the GenBank_prok database has been updated with many new microbial sequences. Therefore, we re-analysed the 16S sequences using the method applied in this study but with an updated database. The comparison with the previous results showed that the updated database allowed us to recover 17 more assignments (out of 44). The taxonomic assignment obtained from TAP using 16S for this environment gave similar results to the previous method in terms of phyla, genera, phylotypes, number of phylotypes and percentage of sequences detected in each phylum. In terms of phylotypes, TAP is able to detect all those detected previously (except one: Coprococcus catus) with few differences in the number of sequences for each phylotype. For the comparison in terms of percentage of sequences in each phylum, a chi-square test was used, which showed no significant differences between the two methods (P = 0.74 for healthy and P = 0.62 for Crohn library). When comparing healthy and Crohn, using TAP on 16S, we obtained significant differences (P = 0.016) as previously shown in (18).
Figure 3.
Taxonomic assignment 16S rDNA sequences. Comparison between 16S analysed with a previous method using ClustalW, a multiple alignment program to align with RDPII (Ribosomal Database Project II) referenced sequences and 16S analysed with TAP, (A) in terms of percentage of sequences and (B) in terms of number of phylotypes. Data were analysed using the chi-square test for two independent sets of samples. Only a P value <0.05 was considered to denote a significant difference. B, Bacteroidetes; F, Firmicutes; P, Proteobacteria; A, Actinobacteria.
Comparison between RSR singletons and RSR doublons from the healthy human gut library
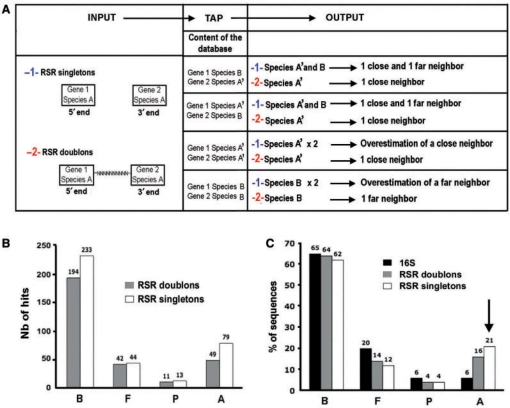
Since we performed one-read sequencing of the two extremities (5' and 3') of each clone insert—and therefore the two end sequences from the same clone could correspond to two different genes—the RSRs could be analysed either as singletons or as doublons (Figure 1). Figure 4A shows theoretical outputs of TAP processing depending on whether RSR singletons or doublons are used and on different possible contents of the database. In the case of RSR singletons, we can end up with an additional far neighbour or an overestimation of the number of a particular species. We used TAP to analyse both sets of sequences (RSR singletons and doublons) from the healthy metagenomic library. The results gave a higher sensitivity with singletons (Figure 4B), with a total of 376 hits for singletons and 303 for doublons, but a lower specificity as shown by the recovering of two genera not found when analysing 16S sequences (Supplementary Tables S3 and S4). Also as expected, the overestimation of one phylum (Actinobacteria) was found in the case of singletons, when compared with 16S assignment (Table S3). The low number of hits recovered is mainly due to the very stringent cut-off parameters we used in TAP. Therefore, doublons instead of singletons seem to be more appropriate for this taxonomic assignment based on random reads and TAP processing.
Figure 4.
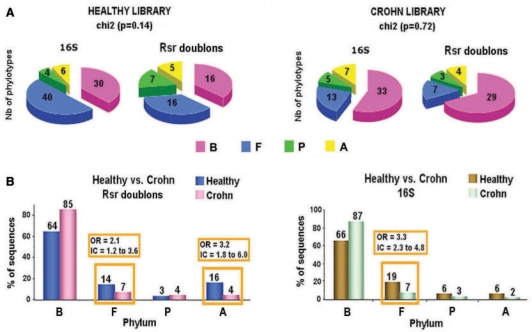
Comparison between RSR singletons and doublons from the healthy library. (A) Theoretical comparison between RSR singletons and doublons depending on different possible contents of the database used in TAP where species A is a closer neighbour of species A’ than of species B. RSR singletons are represented by two queries (5′ and 3′ ends) whereas RSR doublons are represented by only one query (5′ attached to 3′ end by 10 nt). We show possible outputs from these different combinations. (B) Results of TAP processing with the RSR singletons and doublons from the healthy metagenomic library. B, Bacteroidetes; F, Firmicutes; P, Proteobacteria; A, Actinobacteria. (C) Comparison of results with RSR singletons and doublons with those of 16S analysis from the healthy library in terms of percentage of sequences having a hit. The arrow shows the overestimation of Actinobacteria phylum when using RSR doublons compared with RSR singletons.
Biodiversity with RSRs from the healthy human gut library
As shown in Figure 4C, RSR doublons from the healthy metagenomic library processed with TAP allowed us to detect a similar bacterial diversity to that identified by 16S sequence analysis with the same computational method, in terms of proportion of sequences found in the four different bacterial phyla of intestinal microbiota. Using RSRs we detected 45% of the diversity found by 16S at the genus level and 44% at the species level. When considering species representing >1% of the 16S sequences, we reached 84% at the genus level and 54% at the species level (Tables 1 and S4). At the phylotype level, the biodiversity is reduced with RSRs (Figure 5) when compared with that of the 16S. The differences observed at the phylotype level reflect the limitation in the diversity of genes present in GenBank_prok.
Table 1.
Comparison at the genus level: 16S rDNA sequences versus RSR doublons
| Phylum | Genus | 16S |
RSR |
||
|---|---|---|---|---|---|
| Healthy | Crohn | Healthy | Crohn | ||
| Bacteroidetes | Alistipes | 0.7 | 1.1 | ND | 0.3 |
| Bacteroidales | 0.7 | 0.3 | ND | ND | |
| Bacteroides | 50.3 | 43.2 | 63.7 | 80.9 | |
| Prevotella | 6.1 | 11.9 | 0.3 | 1.4 | |
| Tannerella | ND | 8.7 | ND | 1.4 | |
| Firmicutes | Anaerotruncus | 0.6 | ND | ND | ND |
| Anaerovorax | 0.2 | ND | ND | ND | |
| Clostridium | 1.5 | 10.4 | 5.6 | 0.8 | |
| Dialister | 0.2 | ND | ND | ND | |
| Enterococcus | ND | 1.1 | 4.0 | 5.5 | |
| Eubacterium | 1.8 | ND | ND | ND | |
| Faecalibacterium | 1.7 | 0.5 | 0.7 | ND | |
| Lachnospira | 0.2 | ND | ND | ND | |
| Lachnospiraceae | 0.2 | ND | ND | ND | |
| Lactobacillales | 0.2 | 0.3 | ND | ND | |
| Oscillospira | 0.2 | ND | ND | ND | |
| Papillibacter | 0.6 | 0.2 | ND | ND | |
| Phascolarctobacterium | 0.2 | ND | ND | ND | |
| Roseburia | 1.3 | 0.3 | 0.3 | ND | |
| Ruminococcus | 4.2 | 1.4 | 0.7 | ND | |
| Streptococcus | ND | 0.5 | ND | 0.3 | |
| Subdoligranulum | 2.0 | 0.5 | 0.3 | ND | |
| unclassified_ Clostridiaceae | 3.5 | ND | ND | ND | |
| unclassified_ Lachnospiraceae | 0.2 | ND | ND | ND | |
| Proteobacteria | Bilophila | 0.7 | ND | ND | ND |
| Desulfovibrio | ND | 0.2 | ND | ND | |
| Escherichia | 3.7 | 2.8 | 0.3 | 3.0 | |
| Shigella | 0.2 | ND | ND | 0.3 | |
| Methylophilus methylotrophus | 0.9 | ND | ND | ND | |
| Actinobacteria | Bifidobacterium | 2.4 | 1.4 | 14.2 | 4.2 |
| Brachybacterium | 0.2 | ND | ND | ND | |
| Coriobacterium | 3.1 | 0.2 | 1.7 | ND | |
| Corynebacterium | 0.6 | ND | ND | ND | |
Values in bold indicates that each genus represented by at least 1% of the clones analysed by using 16S is also detected when using RSRs. ‘ND’ indicates an absence of detection.
Figure 5.
Biodiversity with RSRs. (A) for the healthy and Crohn libraries, we applied our computational method to the RSR doublons and compared the results to those of 16S rDNA sequences using the same method. Data were analysed using the chi-square test for two independent sets of samples. Only a P value <0.05 was considered to denote a significant difference. (B) Comparison of two metagenomic libraries using TAP and RSR doublons or 16S. An odds ratio of one indicates that the condition or event under study is equally likely in both groups. B, Bacteroidetes; F, Firmicutes; P, Proteobacteria; A, Actinobacteria.
We did not detect Archaea, neither with 16S nor with RSR sequences. This observation is not concordant with other molecular analyses of the gut microbiota (10,17), which show that archaeal species, in particular Methanobrevibacter smithii, are also major players in the human distal gut ecosystem. This discrepancy may have been caused by the biases associated with our bacterial DNA recovery methods, which is a well-known problem (17).
Comparison of RSRs from two human gut metagenomic libraries (healthy and Crohn)
As we did with 16S sequences and the RSR doublons from the healthy metagenomic library, we applied TAP to the RSR doublons obtained from the Crohn metagenomic library (Figure 5A and B). Figure 5A shows that the microbial diversity, at the phylotype level, obtained using RSR doublons is very similar to that of the 16S, indicating that most of the species present in this library are known, that is, present in the database except the case of false positives which should be reduced using RSR doublons instead of singletons. However, the healthy library seems to contain more unknown microbial species especially in both Bacteroidetes and Firmicutes phyla (Tables S4 and S5) compared to the Crohn library. The comparison of the TAP results of the two metagenomic libraries (healthy and Crohn), showed a difference in the percentage of sequences between the two libraries in two bacterial phyla: Firmicutes (as shown with 16S) and Actinobacteria. This difference in the Actinobacteria phylum, not significant when using the 16S sequences, shows another advantage of using RSRs instead of 16S. Indeed, by using RSRs, we are less limited in the number of sequences to be analysed. However, this observation needs to be confirmed by experiment. Here, we show that two metagenomic libraries, built with the same molecular methodology and from the same kind of environment, but presenting two different conditions, can be compared in terms of microbial composition using RSRs.
Analyses of 16S and RSRs from the Sargasso Sea shotgun dataset
To test our computational approach in metagenomic libraries other than those from human gut, we selected randomly 0.25% (5000 RSRs) of the Sargasso Sea dataset and performed a TAP analysis. Then, we compared the results with those reported after the analysis of 1164 distinct 16S gene (or gene fragments) identified in the same shotgun dataset (16). Due to the available 16S data for this library, the results were comparable only at the phylum and class ranks. From the 5000 RSRs, 976 sequences (19.5%) passed our cut-off parameters. At the phylum level, we obtained similar results regarding the number of main phyla and the proportion of sequences in each phylum (Table 2). At the class level, we observed also a similar pattern of class representation for the main phylum (Proteobacteria), which accounted for 85% of the sequences, although there were differences in the proportion of sequences in some classes.
Table 2.
Comparison between 16S and RSRs from the Sargasso Sea shotgun dataset
| Phylum | Class | 16S (%)a | 5000 RSRs_ TAP (%) |
|---|---|---|---|
| Bacteroidetes | 0.5 | 1 | |
| Cyanobacteria | 5 | 10.7 | |
| Proteobacteria | 85 | 88 | |
| Alphaproteobacteria | 41 | 1.4 | |
| Betaproteobacteria | 13 | 56.6 | |
| Gammaproteobacteria | 29 | 30 | |
| Deltaproteobacteria | 1 | 0 | |
| Epsilonproteobacteria | 1 | 0 |
aData extracted from figure 6 of Venter et al. (16).
DISCUSSION
Although PCR-based studies are inherently biased, because of ‘universal’ primers that may not be as universal as they should be, most phylogenetic surveys of microbial ecosystems had relied on analyses of amplified rRNA genes and subsequent comparison with more than 500 000 small subunit rRNA sequences reported in the RDPII (5). Nowadays, metagenomic projects are quickly becoming the default approach for the study of environmental samples. The best results are obtained by the complete sequencing of all genomes in an environmental sample, the approach taken by C. Venter (16). However, most researchers do not have the resources to undertake such an exhaustive analysis.
The goal of our study was to show that using Random Sequence Reads is a cost-effective alternative to the typical approach based on 16S library screening in order to characterize the diversity of a metagenomic community. Although RSRs might not give as accurate an evaluation of the 16S library screening or the sequencing and assembly of entire genomes, it does provide a reliable estimate of the diversity found in a given metagenomic library.
We have chosen not to use more complex taxonomic assignment approaches, which exist for both 16S sequences and Random Sequence Reads. Since these more complex pipelines are specifically tailored to the specific characteristics of the sequence data they are dealing with (16S Sequences versus RSRs), differences observed between 16S and RSRs could be attributed to the differences in the efficiency between the computational pipelines used for taxonomic assignment, rather than intrinsic differences in the taxonomic information in these two sources of sequence data. We have instead used the simplest of all assignment methods possible for both 16S and RSRs: we have assigned a sequence (16S or RSRs) to the taxon to which the closest known sequence belongs (given a minimum degree of sequence conservation). We understand that this is far from ideal for taxonomic assignment, but we believe it is the most appropriate method to compare the taxonomic information contained in 16S sequences versus RSR sequences. In practice, a researcher using RSR sequences should use a more sophisticated taxonomic assignment method, such as MEGAN (36), or the one proposed by Kraus et al. (37). Also, it would be very easy to integrate MEGAN or any other similar tool into our pipeline.
The very low number of hits (13% for the healthy and 11% for the Crohn library) obtained using RSRs is largely due to the specificity resulting from our BLASTN cut-off parameters. We decided to keep these criteria (expect value <e−15, HSP length >150 nt and IS >90%) because the resulting microbial diversity was the most similar in terms of number of phylotypes (P = 0.14) found in each phylum to that obtained from the 16S phylogenetic analysis (18). When analysing diversity using 16S sequencing in (18), we used the cut-off of 98% of identity. According to our results, the analysis of 16S compared with a database of 16S has shown that only 50% of the sequences belong to known species. Therefore, it is not surprising that the comparison of random reads with a general database such as GenBank_prok gives such a low number of hits.
To determine the best database to use with TAP, we compared the potential for recovering the microbial diversity of three different databases: GenBank_prok (without environmental sequences), RefSeq and the 577 complete NCBI microbial genomes. For that, we analysed RSR doublons from the healthy library, using TAP with each of the three databases. The results showed a higher sensitivity with GenBank_prok with 50 more hits and 16 more phylotypes than with RefSeq and 66 more hits and 26 more phylotypes than with ‘complete NCBI genomes’ (Tables S6–S7). We then compared the results in terms of number of phylotypes recovered in the different phyla with those obtained in (18) using the chi-square test. The comparison showed that GenBank_prok did not provide significant differences in diversity compared with the previous results obtained with 16S (P = 0.25), whereas RefSeq and the complete genome databases showed significant differences (P = 0.000007 and P = 0.0001, respectively). For this reason, we chose the less well-annotated and more redundant GenBank_prok for further analyses.
There are, however, certain biases that should be taken into account. First of all, any analysis that depends on a specific database is limited by the contents of that database. Obviously, as long as the databases are not complete no analysis can be perfect. Another problem is the bias in favour of certain species. Sequences from certain pathologically important species and those, which have been completely sequenced are greatly overrepresented in GenBank. This means that our results will be biased in favour of such species. To overcome this bias we can either completely remove these species from the database, resulting in a loss of true positives or we can use stricter cut-offs and thereby lose sensitivity. We believe that the balance struck by our parameters gives a suitable sensitivity to specificity ratio given the limitations of the database. In the coming years as more species are sequenced these biases will gradually decrease and, eventually, disappear altogether.
At present, and taking into account the aforementioned limitations of existing databases, we believe the method presented here is the fastest (a few months for the sequencing and a few weeks for the sequence analyses), easiest (without 16S screening experiments), and cheapest (only the sequencing step) available for the quick estimation of phylogenetic diversity in a metagenomic library.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We acknowledge Dr Robert Castelo and Anne Bergeron for useful comments and discussions. This work was supported by the Research grant no. BIO2006-03380 from the Spanish Ministry of Education and Science and by a pre-doctoral fellowship from the Spanish Ministry of Education and Science. Funding to pay the Open Access publication charges for this article was provided by INRA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 3.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 7.Head IM, Saunders JR, Pickup RW. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 8.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beja O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, Villacorta R, Amjadi M, Garrigues C, Jovanovich SB, et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2000;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- 16.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 17.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 20.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 21.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 23.Abe T, Kanaya S, Kinouchi M, Ichiba Y, Kozuki T, Ikemura T. Informatics for unveiling hidden genome signatures. Genome Res. 2003;13:693–702. doi: 10.1101/gr.634603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe T, Sugawara H, Kinouchi M, Kanaya S, Ikemura T. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 2005;12:281–290. doi: 10.1093/dnares/dsi015. [DOI] [PubMed] [Google Scholar]
- 25.Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, Yeates C, He S, Salamov AA, Szeto E, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 26.Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 27.Teeling H, Waldmann J, Lombardot T, Bauer M, Glockner FO. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 29.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 33.Massana R, Castresana J, Balague V, Guillou L, Romari K, Groisillier A, Valentin K, Pedros-Alio C. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 2004;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Seattle: Department of Genetics, University of Washington; 1993. PHYLIP Phylogeny Inference Package version 3.5c. [Google Scholar]
- 36.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krause L, Diaz NN, Goesmann A, Kelley S, Nattkemper TW, Rohwer F, Edwards RA, Stoye J. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.