Abstract
RNase E of Escherichia coli is an essential endoribonuclease that is involved in many aspects of RNA metabolism. Point mutations in the S1 RNA-binding domain of RNase E (rne-1 and rne-3071) lead to temperature-sensitive growth along with defects in 5S rRNA processing, mRNA decay and tRNA maturation. However, it is not clear whether RNase E acts similarly on all kinds of RNA substrates. Here we report the isolation and characterization of three independent intragenic second-site suppressors of the rne-1 and rne-3071 alleles that demonstrate for the first time the dissociation of the in vivo activity of RNase E on mRNA versus tRNA and rRNA substrates. Specifically, tRNA maturation and 9S rRNA processing were restored to wild-type levels in each of the three suppressor mutants (rne-1/172, rne-1/186 and rne-1/187), while mRNA decay and autoregulation of RNase E protein levels remained as defective as in the rne-1 single mutant. Each single amino acid substitution (Gly→Ala at amino acid 172; Phe → Cys at amino acid 186 and Arg → Leu at amino acid 187) mapped within the 5′ sensor region of the RNase E protein. Molecular models of RNase E suggest how suppression may occur.
INTRODUCTION
Ribonuclease E (RNase E, encoded by the rne gene) is an essential endonuclease in Escherichia coli that plays a major role in many aspects of RNA metabolism (1,2). It is involved in the decay of a large number of mRNAs (3–5), the processing of 5S rRNA (3,6) and the maturation of many tRNA precursors (7,8). RNase E, a 1061 aa protein, is comprised of two distinct regions of defined function: a highly conserved amino-terminal portion of the polypeptide that contains the substrate-binding and catalytic domains (9); and a non-catalytic, largely unstructured, less conserved carboxy-terminal region that also acts as a scaffold for a multienzyme complex called the degradosome (10) (Figure 1). The degradosome includes the 3′→5′ exonuclease polynucleotide phosphorylase (PNPase), the ATP-dependent DEAD-box RNA helicase (RhlB) and the glycolytic enzyme enolase (11–14).
Figure 1.
Diagrammatic representation of RNase E protein. The upper portion of the figure indicates the two major regions of the protein: the N-terminal catalytic domain; and the C-terminal degradosome scaffolding region. The C-terminal region, composed of an arginine-rich binding domain (RBD) (10), a second arginine-rich noncatalytic region (AR2) that binds RNA independent of the longer RBD (16) and the binding regions of polynucleotide phosphorylase (PNPase), RNA helicase (RhlB) and enolase, is based on the work of Vanzo et al. (10). The C-terminal region is drawn after Chandran et al. (67). The lower portion of the figure is an expanded view of the N-terminal catalytic domain showing the five subdomains identified from the X-ray crystallographic analysis of Callaghan et al. (23). The locations of the various rne mutations are indicated. rne-1 (Gly to Ser); rne-3071 (Leu to Phe); rne-172 (Gly to Ala); rne-186 (Phe to Cys) and rne-187 (Arg to Leu). The figure is not drawn to scale.
The degradosome has been implicated in the degradation of untranslated mRNAs (15–18), the sensing of intracellular levels of poly(A) (19), and autoregulation of RNase E synthesis (3,20). However, general mRNA decay is not significantly impaired in the absence of degradosome assembly (3). Autoregulation of RNase E levels is achieved by cleaving specific sequences within the 5′ untranslated region (UTR) of the rne transcript (21,22), leading to its rapid decay. Defects in the activity of RNase E impair autoregulation by increasing the stability of the rne transcript and result in higher intracellular levels of the protein (3,21).
Because of its essential nature, RNase E has been the subject of extensive genetic, biochemical and structural studies aimed at understanding its precise biological role and mechanism of action. According to the three-dimensional crystal structure of the catalytic domain (23), the N-terminal 415-residue polypeptide folds into five distinct regions: (i) the previously described S1 RNA-binding domain (24), (ii) a 5′ sensor region, (iii) an RNase H domain, (iv) a DNase I domain and (v) a ‘Zn-link’ that forms a metal-sharing interface required for oligomerization (25). RNase E binding to the 5′-monophosphate group of an RNA substrate leads to stimulation of catalytic activity in vitro (26,27) and is facilitated by the interaction between the 5′ sensor region and the RNase H domain (23).
Since the catalytic site and the 5′ sensing region of RNase E are physically separated as shown by the crystal structure (23), the question arises as to how the status of the 5′ end of the substrate is communicated to the active site. Callaghan et al. (23) have proposed a model in which 5′ sensing is linked to catalysis through an allosteric change in the protein. According to this induced-fit hypothesis for catalysis, binding to the 5′ monophosphate of an RNA substrate rearranges the 5′ sensor pocket, which then interacts with the S1 domain causing it to clamp onto the RNA. This structural reorganization orients the phosphate backbone for an in-line attack on the scissile phosphate by a nucleophilic hydroxyl group. Based on the crystal structure and the proposed model for catalysis (23), S1 domain mutants could potentially be rescued by compensatory amino acid changes in the 5′ sensor region.
In fact, genetic studies have demonstrated an important role for the S1 domain in the biological activity of RNase E. Specifically, the two well-characterized rne alleles (rne-1 and rne-3071) map within the S1 domain of the protein (at amino acids 66 and 68, respectively, Figure 1) (6,28,29). The phenotypic properties of these mutants include inviability at elevated temperature along with defects in 5S rRNA maturation, tRNA processing, mRNA decay and processing of both tmRNA and the RNA subunit (M1) of RNase P (3,4,6–8,30,31).
E. coli also encodes a nonessential endoribonuclease, RNase G (product of rng), which is homologous to the N-terminal catalytic domain of RNase E (32,33). While both proteins share a 5′-end-dependent catalytic mechanism (34), RNase G has much more limited in vivo substrate specificity (35,36). It is involved in the maturation of the 5′ end of 16S rRNA and cleaves a very limited number of mRNA substrates (37–39). However, it appears to play a very minor role, if any, in tRNA maturation (8,35,40). In fact, it was during experiments designed to determine if RNase G could complement the loss of RNase E activity in an rne-1 mutant that we isolated temperature-resistant revertants at frequencies ranging from 0.3 × 10−8 to 7.0 × 10−8 (36).
While such isolates were previously presumed to be true revertants and had never been systematically studied, here we show that the majority of these temperature-resistant revertants actually arise from intragenic second-site suppressor mutations that map within the rne sequence for the 5′ sensor region of RNase E. These suppressors complement the conditional lethality of both the rne-1 and rne-3071 alleles. A unique feature of these intragenic second-site suppressor alleles is that they restore tRNA maturation and 9S rRNA processing to wild-type levels but do not rescue the defect in mRNA decay that is associated with the rne-1 allele. These mutants are thus distinct from the rne131 (20), rneΔ610 (3) and rneΔ645 (8) truncation alleles in which both tRNA maturation and mRNA decay are defective but 5S rRNA maturation is normal. Hence, these findings represent the first reported case in which the activity of RNase E on tRNA and mRNA substrates has been dissociated in vivo. In addition, computer modeling of the altered protein structures suggests a complex interaction between the S1 domain and the 5′ sensor region of the protein.
MATERIALS AND METHODS
Bacterial strains, plasmids and plasmid displacement
The E. coli strains used in the study (Table 1) were all derivatives of MG1693. pSBK29 (Cmr Aps) was constructed by cleaving pWSK29 (41) with ScaI and inserting a 0.8 kb Cmr cassette derived by cleavage of pKRP10 (42) with HincII. In many cases strains were constructed using the plasmid displacement technique described by Ow et al. (3).
Table 1.
Bacterial strains and plasmids used in the study
| Strain | Genotype | Source or Reference |
|---|---|---|
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 Δ(lacZYA-argF) rfbD1 spoT1 F− | (68) |
| MG1693 | thyA715 rph-1 | E. coli Genetic stock center |
| SK549 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK36 (rne-187 Kmr) | This study |
| SK2541 | rne-1 rng::cat thyA715 rph-1 Cmr | (36) |
| SK2566 | thyA715 rne-1 rph-1 recA56 srlD300::Tn10 Tcr/pUGK24 (rng+ Kmr) Derived from SK2585 | This study |
| SK2580 | thyA715 rne-1 rph-1 recA56 srlD300::Tn10 Tcr/pSBK29 (Cmr) Derived from SK2566 | This study |
| SK2585 | rne-1 thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK24 (rng+ Kmr) | This study |
| SK2586 | rneΔ1018 thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK19 (rne+ Kmr) | This study |
| SK2593 | rne-1 rng::cat thyA715 rph-1 Cmr/pUGK31 (rng+ Apr) | (36) |
| SK2594 | rne-1 thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK31 (rng+ Apr) | (36) |
| SK2741 | DH5α/pHK2 (rne-1/187 Kmr) | This study |
| SK2742 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK2 (rne-1/187 Kmr) | This study |
| SK2747 | rne-1 rng::cat thyA715 rph-1 Cmr/pUGK31 (rng+ Apr) Derived from SK2595 | This study |
| SK2748 | rne-1 thyA715 rph-1 /pUGK31 (rng+ Apr). Temperature resistant derivative of SK2594 | This study |
| SK3169 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK20 (rne-1 Kmr) | This study |
| SK3193 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK34 (rne-1/187 Kmr) | This study |
| SK3208 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK4 (rne-1/187 Spr)/pWSK129 (Kmr) | This study |
| SK3209 | DH5α/pHK5 (rne-1/186 Kmr) | This study |
| SK3210 | DH5α/pHK6 (rne-1/172 Kmr) | This study |
| SK3221 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK5 (rne-1/186 Kmr) | This study |
| SK3222 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK6 (rne-1/172 Kmr) | This study |
| SK3223 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK7 (rne-1/186 Spr)/pWSK129 (Kmr) | This study |
| SK3224 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK8 (rne-1/172 Spr)/pWSK129 (Kmr) | This study |
| SK3705 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK44 (rne-187 Spr)/pWSK129 (Kmr) | This study |
| SK3707 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK42 (rne-1/186 Kmr) | This study |
| SK3708 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK43 (rne-186 Kmr) | This study |
| SK3742 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK46 (rne-3071 Kmr) | This study |
| SK3743 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pUGK43 (rne-3071/187 Kmr) | This study |
| SK6610 | thyA715 rph-1 rne-1 recA56 srlD300::Tn10 Tcr | (69) |
| SK7669 | rne-3071 thyA715 | (55) |
| SK9714 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pSBK1 (rne+ Cmr) | (3) |
| SK9789 | rne-1 rng::cat thyA715 rph-1 Cmr/pUGK31 (rng+ Apr) Derived from SK2595 | This study |
| SK9792 | DH5α/pHK1 (rne-1/187 Kmr) | This study |
| SK9794 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pHK1 (rne-1/187 Kmr) | This study |
| SK9937 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pMOK13 (rne-1 Cmr) | (3) |
| SK10143 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pMOK44 (rne+ Spr)/pWSK129 (Kmr) | (8) |
| SK10144 | rneΔ1018::bla thyA715 rph-1 recA56 srlD300::Tn10 Tcr/pMOK45 (rne-1 Spr)/pWSK129 (Kmr) | (8) |
| Plasmids | ||
| pHK2 | 6–8 copy plasmid with rne-1/187 Kmr | This study |
| PHK4 | Single-copy plasmid (mini-F) with rne-1/187 Smr/Spr | This study |
| pHK5 | 6–8 copy plasmid with rne-1/186 Kmr | This study |
| pHK6 | 6–8 copy plasmid with rne-1/172 Kmr (Derived by site-directed mutagenesis) | This study |
| pHK7 | Single-copy plasmid (mini F) with rne-1/186 Smr/Spr | This study |
| pHK8 | Single copy plasmid (mini F) with rne-1/172 Smr/Spr (mini-F) | This study |
| pMOK40 | Single copy plasmid Smr/Spr | (8) |
| pSBK29 | Cmr Aps derivative of pWSK29 | S. Bundy, M. Ow and S.R. Kushner, unpublished data |
| pUGK19 | 6–8 copy plasmid with rne+ Kmr | This study |
| pUGK20 | 6–8 copy plasmid with rne-1 Kmr | This study |
| pUGK24 | 6–8 copy plasmid with rng+ Kmr | (36) |
| pUGK31 | 30–50 copy plasmid with rng+ Apr | (36) |
| pUGK34 | 6–8 copy plasmid with rne-1/187 Kmr (Derived by site-directed mutagenesis) | This study |
| pUGK36 | 6–8 copy plasmid with rne-187 Kmr (Derived by site-directed mutagenesis) | This study |
| pUGK42 | 6–8 copy plasmid with rne-1/186 Kmr (Derived by site-directed mutagenesis) | This study |
| pUGK43 | 6–8 copy plasmid with rne-186 Kmr (Derived by site-directed mutagenesis) | This study |
| pUGK44 | Single copy plasmid with rne-187 Smr/Spr | This study |
| pUGK46 | 6–8 copy plasmid with rne-3071 Kmr | This study |
| pUGK47 | 6–8 copy plasmid with rne-3071/187 Kmr (derived by site-directed mutagenesis) | This study |
| pWSK129 | pSC101 replicon Kmr | (41) |
Growth of bacteria
All strains were grown in Luria broth supplemented with thymine (50 μg/ml). When needed, antibiotics were added to the cultures at the following concentrations: kanamycin, 25 μg/ml; chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml and spectinomycin, 25 μg/ml. Growth curves were carried out either at 37°C or by shifting to 44°C cultures initially grown at 30°C, after they had reached approximately 40 Klett units (No. 42 green filter) above background. For growth curves, cultures were maintained in exponential phase by periodic dilutions with fresh pre-warmed medium.
Isolation and identification of intragenic suppressor alleles
Strains SK2585 [rne-1/pUGK24(rng+)], SK2593 [rne-1/pUGK31 (rng+)], SK2594 [rne-1 recA56/pUGK31(rng+)] and SK6610 (rne-1 recA56) were grown for 30–50 generations in Luria broth at 37°C. Subsequently, ∼108 cells from each culture were spread onto individual Luria agar plates and incubated at 44°C for 36 h. Colonies that grew at the nonpermissive temperature were purified and retested by replica plating for their ability to grow at 44°C. In the case of SK2585, SK2593 and SK2594, temperature-resistant isolates were transformed either with pSBK29 (Cmr) to displace pUGK24 or with pBR325 (Cmr) to displace pUGK31, the resident plasmids carrying the rng+ gene. The ability of the transformants to grow at 44°C in the absence of either pUGK24 or pUGK31 indicated that the potential suppressor mutations were on the chromosome. To rule out the possibility that the rne-1 allele had reverted to wild-type, we initially sequenced the region of rne gene spanning the site of rne-1 mutation from six independent temperature-resistant isolates. In all cases, the rne-1 mutation was retained.
Subsequently, the entire rne-coding sequence along with its promoter region was amplified from eleven independent isolates (four derived from either SK2585, SK2593 or SK2594 and seven obtained from SK6610) using PCR and cloned into pWSK129 (41) as described in the next section. Each new plasmid was then transformed into SK6610 (rne-1 recA56) and the ability of the resulting transformants to grow at 44°C was determined by replica plating. In every case, the transformants formed colonies at the non-permissive temperature suggesting that the suppressor mutations were on the cloned DNA segment. To precisely identify the mutations, we amplified the rne gene along with its promoter region from genomic DNA of all eleven independent isolates and subjected them to automated sequencing using an Applied Biosystem 3730xl DNA Analyzer.
Molecular cloning and site-directed mutagenesis
Genomic DNA from strains SK2580, SK2747, SK2748 and SK9789 was used as templates in PCR reactions to amplify the rne gene along with its promoters. Amplification was done with primers rne1014 (5′-CGGTGCTTGAATTCTCTTCACATGCGACTG) and rne3628 (5′-CATTCTAGATTAGCAAGGATGCCATTC) to introduce EcoRI and XbaI restriction sites (in bold), and Pfu DNA polymerase (Stratagene). Subsequently, the amplified DNA fragments were cleaved with EcoRI and XbaI and cloned into pWSK129 [Kmr, (41)] linearized with the same enzymes. The resulting plasmids (pHK1, pHK5, pHK6 and pHK2, respectively) were transformed into SK9714 [rneΔ1018/pSBK1(rne+ Cmr)] and, by use of plasmid incompatibility (3), Kmr Cms transformants were selected to generate SK9794 (rneΔ1018/pHK1), SK3221(rneΔ1018/pHK5), SK3222(rneΔ1018/pHK6) and SK2742(rneΔ1018/pHK2). Similarly, genomic DNA from MG1693 (rne+), SK6610 (rne-1) and SK7669 (rne-3071) was also amplified by PCR as above and cloned into pWSK129 (41) to produce pUGK19 (rne+), pUGK20 (rne-1) and pUGK46 (rne-3071), respectively. Subsequent transformation of SK9714 [rneΔ1018/pSBK1(rne+ Cmr)] with these plasmids and displacement of pSBK1 produced SK2586, SK3169 and SK3742, respectively.
The megaprimer PCR method for in vitro mutagenesis (43) was used to introduce the rne-172, rne-186 or rne-187 point mutations into plasmids pUGK19 (rne+), pUGK20 (rne-1) or pUGK46 (rne-3071). The PCR products were subsequently cleaved with EcoRI and SphI and cloned into pUGK19, pUGK20 or pUGK46, respectively, which had been linearized with the same enzymes to generate pHK6 (rne-1/172), pUGK36 (rne-187), pUGK43 (rne-186), pUGK34 (rne-1/187), pUGK 42 (rne-1/186) and pUGK47 (rne-3071/187). The resulting plasmids were then transformed into SK6610 (rne-1 recA56). Subsequently, these plasmids were also transformed into SK9714 [rneΔ1018/pSBK1 (rne+ Cmr)] and the resident pSBK1 (rne+ Cmr) plasmid was displaced to generate the strains used in this study (Table 1). The nucleotide sequences of the mutagenic primers used to generate point mutations are shown in Table 2.
Table 2.
Mutagenic primers used in this study
| Primer | Sequence | Amino acid change |
|---|---|---|
| phk1-mut | 5′ GTGTTCAGAAGGAAGCTTAAATCC 3′ | Arg187Leu |
| phk5-mut | 5′ GTTTCAGACGGCAGCTTAAATCCC 3′ | Phe186Cys |
| Phk6-mut | 5′ ATTTGCCGACGGCAGCGGTGCGCAC 3′ | Gly172Ala |
The mismatched nt in each mutagenic primer is shown in bold.
To construct single-copy plasmids carrying various rne alleles used in this study, the multiple-copy plasmids, pHK2, pHK5, pHK6 and pUGK36 were cleaved with EcoRI and NotI and each rne-containing fragment was cloned into pMOK40 that had also been digested with these two enzymes (8). The resulting plasmids, pHK4, pHK7, pHK8 and pUGK44, respectively, were transformed into SK9714 [rneΔ1018/pSBK1 (rne+ Cmr)]. The resident pSBK1 plasmid was subsequently displaced with pWSK129 (Kmr), based on plasmid incompatibility, to generate the strains used in this study.
RNA preparation and northern analysis
Total RNA was extracted and northern blot analysis carried out according to the procedures of O’Hara et al. (44) and Burnett (45). To visualize tRNA and 9S rRNA processing, 5.0 μg of total RNA from each sample (except for 7.5–10 μg for the rne-1 strains in the tRNA northerns) were electrophoresed in 8% polyacrylamide/7 M urea gels. For mRNA half-life determinations, either 6% polyacrylamide/7 M urea or 1.2% agarose/glyoxal gels were used. For assays of tRNA maturation, total RNA was extracted from cultures that had been shifted from 37°C (at a cell density of 1.0 × 108 cells/ml) to 44°C for 2 h. Probes for the 5S rRNA (46) and the tRNAs (8) were oligonucleotides complementary to the mature species and were 5′-end-labeled with γ-32P-ATP and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA). The DNA probes used for the mRNA half-life experiments were 32P-labeled using random primers as specified by the supplier (Ambion, Austin, TX, USA). Northern blots were scanned with a PhosphorImager scanner (Storm 840, GE Healthcare, Piscataway, NJ, USA) and quantified using ImageQuant 5.2 software. Half-lives were determined by linear regression analysis.
Western blot analysis
Western blot analysis was performed essentially as described in Ow et al. (3). Cell lysates were prepared from cultures growing exponentially at 37°C or shifted at mid-log from 37°C to 44°C for 1 h. Total protein (10 μg/lane) was electrophoresed in 6% SDS–PAGE gels and electrotransferred onto Immobilon-P membranes (Millipore). The RNase E protein was detected using ECL Plus western analysis system (GE Healthcare) with a rabbit anti-RNase E MAP antibody (1:2000 dilution) raised against the first 20 amino acids of the N-terminus (3).
Computer modeling of the suppressor mutations
Three PDB entries (2bx2, 2c4r and 2c0b) derived from the work of Callaghan et al. (23) with coordinate information for the catalytic domain of RNase E were selected as homologous templates for molecular modeling using Geno3D (http://geno3d-pbil-ibcp.fr), an online homology modeling tool (47). For the altered RNase E proteins, the inter/intra molecular (dihedral angle and distance) restraints ratios were set to 0.9. The margins in distance restraints and angle restraints were set at 0.5 Å and 1.0°, respectively. The maximum number of distance restraints was 20 000. Three models were generated and the model with the lowest free energy was used in this work. In this model, 0.4% of the residues occupied disallowed regions of the Ramachandran plot. The root mean square deviation (RMSD) between this model and the templates ranged from 1.46 to 1.57 Å. The lowest free-energy model for the altered RNase E proteins and the 3D structure of wild-type RNase E were analyzed and viewed with PyMol (http://pymol.sourceforge.net/).
RESULTS
Isolation of second-site suppressors of the rne-1 allele
In previous experiments, we attempted to determine if overexpression of RNase G [a homologue of RNase E in E. coli (32)] could complement the loss of RNase E activity (36). Since SK6610 (rne-1 recA56) and SK5665 (rne-1) are both temperature sensitive for growth, we transformed them with either pUGK24 (rng+, 6–8 copies/cell) or pUGK31 (rng+, 30–50 copies/cell) and tested for the ability of transformants to grow at 44°C. When neither plasmid complemented the conditional lethality associated with the rne-1 allele, we tried a different approach. We transformed a strain that carries the rneΔ1018 deletion allele (SK9714) and a pSC101 derivative containing rne+ (3) with pUGK24 (pSC101) and tried to displace the resident rne+ plasmid, based on the incompatibility of the two plasmids.
Although neither approach led to complementation, we hypothesized that it might be possible to obtain spontaneously arising suppressors of the rne-1 allele that would allow cells to grow at 44°C. When we spread ∼108 cells/plate from cultures of SK2585 [rne-1/pUGK24 (rng+)], SK2593 [rne-1/rng:cat pUGK31 (rng+)] and SK2594 [rne-1/pUGK31 (rng+)] at 44°C, we obtained a small number of colonies that grew after 24–48 h (frequency of 0.3–7.0 × 10−8) (36). When purified and retested, these isolates grew at 44°C (‘Materials and methods’ section). We subsequently chose four independently identified isolates for further analysis.
Since we hypothesized that at least some of the temperature resistant revertants arose from intragenic suppression of the rne-1 allele, we amplified the entire rne gene from four isolates using PCR and cloned them into pWSK129, a low-copy number vector carrying Kmr (41) (See ‘Materials and methods’ section) to generate plasmids pHK1, pHK2, pHK5 and pHK6. After determining via replica plating that each of these plasmids complemented the rne-1 allele in SK6610 (rne-1 recA56) at 44°C, we sequenced the entire rne regulatory and coding region from each plasmid. To our surprise, plasmids pHK1 and pHK2 each contained a G to T single nucleotide change resulting in an Arg to Leu substitution at amino acid 187 (Figure 1). Plasmid pHK5 carried a T to G nucleotide change causing a Phe to Cys substitution at amino acid 186, while pHK6 contained a G to C mutation that resulted in a Gly to Ala substitution at amino acid 172 (Figure 1). All four clones also retained the original rne-1 mutation.
In order to establish that the isolation of these apparent intragenic second-site suppressor alleles was not dependent on the presence of a plasmid carrying the rng+ gene, we repeated the experiment using SK6610 (rne-1 recA56). The entire rne+ gene and promoter region was cloned and sequenced from seven independently isolated temperature resistant revertants as described above. From these seven independent isolates we identified one true revertant of the rne-1 allele as well as the same three suppressor alleles described above (data not shown).
For detailed in vivo analysis of the double mutants, plasmids pHK2, pHK5 and pHK6 were transformed into SK9714 (rneΔ1018/rne+) to determine if they could support cell viability in the complete absence of RNase E activity. All three plasmids displaced the resident plasmid pSBK1 carrying the wild-type rne+ gene, generating strains SK2742 (rneΔ1018/rne-1/187), SK3221 (rneΔ1018/rne-1/186) and SK3222 (rneΔ1018/rne-1/172). Subsequently, the inserts containing the entire rne gene along with its promoter sequences were transferred from pHK2, pHK5 and pHK6 (6–8 copies/cell) and cloned into a single copy vector (pMOK40) (8) to generate pHK4 (rne-1/187), pHK7 (rne-1/186) and pHK8 (rne-1/172), respectively. These plasmids were also able to support cell viability in the rneΔ1018 deletion strain at both 37 and 44°C.
To verify that the rne-172, rne-186 and rne-187 alleles were solely responsible for suppression of the growth phenotype associated with the rne-1 mutation, we introduced the same point mutations individually into pUGK20 (rne-1) using site-directed mutagenesis. The resulting plasmids were then transformed into SK6610 (rne-1 recA56). The plasmids generated by site-directed mutagenesis containing the rne-1/187, rne-1/186 and rne-1/172 double mutations all supported cell viability at 44°C (data not shown).
Growth properties of strains containing the rne-1/172, rne-1/186 and rne-1/187 double mutations
In order to establish how well the rne-172, rne-186 and rne-187 alleles suppressed the growth deficiency of an rne-1 mutant at 44°C when present in 6–8 copies/cell, we grew cultures of SK9714 (rne+), SK3222 (rne-1/172), SK3221 (rne-1/186), SK2742 (rne-1/187) and SK9937 (rne-1) at 30°C in Luria broth and then shifted the exponentially growing cultures to 44°C. While the rne-1 mutant stopped growing within 2 h after shift to 44°C as previously observed (3), the three strains containing the different double mutants continued to grow, albeit with slightly longer generation times than the wild-type control (Figure 2A, Table 3). SK3221 (rne-1/186) consistently grew slower than either SK3222 (rne-1/172) or SK2742 (rne-1/187) (Table 3). When we performed a similar experiment with the double mutations present on single copy plasmids, the generation time of the rne-1/186 double mutant (SK3223) was 68 min at 44°C compared with 32 min for the rne+ control (SK10143, Figure 2B, Table 3). In contrast, the rne-1/172 (SK3224) strain was intermediate in growth, while the rne-1/187 (SK3208) isolate grew at a rate almost identical to the wild-type control strain (SK10143) (Table 3).
Figure 2.
Growth curves of rneΔ1018 strains carrying various rne alleles. Cultures were grown in Luria broth as described in ‘Materials and methods’ section. (A) 6–8 copies of various rne alleles. Filled triangle, rne+ (SK9714); Open circle, rne-1/172 (SK3222); filled square, rne-1/187 (SK2742); filled diamond, rne-1/186 (SK3221); open square, rne-1 (SK9937); open triangle, rne-3071 (SK3742); open diamond, rne-3071/187 (SK3743). (B) single copies of the various rne alleles. Filled triangle, rne+ (SK10143); open circle, rne-1/172 (SK3224); filled square, rne-1/187 (SK3208); filled diamond, rne-1/186 (SK3223); open square, rne-1 (SK10144).
Table 3.
Generation times (min) of strains carrying various rne alleles
| Allele | 6–8 copies | Single copy |
|---|---|---|
| rne+ | 31 | 32 |
| rne-1 | TSa | TSa |
| rne-1/172 | 36 | 47 |
| rne-1/186 | 45 | 68 |
| rne-1/187 | 35 | 33 |
Cultures were grown as described in Materials and methods section and shifted to 44°C for the determination of their generation times.
aTemperature sensitive at 44°C.
We used site-directed mutagenesis (see ‘Materials and methods’ section) to construct plasmids carrying either rne-186 or rne-187 single mutants. Somewhat surprisingly, the growth of strains carrying these alleles in multiple copies was indistinguishable from that of SK9714 (rneΔ1018/rne+) at 44°C (data not shown). In addition, the growth rates at 37°C of strains carrying single copies of the individual suppressor alleles were also comparable to those of the corresponding strains carrying rne+ (data not shown), suggesting that the rne-186 and rne-187 alleles did not significantly affect RNase E activity. We did not test the rne-172 allele.
The suppressor alleles do not restore autoregulation of RNase E at 44°C
Since RNase E autoregulates its own synthesis via a feedback loop that involves controlling the rate of degradation of its mRNA, alterations in RNase E activity caused by mutations such as rne-1 lead to increased protein levels at both 37 and 44°C (3,21). Accordingly, we tested if autoregulation of RNase E levels at 44°C was restored by the intragenic second-site suppressor alleles. Quantitative western blot analysis of RNase E protein levels at both 44 and 37°C was performed in strains carrying the rne+, rne-1, rne-1/187, rne-1/186 and rne-1/172 alleles in 6–8 copies (data not shown) or in single copy/cell (Figure 3). At 44°C there was a 6.9-fold increase in the amount of the full-length protein in the rne-1 strain compared to rne+ strain (Figure 3, lanes 1 and 2). Strikingly, the RNase E levels in all three double mutants were similar to those observed in the rne-1 single mutant (Figure 3, lanes 2–5). In contrast, the protein level in the rne-187 and rne-186 single mutants were experimentally equivalent to what was observed in the wild-type strain, suggesting that autoregulation was unaffected in either single mutant (data not shown). When we carried out an identical experiment at 37°C, the levels of the RNase E protein were equivalent for all the strains except rne-1, suggesting that autoregulation was restored by the intragenic second-site suppressors at 37°C (data not shown). In the case of rne-1 strain there was a small, but reproducible increase in the amount of both full-length and breakdown products (data not shown).
Figure 3.
Western blot analysis of RNase E levels in various strains. Total protein was isolated from each of the strains after they had been shifted to 44°C for 60 min as described in ‘Materials and methods’ section. RNase E was detected with a MAP antibody directed against the first 20 amino acids of protein (3) and was detected using the ECL Plus system (GE Healthcare). Quantitation was obtained using a Storm 840 PhosphorImager (GE Healthcare). The relative amount of RNase E in each strain as compared to the rne+ control represents the average ± SEM (n ≥ 3). All alleles were in single copy/cell in a strain carrying the rneΔ1018 deletion allele in the chromosome. Lane 1, SK10143; Lane 2, SK10144; Lane 3, SK3208; Lane 4, SK3224 and Lane 5, SK3223.
mRNA decay remains defective in the double mutants
RNase E plays a central role in the endonucleolytic initiation of decay of a wide variety of cellular mRNAs (3,5), a role that has historically been viewed as its quintessential in vivo function (48). The failure of the suppressor mutations to restore normal autoregulation of RNase E protein levels at 44°C (Figure 3) suggested that the altered enzymes were probably still defective in their ability to initiate the endonucleolytic decay of specific mRNAs. To test this hypothesis directly, we measured the half-lives of three known RNase E-dependent transcripts, cpsE, rpsT and rpsO (49–51) at 44°C in strains carrying single copies of the rne+, rne-1, rne-1/187, rne-1/186 and rne-1/172 alleles. In agreement with previously reported results (3,36), the half-life of the major rpsOP1-t1 transcript (52) increased significantly from 2.8 min in the rne+ control to 10.9 min in the rne-1 single mutant (Table 4). Surprisingly, the half-life of this transcript increased further in the rne-1/187 double mutant to 16.3 min. In contrast, the half-life of the rpsOP1-RIII transcript was almost the same in the two mutant strains and was higher than in the rne+ control. The half-lives of the cspE and rpsT mRNAs in the rne-1/187 double mutant were also similar to the rne-1 single mutant (Table 4) and were 2–4-fold longer than in the rne+ control. In the rne-1/186 and rne-1/172 double mutants there was a marginal reduction in the half-lives of the rpsT and cspE transcripts relative to the rne-1 single mutant (Table 4), but the half-lives were still 2- to 3-fold longer than in the rne+ control. Although the mRNA half-life data directly correlated with the failure of the suppressor alleles to restore normal autoregulation of RNase E protein levels in all three double mutant strains (Figure 3), it did not correlate with the generation times reported in Table 3.
Table 4.
mRNA decay in various rne mutants
| Half-live (min)a |
|||||
|---|---|---|---|---|---|
| Transcript/Alleleb | rne+ | rne-1 | rne-1/172 | rne-1/186 | rne-1/187 |
| cspE | 2.5 ± 0.1 | 9.2 ± 0.2 | 6.7 ± 0.8 | 7.2 ± 0.6 | 10.4 ± 0.4 |
| rpsTP1 | 2.1 ± 0.3 | 6.1 ± 0.2 | 4.3 ± 0.5 | 4.6 ± 0.3 | 5.9 ± 0.1 |
| rpsTP2 | 2.0 ± 0.1 | 4.6 ± 0.3 | 4.1 ± 0.3 | 4.4 ± 0.7 | 5.1 ± 0.2 |
| rpsOP1-RIII | 1.3 ± 0.1 | 2.0 ± 0.4 | ND | ND | 1.9 ± 0.2 |
| rpsOP1-t1 | 2.8 ± 0.3 | 10.9 ± 0.8 | ND | ND | 16.3 ± 0.6 |
aHalf-lives were determined as described in the Materials and methods section. Each value is the average of at least two independent determinations.
bAll alleles are present in single copy.
5S rRNA processing is restored to wild-type levels in the double mutants
RNase E was initially discovered based on its role in the maturation of 5S rRNA from a larger 9S rRNA precursor (6), such that unprocessed 5S rRNA precursors accumulate in an rne-1 mutant at the nonpermissive temperature (3,6). To determine if 5S rRNA precursors were processed with increased efficiency in the presence of the intragenic second-site suppressor mutations, we prepared total RNA from strains carrying single copies of the rne+, rne-1, rne-1/172, rne-1/186 and rne-1/187 alleles after growth at 44°C for 2 h and used northern analysis to measure the processed fraction (PF) for 5S rRNA. As shown in Figure 4, the Processed Fraction (PF) values of mature 5S rRNA in all three double mutants were essentially identical to that observed in the rne+ control.
Figure 4.
Analysis of 9S rRNA processing in various strains. Total steady-state RNA was isolated from the various strains after the cultures had been shifted to 44°C for 120 min as described in ‘Materials and methods’ section. Five micrograms of total RNA was loaded in each lane of an 8% polyacrylamide/7 M urea gel. The northern blot was subsequently probed with an oligonucleotide specific for the mature 5S rRNA (55). PF (processed fraction) denotes the fraction of the mature 5S rRNA relative to the total amount of both processed and unprocessed species in each specific genetic background. All alleles were in single copy/cell in a strain carrying the rneΔ1018 deletion. The data shown represents the average ± SEM (n ≥ 3). Lane 1, SK10143; Lane 2, SK10144; Lane 3, SK3208; Lane 4, SK3224 and Lane 5, SK3223.
tRNA maturation is restored in the double mutants
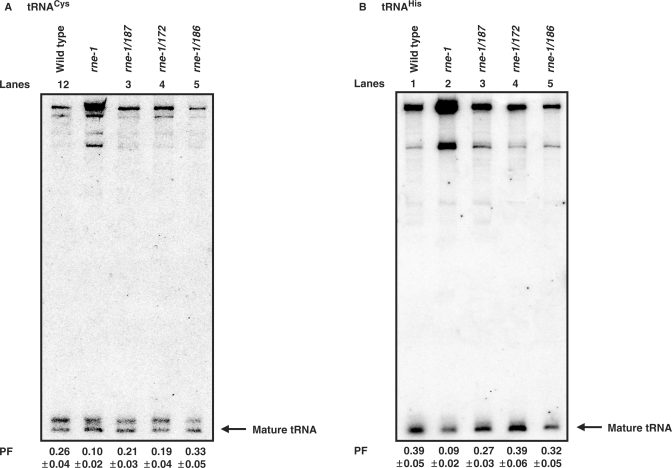
RNase E is required for catalyzing the conversion of some polycistronic tRNA transcripts into pre-tRNAs that undergo further processing by RNase P and various 3′ → 5′ exonucleases (7,8). In fact, it has been hypothesized that RNase E is essential for the cell because of its role in tRNA maturation (8). To determine if the altered RNase E proteins were able to process precursor tRNAs, we examined the maturation of tRNAPro, tRNAHis and tRNACys in strains carrying single copies of the various rne alleles. We chose these tRNAs because their maturation has been shown to be highly dependent on RNase E, and there is significant accumulation of their precursor species in an rne-1 strain (8). Total RNA, from rne+, rne-1, rne-1/172, rne-1/186 and rne-1/187 strains was obtained after shifting exponentially growing cultures to 44°C for 2 h. As expected, northern blot analysis demonstrated that the maturation of each tRNA species was significantly impaired in the rne-1 mutant compared to the wild-type control (Figure 5A and B and data not shown). For example, the PF values of tRNACys and tRNAHis decreased 2.6- and 4.3-fold, respectively, in the rne-1 strain (Figure 5A and B). In contrast, all three intragenic second-site suppressor alleles led to an almost complete restoration of rne+ tRNA maturation levels as measured by the PF value of each transcript (Figure 5A and B and data not shown).
Figure 5.
Analysis of tRNA maturation in various strains. Total steady-state RNA was isolated from various strains after the cultures had been shifted to 44°C for 120 min as described in ‘Materials and methods’ section. Typically 5 μg of total RNA was loaded in each lane (except for the rne-1 mutants where 7.5 μg of RNA were loaded for better visualization of mature species) of an 8% polyacrylamide/7 M urea gel. The northern blot was subsequently sequentially probed with oligonucleotides (8) specific for either (A) tRNACys, (B) tRNAHis or tRNAPro (data not shown). PF (processed fraction) denotes the fraction of the mature tRNA relative to the total amount of both processed and unprocessed species in each genetic background. All alleles were in single copy/cell in an rneΔ1018 deletion strain. The PF values are the average ± SEM (n ≥ 3). Leftward arrow indicates the position of the mature tRNA species. Lane 1, SK10143; Lane 2, SK10144; Lane 3, SK3208; Lane 4, SK3224 and Lane 5, SK3223.
The rne-187 mutation also suppresses the conditional lethality associated with the rne-3071 allele
To determine if the suppression of the growth deficiency at 44°C associated with the rne-1 mutation was an allele-specific effect rather than a more general phenomenon, we examined the ability of rne-187 to suppress the conditional lethality of the rne-3071 allele, a different temperature-sensitive mutation that was originally identified by Apirion and coworkers (53,54). This particular allele arose from a Leu to Phe amino acid substitution at position 68 (29), which also lies within the S1 RNA-binding domain of RNase E (23,24), and is two amino acids downstream of the change associated with the rne-1 allele (Figure 1). To carry out this experiment, plasmids pUGK46 (rne-3071) and pUGK47 (rne-3071/187) were constructed as described in the ‘Materials and methods’ section. Both plasmids were subsequently introduced into the rneΔ1018 genetic background (SK9714). As expected, when a culture of SK3742 (rneΔ1018/rne-3071) was shifted to 44°C, growth ceased within 180 min (Figure 2A). In contrast, SK3743 (rneΔ1018/rne-3071/187) continued to grow with a generation time comparable to a wild-type control, demonstrating that the rne-187 allele suppressed both the rne-1 and rne-3071 mutations (Figure 2A).
Predicted molecular interactions among the rne-1, rne-172, rne-186 and rne-187 alleles
The existence of a three-dimensional structural model for the catalytic region of RNase E based on X-ray crystallography (23) allowed us to generate models of the variant RNase E proteins using Geno3D (http://geno3d-pbil.ibcp.fr), an online homology modeling tool (47). By using distance geometry, simulated annealing and energy minimization, this algorithm builds protein 3D models to satisfy spatial restraints derived from the empirically determined structures of homologous proteins. Three experimentally determined structures of the E. coli RNase E catalytic domain (23) were selected as homologous templates for molecular modeling (see ‘Materials and methods’ section). The overall predicted structures of RNase E from various double mutants were very similar to that of the native protein (data not shown). Structural differences among wild-type and the three altered proteins appeared to be localized to regions of the respective amino acid substitutions (Figure 6A). All three second-site suppressor alleles identified here (rne-172, rne-186 and rne-187) mapped within the 5′ sensor region of the protein. Interestingly, the affected residues were not located close enough to amino acid 66 (Ser) in the S1 domain to have any direct interactions (Figure 6A). Furthermore, none of the altered residues appeared to be directly involved in physical interaction with the 5′ termini of RNA substrates (Figure 6B).
Figure 6.
Structural model of the RNase E protein bound to an RNA molecule. (A) Locations of the rne-1 (Gly66), rne-172 (Gly172), rne-186 (Phe186) and rne-187 (Arg187) alleles. The model is based on the three-dimensional structure generated by Callaghan et al. (23) and the modeling program of Combet et al. (47). The four major subdomains (S1 RNA-binding domain, magenta, 5′ sensor region, yellow, RNase H domain, cyan and DNase I domain, brown) are indicated. (B) Predicted changes in the interaction of RNase E with an RNA substrate in rne-1/172 double mutant. The wild-type protein is shown in green, the rne-1/172 mutant in magenta.
DISCUSSION
We have described here for the first time intragenic second-site suppressor mutations within the RNase E gene that lead to complementation of the conditional lethality associated with both the rne-1 and rne-3071 alleles (4,6,28,29,55). Strikingly, however, only a subset of the phenotypic defects associated with the loss of RNase E activity were complemented. Specifically, the maturation of tRNACys, tRNAHis and tRNAPro and the processing of the 9S rRNA precursor into p5S rRNA returned to wild-type levels in all three mutants (rne-1/rne-172, rne-1/186 and rne-1/187, Figures 4 and 5), while mRNA decay remained as defective as observed in the rne-1 single mutant (Table 4). In fact, in some cases the half-lives of the three RNase E-dependent mRNAs tested (cspE, rpsO and rpsT) were longer than those observed for the rne-1 single mutant (rne-1/187, Table 4). These results, taken together with the failure to restore normal autoregulation of intracellular RNase E levels (Figure 3), demonstrate that mRNA decay remains very defective even when tRNA maturation and 9S rRNA processing have been restored to wild-type levels.
These observations are in contrast to results derived from a series of rne truncation alleles (rne131, rneΔ610 and rneΔ645) that encode proteins that lack 477, 609 and 644 amino acids, respectively, from their carboxy termini (3,8,20). In strains carrying these alleles, 9S rRNA processing is normal but both mRNA decay and tRNA maturation are very defective (3,8,20). Thus, the rne-1/rne-172, rne-1/186 and rne-1/187 mutants represent the first example of strains in which the activity of RNase E on mRNA and tRNA substrates can be distinguished in vivo. Even though we tested only the growth properties of the rne-3071/187 (Figure 2A), we hypothesize that the rne-187 allele works similarly in both cases by restoring in part the ability of the S1-binding domain to interact normally with tRNA and rRNA substrates. This conjecture is based on the fact that both the rne-1 and rne-3071 alleles fall within the S1-binding domain and are only two amino acids apart.
Although the mRNAs examined here were only a small sample of the E. coli transcriptome, they represent well-characterized in vivo substrates of RNase E. Furthermore, the fact that the rne-1/187 strain had a generation time at 44°C that was almost identical to the wild-type control (Table 3), but was still very defective in mRNA decay, suggests that defects in the initiation of mRNA decay by RNase E do not have a major impact on cell viability. Further support for this conclusion comes from the recent identification in E. coli of RNA 5′ pyrophosphohydrolase (RppH), encoded by the rppH gene (56). RNase E activity is known to be significantly stimulated by the presence of a 5′ phosphomonoester on the substrate, and conversely is strongly inhibited by a 5′ triphosphate terminus. In vivo, RppH catalyzes the conversion of 5′ triphosphorylated transcripts into 5′ phosphorylated species, which are then efficiently degraded via an RNase E-mediated decay pathway. As expected, in an rppH deletion mutant, where removal of the 5′ triphosphate from primary transcripts is blocked, the decay of hundreds of mRNAs was defective yet the strain grew as well as the rne+ control (56).
In contrast, the growth rate of E. coli has been shown to be highly dependent on the availability of major tRNAs cognate to the preferred codons of the genes encoding the highly expressed proteins in rapidly growing bacteria (57,58). In fact, it has been argued that initiation of the processing of polycistronic tRNA precursors is the critical function of RNase E (7,8). Furthermore, recent experiments examining tRNA maturation in an rne deletion strain have shown that for at least some tRNAs, such as tRNAHis, tRNAPro and tRNACys, little if any of these mature species would be produced in the absence of RNase E (59). Clearly the cell would not survive under these circumstances. Thus, the observation that the rne-172, rne-186 and rne-187 alleles suppressed the growth defect at 44°C associated with the rne-1 allele and concomitantly restored normal tRNA maturation (Figures 2 and 5, data not shown), provides the best direct support to date for the hypothesis that tRNA maturation is an essential biological activity of RNase E, even though one report has argued that this is not the case (35).
It is important to note that there are other possible essential function(s) for RNase E that cannot be unequivocally ruled out at this time. For example, the inability of RNase E to process or degrade one of the many small regulatory RNAs in the cell (60,61) could either inhibit an essential pathway or result in toxicity due to a protein whose intracellular level is normally under tight regulation. Another potential explanation relates to a recently implied structural role for RNase E. The full-length RNase E was identified immunologically by an antibody against the yeast actin-like protein Hmp-1 (62). Furthermore, it has been suggested that full-length RNase E forms helical-like structures as part of the cell's cytoskeleton (63). Thus it is conceivable that RNase E is necessary for some aspect of cytoskeleton formation that is essential for cell viability, although the existence of extensive RNase E truncation alleles (3,8,20) that are viable tends to discount this hypothesis. Finally, it should be emphasized that the essential role of RNase E in tRNA maturation is not mutually exclusive with some other additional critical function(s) for the enzyme. Further experiments will be required to unequivocally determine the primary requirement for RNase E in cell viability, but clearly these mutants provide an important new tool for examining the substrate specificity of RNase E.
Is it possible to explain the second-site suppressors by employing molecular modeling based on the crystal structure (23) of the catalytic region of RNase E? RNase E prefers RNA substrates containing 5′ phosphomonoesters over those with a 5′ triphosphate both in vivo and in vitro (27,34,56,64,65). The terminal monophosphate of the substrate forms hydrogen bonds with the side chain and peptide amide of Thr 170 and the guanidino group of Arg 169, the latter of which is then consolidated by a hydrogen bond in the neighboring strand to the peptide carboxyl of Gly 124 (23). Concomitantly, the side chain of Val 128 engages in hydrophobic interaction with the terminal base of the RNA substrate. These interactions organize the 5′ sensing pocket allowing it to interact productively with the S1 domain.
The S1 domain in turn clamps down on the RNA downstream from the region of initial binding to the 5′ terminus for the subsequent cleavage at the scissile phosphate bond by the active hydroxyl group (23). Within the S1 domain of RNase E, Phe 67 makes a stacking contact with the aromatic face of the first base after the cleavage site. The computer model suggests that with a Gly 66 to Ser substitution in the S1 domain (rne-1 allele), the Ser moiety pushes Phe 67 closer to the RNA substrate (data not shown). This alteration changes the position of the RNA molecule within the binding site at 44°C resulting in altered interactions of the substrate with other amino acids leading to a loss of catalytic activity at the elevated temperature. Alternatively, since the Φ,Ψ backbone angles of glycine are not permitted for serine, a Gly → Ser substitution can potentially result in destabilization of a protein fold if the glycine occurs at the region of a turn. Whether this substitution indeed destabilizes the S1 fold of RNase E, rather than merely changing its interaction with RNA, will require further experimental verification. However, we feel that any gross destabilization of the S1 domain, due to the rne-1 mutations, seems less plausible since this tends to make suppression with single amino acid substitutions much less likely.
Our computer model suggests that with Gly 172 replaced by Ala (rne-1/172), the spatial relationship of Thr 170 and Arg 169 changes such that the proximity of the 5′ phosphate to the Arg side chain is closer to what is found in the wild-type protein (Figure 6B). This modification probably compensates for the effect of the original mutation in the S1 RNA-binding domain. With the Arg 187 to Leu substitution (rne-1/187), Leu 187 can engage in hydrophobic contacts with Leu 122 and 129, potentially resulting in a more stable structure than is possible with Arg in this position (data not shown). This may indirectly affect the interaction of Val 128 with the RNA substrate and the function of Gly 124. Since Phe 186 is located on the outer surface of a helix that is on the outside of the protein (Figure 6A), the Phe 186 to Cys substitution may change the surface entropy and the interaction of the protein with the solution (data not shown).
Interestingly, all of the altered residues are located in the 5′ sensing region of the enzyme (Figure 6A) and none of them directly interact with the 5′ terminus of an RNA substrate. Nevertheless, since the 5′ sensor domain engages in interactions with the RNA substrate, these substitutions may cause residues in direct contact with substrates to subtly alter their positions relative to the RNA. Such structural modulation in turn can compensate for the altered interaction with the RNA substrates that occur at 44°C in the rne-1 and rne-3071 mutant proteins. One puzzling feature of these mutants is that activity of RNase E on tRNA and rRNA precursors is restored to almost wild-type levels, while mRNA decay remains as defective as observed in the rne-1 single mutant. We hypothesize that the more highly structured nature of the tRNA and rRNA substrates may explain the difference in activity, but the actual mechanism of action of these second-site suppressors will require structural analysis of the mutant proteins bound to either a tRNA or mRNA substrate.
Finally, any discussion of RNase E activity needs to take into account the RraA protein, an inhibitor of RNase E activity (66). Protein–protein interactions between RraA and RNase E affect the half-lives of a large number of E. coli mRNAs (66). Conceivably, the defective mRNA decay observed here with the various double mutants could arise from a change in the interaction of RraA and RNase E. However, this does not appear likely since the interaction of these two proteins has been shown to only require the C-terminal portion of the RNase E protein (66) and all three intragenic suppressor alleles map in the 5′ sensor region and do not appear to affect the global structure of RNase E.
ACKNOWLEDGEMENTS
The authors thank Nan Chen, Vimary Bermúdez and Stephanie Bundy for valuable technical assistance and B. Mohanty for reading the manuscript. This work was supported in part by a grant from the National Institute of General Medical Sciences (GM57220) to S.R.K. Funding to pay the Open Access publication charges for this article was provided by 6M57220.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carpousis AJ. The Escherichia coli degradosome: Structure, function and relationship to other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- 2.Kushner SR. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ow MC, Liu Q, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- 4.Arraiano CM, Yancey SD, Kushner SR. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein JA, Lin P.-H, Cohen SN, Lin-Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl Acad. Sci. USA. 2004;101:2748–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apirion D, Lasser AB. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J. Biol. Chem. 1978;253:1738–1742. [PubMed] [Google Scholar]
- 7.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in Escherichia coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaberdin VR, Miczak A, Jakobsen JS, Lin-Chao S, McDowall KJ, von Gabain A. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl Acad. Sci. USA. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 12.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 13.Py B, Causton H, Mudd EA, Higgins CF. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 14.Miczak A, Kaberdin VR, Wei C.-L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 16.Leroy A, Vanzo NF, Sousa S, Dreyfus M, Carpousis AJ. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 2002;45:1231–1243. doi: 10.1046/j.1365-2958.2002.03104.x. [DOI] [PubMed] [Google Scholar]
- 17.Coburn GA, Mackie GA. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 1998;279:1061–1074. doi: 10.1006/jmbi.1998.1842. [DOI] [PubMed] [Google Scholar]
- 18.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3' exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohanty BK, Kushner SR. Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E. Mol. Microbiol. 2002;45:1315–1324. doi: 10.1046/j.1365-2958.2002.03097.x. [DOI] [PubMed] [Google Scholar]
- 20.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 21.Jain C, Belasco JG. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- 22.Diwa A, Belasco JG. Critical features of a conserved RNA stem-loop important for feedback regulation of RNase E synthesis. J. Biol. Chem. 2002;277:20415–20422. doi: 10.1074/jbc.M202313200. [DOI] [PubMed] [Google Scholar]
- 23.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 24.Bycroft M, Hubbard TJP, Proctor M, Freund SMV, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 25.Callaghan AJ, Redko YU, Murphy LM, Grossman JG, Yates D, Garman E, Ilag LL, Robinson CV, Symmons MF, McDowall KJ, et al. “Zn-Link”: a metal sharing interface that organizes the quaternary structure and catalytic site of endoribonuclease, RNase E. Biochem. 2005;44:4667–4775. doi: 10.1021/bi0478244. [DOI] [PubMed] [Google Scholar]
- 26.Callaghan AJ, Grossman JG, Redko YU, Ilag LL, Moncrieffe MC, Symmons MF, Robinson CV, McDowall KJ, Luisi BF. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochem. 2003;42:12848–13855. doi: 10.1021/bi0351099. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5'-monophosphorylated RNA. Proc. Natl Acad. Sci. USA. 2004;101:9211–9216. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J. Mol. Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 29.McDowall KJ, Hernandez RG, Lin-Chao S, Cohen SN. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli rne locus. J. Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg U, Altman S. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1995;1:327–334. [PMC free article] [PubMed] [Google Scholar]
- 31.Lin-Chao S, Wei C.-L, Lin Y.-T. RNase E is required for the maturation of ssrA and normal ssrA RNA peptide-tagging activity. Proc. Natl Acad. Sci. USA. 1999;96:12406–12411. doi: 10.1073/pnas.96.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wachi M, Umitsuki G, Nagai K. Functional relationship between Escherichia coli RNase E and the CafA protein. Mol. Gen. Genet. 1997;253:515–519. doi: 10.1007/s004380050352. [DOI] [PubMed] [Google Scholar]
- 33.Okada Y, Wachi M, Hirata A, Suzuki K, Nagai K, Matsuhashi M. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J. Bacteriol. 1994;176:917–922. doi: 10.1128/jb.176.3.917-922.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 35.Deana A, Belasco JG. The function of RNase G in Escherichia coli is constrained by its amino and carboxyl termini. Mol. Microbiol. 2004;51:1205–1217. doi: 10.1046/j.1365-2958.2003.03905.x. [DOI] [PubMed] [Google Scholar]
- 36.Ow MC, Perwez T, Kushner SR. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 2003;49:607–622. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- 37.Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Comm. 1999;259:483–488. doi: 10.1006/bbrc.1999.0806. [DOI] [PubMed] [Google Scholar]
- 38.Lee K, Bernstein JA, Cohen SN. RNase G complementation of rne null mutation identified functional interrelationships with RNase E in Escherichia coli. Mol. Microbiol. 2002;43:1445–1456. doi: 10.1046/j.1365-2958.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohanty BK, Kushner SR. Rho-independent transcription terminators inhibit RNase P processing of the secG leuU and metT tRNA polycistronic transcripts in Escherichia coli. Nucleic Acids Res. 2008;36:364–375. doi: 10.1093/nar/gkm991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 42.Reece KS, Phillips GJ. New plasmids carrying antibiotic-resistance cassettes. Gene. 1995;165:141–142. doi: 10.1016/0378-1119(95)00529-f. [DOI] [PubMed] [Google Scholar]
- 43.Barik S. Mutagenesis by Megaprimer PCR. In: Horton RM, Tait RC, editors. Genetic Engineering with PCR. Wymondham: Horizon Scientific Press; 1998. pp. 22–37. [Google Scholar]
- 44.O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burnett WV. Northern blotting of RNA denatured in glyoxal without buffer recirculation. BioTechn. 1997;22:668–671. doi: 10.2144/97224st01. [DOI] [PubMed] [Google Scholar]
- 46.Babitzke P, Granger L, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J. Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Combet C, Jambon M, Deleage G, Geourjan C. Geno3D: Automatic comparative molecular modelling of protein. Bioinformatics. 2002;18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 48.Regnier P, Arraiano CM. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 49.Mackie GA. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the products of the ams gene in vivo and in vitro. J. Bacteriol. 1991;173:2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol. Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 51.Regnier P, Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J. Mol. Biol. 1991;187:23–32. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- 52.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–3377. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apirion D, Lassar AB. Conditional lethal mutant of Escherichia coli which affects processing of ribosomal RNA. J. Biol. Chem. 1978;253:1738–1742. [PubMed] [Google Scholar]
- 54.Ghora BK, Apirion D. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 55.Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc. Natl Acad. Sci. USA. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 57.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 58.Berg OG, Kurland CG. Growth rate optimised tRNA abundance and codon usage. J. Mol. Biol. 1997;270:544–550. doi: 10.1006/jmbi.1997.1142. [DOI] [PubMed] [Google Scholar]
- 59.Chung D-H, Min Z, Wang B.-C, Kushner SR. Single amino acid changes in the predicted RNase H domain of E. coli RNase G lead to the complementation of RNase E mutants. J. Mol. Biol. 2008 doi: 10.1261/rna.2104810. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gullier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 61.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casarégola S, Jacq A, Laoudj D, McGurk G, Margarson S, Tempête M, Norris V, Holland IB. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp-1 and encodes a 114 kDA protein that migrates as a 180 kDa protein. J. Mol. Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- 63.Taghbalout A, Rothfield L. RNase E and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl Acad. Sci. USA. 2007;104:1667–1672. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 65.Mackie GA. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 2000;275:25069–25072. doi: 10.1074/jbc.C000363200. [DOI] [PubMed] [Google Scholar]
- 66.Lee K, Zhan X, Gao J, Feng Y, Meganathan R, Cohen SN, Georgiou G. RraA: a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- 67.Chandran V, Poljak L, Vanzo NF, Leroy A, Miguel RN, Fernandex-Recio J, Parkinson J, Burns C, Carpousis AJ, Luisi BF. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J. Mol. Biol. 2007;367:113–132. doi: 10.1016/j.jmb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanahan D, Jessee J, Bloom F. Techniques for transformation of E. coli. In: Glover DM, Hames BD, editors. DNA Cloning: A Practical Approach. 2005. Vol. 1, 2nd edn, IRL Press, Oxford, pp. 1–35. [Google Scholar]
- 69.Claverie-Martin F, Diaz-Torres MR, Yancey SD, Kushner SR. Cloning of the altered mRNA stability (ams) gene of Escherichia coli K-12. J. Bacteriol. 1989;171:5479–5486. doi: 10.1128/jb.171.10.5479-5486.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]