Abstract
Group I intron-derived ribozymes can catalyze a variety of non-native reactions. For the trans-excision-splicing (TES) reaction, an intron-derived ribozyme from the opportunistic pathogen Pneumocystis carinii catalyzes the excision of a predefined region from within an RNA substrate with subsequent ligation of the flanking regions. To establish TES as a general ribozyme-mediated reaction, intron-derived ribozymes from Tetrahymena thermophila and Candida albicans, which are similar to but not the same as that from Pneumocystis, were investigated for their propensity to catalyze the TES reaction. We now report that the Tetrahymena and Candida ribozymes can catalyze the excision of a single nucleotide from within their ribozyme-specific substrates. Under the conditions studied, the Tetrahymena and Candida ribozymes, however, catalyze the TES reaction with lower yields and rates [Tetrahymena (kobs) = 0.14/min and Candida (kobs) = 0.34/min] than the Pneumocystis ribozyme (kobs = 3.2/min). The lower yields are likely partially due to the fact that the Tetrahymena and Candida catalyze additional reactions, separate from TES. The differences in rates are likely partially due to the individual ribozymes ability to effectively bind their 3′ terminal guanosines as intramolecular nucleophiles. Nevertheless, our results demonstrate that group I intron-derived ribozymes are inherently able to catalyze the TES reaction.
INTRODUCTION
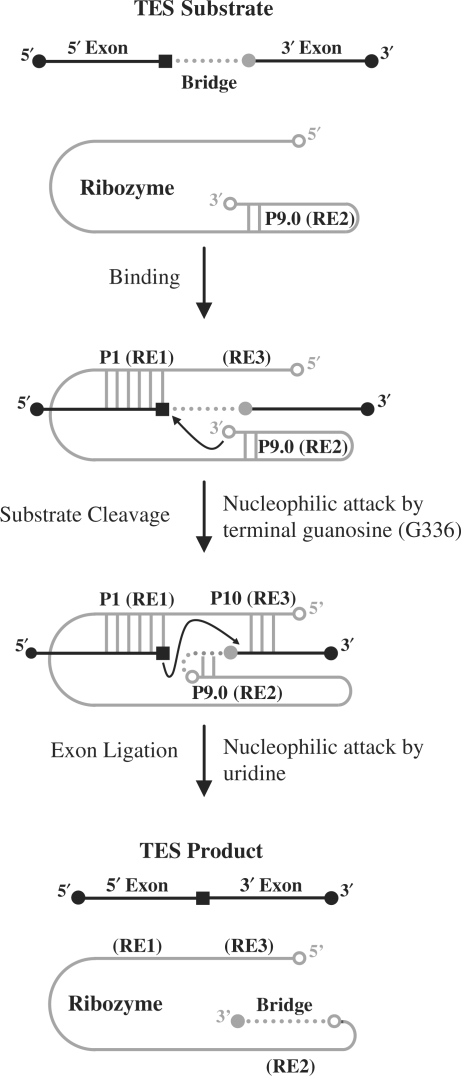
Group I intron-derived ribozymes, which are essentially exonless introns, have previously been shown to catalyze a variety of non-native reactions (1,2). In one such reaction, termed trans-excision splicing (TES), an intron-derived ribozyme from Pneumocystis carinii catalyzes the excision of an internal segment from within an RNA substrate, resulting in the reattachment of the flanking regions (Figure 1) (3). The TES reaction, which is similar to the self-splicing reaction, proceeds through two consecutive transesterification reactions: substrate cleavage followed by exon ligation (Figure 1). In substrate cleavage, the Pneumocystis ribozyme (rPC) catalyzes the intramolecular cleavage of the TES substrate utilizing its 3′ terminal guanosine as an intramolecular nucleophile (Figure 1) (4). In exon ligation, the newly created 3′ terminal uridine of the substrate catalyzes the reattachment of the flanking substrate regions, resulting in formation of the final TES product (Figure 1).
Figure 1.
Diagram of the ribozyme-mediated trans-excision-splicing reaction. The P. carinii ribozyme is denoted in gray and the 5′- and 3′-exon sequences are denoted by black lines. The black square within the 5′-exon region represents a uridine and the gray circle adjacent to the 3′-exon region represents a guanosine. The bridge region to be excised is denoted as a dotted line. The P1 helix is formed through base-pairing of recognition element 1 (RE1) with the 5′-exon. The P10 helix is formed through base-pairing of recognition element 3 (RE3) with the 3′-exon. The first step (substrate cleavage) is catalyzed by G336 (open circle at 3′-end of ribozyme) resulting in the covalent attachment of the 3′-end of the substrate to the 3′-end of the ribozyme. The second step (exon ligation) proceeds through attack of the uridine upon the guanosine of the substrate, resulting in ligation of the flanking sequences.
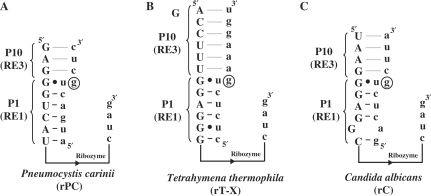
Group I intron-derived ribozymes inherently contain all the structural elements required to catalyze the TES reaction. This suggests that group I intron-derived ribozymes other than P. carinii should be able to catalyze the TES reaction. If true, then the effectiveness of these ‘similar but different’ ribozymes would be useful to know for designing more effective TES ribozymes, as well as to understand intron activity in general. Therefore, intron-derived ribozymes from Tetrahymena thermophila and Candida albicans were constructed and tested for their ability to catalyze TES reactions, using as substrates their native 5′- and 3′-exon sequences (Figure 2) (5,6).
Figure 2.
Diagram of P. carinii (rPC), T. thermophila (rT-X) and C. albicans (rC) ribozyme constructs. The sequence of the Pneumocystis (A), Tetrahymena (B) and Candida (C) ribozyme constructs are shown in uppercase letters with their respective substrate shown in lowercase letters. Only the sequences that are involved with binding the individual substrates are shown, as well as the 3′-end of the ribozymes. Note that the remaining ribozyme sequences, represented with a line, are different for each of the constructs. The nucleotide to be excised (a guanosine) is circled for all three ribozyme constructs.
In this study, the Tetrahymena and Candida ribozymes were shown to catalyze the excision of a single nucleotide from within their ribozyme-specific substrates. Furthermore, like that for Pneumocystis, both ribozymes were shown to catalyze substrate cleavage through an intramolecular transesterification reaction mechanism. The overall yield of TES reactions, however, differed significantly between the individual ribozyme constructs (Pneumocystis, 68%; Tetrahymena, 47% and Candida, 50%). This variation appears at least partly due to both the Tetrahymena and Candida ribozymes producing high-molecular-weight cryptic products, whereas Pneumocystis produces only a single product. Pneumocystis also has a higher rate of reaction, which appears at least partially due to each ribozyme's ability to productively bind its 3′ terminal guanosine, to be used as an intramolecular nucleophile. Nevertheless, our results demonstrate that group I intron-derived ribozymes are inherently able to catalyze the TES reaction.
MATERIALS AND METHODS
Oligonucleotide synthesis and preparation
DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA, USA) and used without further purification. RNA oligonucleotides were obtained from Dharmacon Research, Inc. (Lafayette, CO, USA) and were deprotected using the manufacturer's suggested protocol. Concentrations of individual oligonucleotides were calculated from UV absorption measurements using a Beckman DU 650 spectrophotometer (Beckman Coulter Inc., Fullerton, CA, USA). RNA oligonucleotides were 5′-end radiolabeled by phosphorylation of the 5′-terminal hydroxyl group with (γ-32P) ATP (Amersham Pharmacia Biotech, Piscataway, NJ, USA) using T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA) as previously described (7). RNA oligonucleotides were 3′-end radiolabeled by ligation of radiolabeled pCp (5′-*pCp) to the 3′-end of the RNA oligonucleotide using T4 RNA ligase (New England Biolabs), as previously described (8). Note that 3′-end radiolabeling results in substrates being 1 nt longer than 5′-end radiolabeled substrates.
Plasmid construction
For these studies, two ribozyme constructs were created: pT-X, derived from the plasmid pT7L-21 (5) (kindly provided by Douglas H. Turner, University of Rochester, Rochester, NY, USA), which contains the Tetrahymena ribozyme (5); and pC (6), derived from the plasmid pC10/1x (kindly provided by Douglas H. Turner), which contains the Candida ribozyme (6). The individual ribozyme sequences were modified to include an RE3 sequence so that a P10 helix interaction would form (Figure 2).
The plasmids, pT and pC, were synthesized by altering the parental plasmids, pT7L-21 and pC10/1x, via site-directed mutagenesis. This was done because the formation of the P10 helix interaction is important for the second reaction step of the TES reaction (9,10). Note that the RE3 sequence added was native to each respective intron. The pT plasmid was created using a single round of site-directed mutagenesis using the primer pairs (underlined bases represent inserted nucleotides) 5′CGACTCACTATAGACCTTTGGAGGGAAAAG3′ and 5′CTTTTCCCTCCAAAGGTCTATAGTGAGTCG3′ for RE3 formation. The pC plasmid was created using a single round of site-directed mutagenesis using the primer pairs 5′CGACTCACTATAGGTAAGGGAGGCAAAAGTAGGG3′ and 5′CCCTACTTTTGCCTCCCTTACCTATAGTGAGTCG3′ for RE3 formation. Note that a guanosine was inserted at the 5′-end of each ribozyme construct to ensure efficient transcription by T7 RNA polymerase (11).
Additionally, an XbaI restriction site was inserted at the 3′-end of the ribozyme region in the pT plasmid, which creates a terminal guanosine required for the intramolecular transesterification reaction. This also makes the 3′-end of the Tetrahymena ribozyme the same as those of the Candida and Pneumocystis ribozymes (6,7). The plasmid, pT-X, was created in two rounds using the following primer pairs (underlined bases represent changes to the 3′-end of the ribozyme) 5′GGAGTACTCGATAAGGTAGCCAAATGCC3′ and 5′GGCATTTGGCTACCTTATCGAGTACTCC3′ for the first round. The primer pairs 5′GGAGTACTCTAGATAAGGTAGCCAAATGCC3′ and 5′GGCATTTGGCTACCTTATCTAGAGTACTCC3′ were used for the second round.
Each amplification reaction consisted of 22.5 pmol of each primer, 25 ng parental plasmid (pT7L-21, pT or pC10/1x), 2.5 U Pfu DNA polymerase (Stratagene, La Jolla, CA, USA) and 0.5 mM dNTPs (Invitrogen, Grand Island, NY, USA) in a buffer consisting of 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris–HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100 and 0.1 mg/ml BSA (total reaction volume is 50 μl). After an initial denaturation step at 95°C for 30 s, the mixture was subjected to 20 cycles of 95°C for 30 s, 50°C for 2 min and 68°C for 6 min. Parental plasmid was then digested with 20 units DpnI (New England Biolabs) in 4.1 μl of the manufacturer's supplied buffer (10X NEBuffer 4) for 2 h at 37°C. A 3-μl-aliquot of this mixture was then used to transform Escherichia coli DH5α competent cells (Invitrogen). The plasmids were purified using a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA) and were sequenced for confirmation (Davis Sequencing, Davis, CA, USA).
Ribozyme synthesis and purification
The Pneumocystis plasmid (PC), the Tetrahymena plasmid (pT-X), and the Candida plasmid (pC) were linearized in a 50 uL reaction consisting of 16 μg plasmid, 5 μL 10X REACT 2 buffer and 50 U XbaI (Invitrogen). The reactions were incubated at 37°C for 2 h, followed by plasmid purification with a QIAquick PCR purification kit (Qiagen). The resulting linearized plasmids were subjected to run-off transcription reactions in a 50 μl volume consisting of 1 μg DNA template, 50 U of T7 RNA polymerase (New England Biolabs), 40 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 5 mM spermidine and 1 mM rNTP mix. The reactions were incubated at 37°C for 2 h, at which time each ribozyme construct was purified using a Qiagen Plasmid Midi Kit (Qiagen), as previously described (3). Concentrations of individual ribozymes were calculated from UV absorption measurements using a Beckman DU 650 spectrophotometer (Beckman Coulter Inc.) using the extinction coefficient of 3.5 × 106/M/cm for the Pneumocystis (rPC) ribozyme (7), 3.2 × 106/M/cm for the Tetrahymena (rT-X) ribozyme (5) and 3.72 × 106/M/cm for the Candida (rC) ribozyme (6).
TES reactions
Reactions were performed at 44°C in buffer consisting of 50 mM Hepes (25 mM Na+), 135 mM KCl and various concentrations of MgCl2. TES reactions were optimized for magnesium concentration (0–35 mM MgCl2), ribozyme concentration (0–500 nM) and time (1–45 min). In each case, TES reaction conditions were optimized to give the greatest TES product yield. Briefly, a ribozyme solution lacking substrate in HXMg buffer was preannealed at 60°C for 5 min and then slow cooled to 44°C. Reactions were initiated by the addition of 1 μl of 8 nM 5′-end radiolabeled substrate (10 mer, 5′AUGACUGCUC3′ for rPC; 13 mer, 5′CUCUCUGAAAGGU3′ for rT-X; or 11 mer, 5′GACUCUGCUUA3′ for rC). Note that the italic lettering in the substrates represent the nucleotide to be excised. The final concentration of the substrates in the reaction mixtures were 1.3 nM.
To distinguish between possible substrate cleavage mechanisms, 3′-end radiolabeled substrates were employed at the same concentrations stated above. Reactions were conducted at 44°C for the optimized amount of time and stopped by adding an equal volume of stop buffer (10 M urea, 3 mM EDTA and 0.1X TBE). Reactions were denatured for 1 min at 90°C and substrates, intermediates and products were separated on a 12% polyacrylamide/8 M urea gel. The gel was transferred onto chromatography paper (Whatman 3MM CHR) and dried under vacuum. The bands were visualized and quantified on a Molecular Dynamics Storm 860 Phosphorimager.
Kinetic studies
The observed rate constants (kobs) for the individual steps of the TES reaction were obtained under single-turnover ‘ribozyme excess’ conditions. In these experiments, 35 μl of ribozyme in HXMg buffer (identified in the figure legends) was preannealed at 60°C for 5 min and slow-cooled to 44°C. The reactions were initiated by the addition of 7 μl of 8 nM 5′-end radiolabeled substrate, bringing the final concentrations to 1.3 nM substrate and 100 nM (for rPC), 166 nM (for rT-X) or 75 nM (for rC) ribozyme. Three microliters aliquots were periodically removed and added to an equal volume of stop buffer. The reactants and products were separated on a 12% polyacrylamide/8 M urea gel, and the gel was dried under vacuum. The resulting bands were visualized and quantified as described above. The observed rate constants for the substrate cleavage step and for exon ligation were quantified as previously described (3).
RT–PCR amplification of high–molecular-weight products
TES reactions were conducted under optimized reaction conditions using a 29-mer substrate (5′ACUAUGACUCUCUCUGAAAGGUAGCCAAAUG3′) for rT-X and a 28 mer (5′ACUAUGACUCUGCUUACUAUCAUCUAUC3′) for rC. The reaction intermediates were RT–PCR amplified using an Access RT–PCR kit (Promega, Madison, WI, USA) with primers 5′GGAGGACCGGTACTATGACTCTC3′ and 5′AGTTCGAAGGCATTTGGCTACC3′ for rT-X and 5′GCACAGGTTCGAACATACTATGACTC3′ and 5′GCTGGAACCGGTGATAGATGATAG3′ for rC. Optimal amplifications occurred in a total reaction volume of 50 μl containing 2 μl TES reaction (166 nM rT-X or 75 nM rC ribozyme), 1 mM MgSO4, 45 pmol of each primer, 0.2 mM dNTPs, 5 U AMV reverse transcriptase and 5 U Tfl DNA polymerase. First-strand synthesis occurred at 45°C for 45 min, followed by 2 min at 94°C for deactivation of the AMV reverse transcriptase. The reactions were then subjected to 40 PCR cycles consisting of 94°C for 30 s, 55°C for 60 s and 68°C for 120 s (10 min for the final cycle). RT–PCR products were separated on a 2% agarose gel and excised from the gel matrix using the QIAquick Gel Extraction Kit (Qiagen). The excised products were ligated into a pDrive cloning vector (Qiagen) using the manufacturer's recommended protocol and transformed into DH5α cells (Invitrogen). Colonies were picked and sequenced for identification (Davis Sequencing).
Inhibition studies using exogenous guanosine
TES reactions were conducted at 44°C in HXMg buffer with various concentrations of rGTP (0–10 mM). Briefly, a ribozyme solution lacking substrate in HXMg buffer was preannealed at 60°C for 5 min with X mM rGTP. Reactions were then slow cooled to 44°C and initiated by the addition of 1 μl of 8 nM 5′-end radiolabeled substrate, bringing the final concentrations to 1.3 nM substrate and 100 nM (for rPC), 166 nM (for rT-X) or 75 nM (for rC) ribozyme. TES reactions were conducted under the optimal time for each ribozyme; 30 min (for rPC), 45 min (for rT-X) and 30 min (for rC). Reactions were denatured for 1 min at 90°C and were separated on a 12% polyacrylamide/8 M urea gel. The gel was transferred onto chromatography paper (Whatman 3MM CHR) and dried under vacuum. The bands were visualized and quantified on a Molecular Dynamics Storm 860 Phosphorimager.
RESULTS
To simplify the comparison of the individual ribozyme constructs, the simplest TES model test system was utilized, where a single nucleotide is excised from within their ribozyme-specific substrates. For these studies, the previously constructed Tetrahymena and Candida ribozymes were modified in two ways (Figure 2). First, the initial ribozyme constructs lacked the nucleotides required to form the P10 helix (RE3), and so an RE3 region (native to the ribozyme) was added. As seen for the Pneumocystis ribozyme, P10 helix formation has been shown to aid in binding the 3′-exon region of TES substrates (9,10). Second, the 3′-end of each ribozyme sequence was modified to include the addition of a terminal guanosine, (which is found naturally) as it is required as the intramolecular nucleophile in the substrate-cleavage reaction (4).
TES reactions
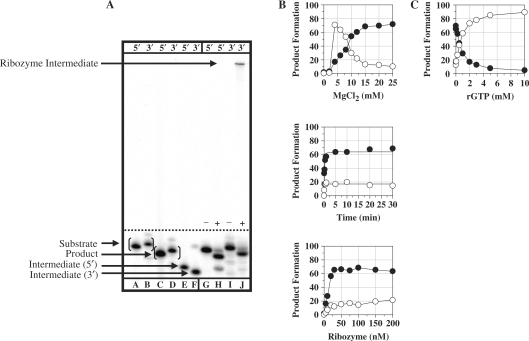
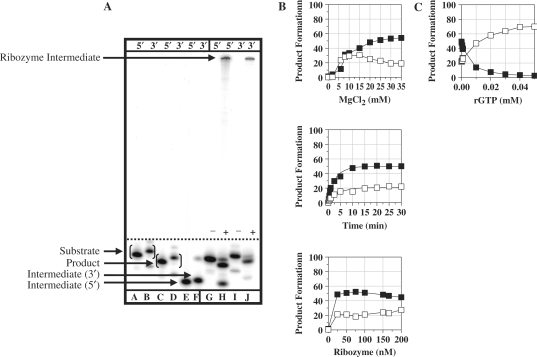
For the Pneumocystis ribozyme (rPC), the TES reaction using the previously characterized Pneumocystis ribozyme (3) was run (Figure 2). TES reactions conducted with 5′-end radiolabeled substrate (5′AUGACUGCUC3′, the nucleotide in italics is to be excised) resulted in the expected 9-mer product (5′AUGACUCUC3′) and 6-mer intermediate (5′AUGACU3′) (Figure 3A, lanes G–H). TES reaction conditions were optimized for magnesium concentration, time and ribozyme concentration for maximum TES product yields (Figure 3B). Optimized TES reaction conditions resulted in 68.5 ± 4.4% TES product yield, with an observed rate constant, kobs, of 5.0 ± 1.1/min for substrate cleavage and 3.2 ± 0.1/min for exon ligation. Note, optimized TES reactions conditions are compiled in Table 1.
Figure 3.
TES reactions conducted with the P. carinii (rPC) ribozyme. (A) Polyacrylamide gel of TES reactions using either 5′ or 3′-end radiolabeled substrates. Reactions were conducted using 100 nM rPC, H0Mg (−) or H15Mg (+) buffer for 30 min at 44°C. Lanes A, C and E contain 5′-end labeled size controls, respectively. Lanes B, D and F contain 3′-end radiolabeled size controls, respectively. Note that the 3′-end labeled size controls are one nucleotide larger than 5′-end labeled size controls. Lanes G–J contain the rPC ribozyme with 5′-end radiolabeled (lanes G and H) or 3′-end radiolabeled (lanes I and J) substrate. (B) Graphs of TES reactions using 5′-end radiolabeled substrate. All reactions were conducted as above except for the changing variable. (C) Graph of exogenous guanosine concentration dependence for TES reactions. All data points represent the average of at least two independent reactions with standard deviations typically below 10%. For all graphs, the TES product is represented by filled circles and the substrate cleavage product is represented as open circles.
Table 1.
Optimized TES reaction conditions, product yields and reaction rates
| Optimized TES reactions conditionsa |
TES product yield and observed rate constants for substrate cleavage and exon ligation reactionsb |
||||||
|---|---|---|---|---|---|---|---|
| Ribozyme | MgCl2 (mM) | Time (min) | Ribozyme (nM) | TES product (%) | Substrate cleavage (kobs/min) | Exon ligation (kobs/min) | |
| Pneumocystis | 15 | 30 | 100 | 68.5 ± 4.4 | 5.0 ± 1.1 | 3.2 ± 0.1 | |
| Tetrahymena | 10 | 45 | 166 | 47.5 ± 0.18 | 0.17 ± 0.05 | 0.14 ± 0.02 | |
| Candida | 25 | 30 | 75 | 50.0 ± 1.5 | 0.31 ± 0.01 | 0.34 ± 0.03 | |
aTES reactions were optimized with regard to magnesium concentration, time and ribozyme concentration for maximum TES product yields.
bThe observed rate constants, kobs, were obtained for the substrate cleavage and exon-ligation reactions under single turnover conditions. Note that ribozyme and substrate sequences are different between the ribozyme constructs.
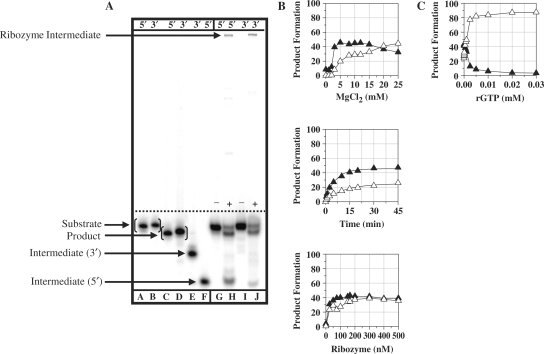
For the Tetrahymena ribozyme (rT-X), TES reactions conducted with 5′-end radiolabeled substrate (5′CUCUCUGAAAGGU3′, the nucleotide in italics is to be excised) resulted in the predicted 12-mer product (5′CUCUCUAAAGGU3′) and 6-mer intermediate (5′CUCUCU3′) (Figure 4A, lanes G and H). Additionally, a high molecular weight product band was observed, which likely reduces the yield of the TES product. This product was found to be due to ribozyme-mediated reverse splicing (see following section) (12,13). Nevertheless, optimized TES reactions resulted in 47.5 ± 0.2% TES product yield (Figure 4B), with an observed rate constant, kobs, of 0.17 ± 0.05/min for substrate cleavage and 0.14 ± 0.02/min for exon ligation (Table 1).
Figure 4.
TES reactions conducted with the T. thermophila (rT-X) ribozyme. (A) Polyacrylamide gel of TES reactions using either 5′ or 3′-end radiolabeled substrates. Reactions were conducted using 166 nM rT-X, H0Mg (−) or H10Mg (+) buffer, for 45 min at 44°C. Lanes A, C and F contain 5′-end radiolabeled size controls, respectively. Lanes B, D and E contain 3′-end radiolabeled size controls, respectively. Note that the 3′-end radiolabeled size controls are one nucleotide larger than 5′-end radiolabeled size controls. Lanes G–J contain the normal rT-X ribozyme with 5′-end radiolabeled (lanes G and H) or 3′-end radiolabeled (lanes I and J) substrate. (B) Graphs of TES reactions using 5′-end radiolabeled substrate. All reactions were conducted as above except for the changing variable. (C) Graph of rGTP concentration dependence for TES reactions conducted with the rT-X ribozyme. All data points represent the average of at least two independent reactions with standard deviations typically below 10%. Note for all graphs, the TES product is represented by filled triangles and the substrate cleavage product is represented as open triangles.
For the Candida ribozyme (rC), TES reactions conducted with 5′-end radiolabeled substrate (5′GACUCUGCUUA3′, the nucleotide in italics is to be excised) resulted in the predicted 10-mer product (5′GACUCUCUUA3′) and 6-mer intermediate (5′GACUCU3′) (Figure 5A, lanes G and H). Similar to the Tetrahymena ribozyme, an additional high-molecular-weight product band was observed, which likely reduces the yield of TES product. Similar to the Tetrahymena ribozyme, the high molecular weight product band was found to be due to ribozyme-mediated reverse splicing (see following section) (12,13). Optimized TES reactions conditions resulted in 50 ± 1.3% TES product yield (Figure 5B), with an observed rate constant, kobs, of 0.31 ± 0.01/min for substrate cleavage reaction and 0.34 ± 0.03/min for exon ligation (Table 1). In conclusion, these results demonstrate that the Tetrahymena and Candida ribozymes can catalyze the excision of a single nucleotide from within their ribozyme-specific substrates through the TES reaction.
Figure 5.
TES reactions conducted with the C. albicans (rC) ribozyme. (A) Polyacrylamide gel of TES reactions using both 5′ and 3′-end radiolabeled substrates. Reactions were conducted using 75 nM rC, H0Mg (−) or H25Mg (+) buffer, for 30 min at 44°C. Lanes A, C and E contain 5′-end radiolabeled size controls, respectively. Lanes B, D and F contain 3′-end radiolabeled size controls, respectively. Note that the 3′-end radiolabeled size controls are one nucleotide larger than 5′-end radiolabeled size controls. Lanes G–J contain the normal rC ribozyme with 5′-end radiolabeled (lanes G and H) or 3′-end radiolabeled (lanes I and J) substrate. (B) Graphs of TES reactions using 5′-end radiolabeled substrate. All reactions were conducted as above except for the changing variable. (C) Graph of rGTP concentration dependence for TES reactions conducted with the rC ribozyme. All data points represent the average of at least two independent reactions, with standard deviations typically below 10%. Note for all graphs, the TES product is represented by filled triangles and the substrate cleavage product is represented as open triangles.
Sequence identification of high molecular weight products
RT–PCR was employed to isolate and subsequently sequence the reverse-splicing ribozyme product. If the reverse-splicing reaction is occurring, we expect to see the ribozyme become embedded between the 5′- and 3′-exon regions of the substrate. For these studies, a 29-mer substrate (rT-X, 5′ACUAUGACUCUCUCUGAAAGGUAGCCAAAUG3′) and a 28-mer substrate (rC, 5′ACUAUGACUCUGCUUACUAUCAUCUAUC3′) were used as the predicted reverse-splicing product could be more easily RT–PCR amplified using these larger substrates. Therefore, if the reverse-splicing reaction is occurring, both the 5′-exon region (5′ACUAUGACUCUCUCU3′ for rT-X or 5′ACUAUGACUCU3′ for rC) and 3′-exon region of the substrate (5′GAAAGGUAGCCAAAUG3′ for rT-X or 5′GCUUACUAUCAUCUAUC3′ for rC) will become covalently attached to the 5′- and 3′-ends of the ribozyme. Our results from sequencing the reaction products (data not shown) show that the expected reverse-splicing product is produced whereby the ribozyme becomes embedded within the 5′- and 3′-exon regions of the substrate.
The mechanism of the substrate cleavage reaction
In the absence of an exogenous guanosine cofactor, the substrate cleavage reaction could possibly be catalyzed through two distinct reaction mechanisms. These include ribozyme-mediated hydrolysis (6,14–16) and intramolecular transesterification (4,6,12,13,17–25). For the Pneumocystis ribozyme, the intramolecular transesterification reaction predominates, with the 3′ terminal guanosine as the nucleophile (4). Given that the Tetrahymena and Candida ribozyme constructs were also synthesized with 3′ terminal guanosines, it is likely that each of these constructs will catalyze their substrate cleavage reactions predominately through intramolecular transesterification. The two substrate cleavage reaction mechanisms can be distinguished by utilizing 3′-end radiolabeled substrates (4), as the size of the TES reaction intermediates and products will differ according to the mechanism.
For the Tetrahymena ribozyme, TES reactions were conducted with 3′-end labeled substrate, (5′CUCUCUGAAAGGUC3′, underlined nucleotide added during the 3′-end labeling procedure), which resulted in the predicted 13-mer product (5′CUCUCUAAAGGUC3′), as well as a higher-molecular-weight product band (Figure 4A, lanes I and J). This high-molecular-weight band is potentially due to the 3′-end radiolabeled 3′-exon region of the substrate becoming covalently attached to the 3′-end of the ribozyme through intramolecular transesterification via ribozyme-mediated reverse splicing. That both the substrate cleavage step of TES and reverse splicing are catalyzed through intramolecular transesterification complicates delineation of the reaction mechanism through identification of the high-molecular-weight product band. Nevertheless, the absence of the predicted hydrolysis product (5′GAAAGGUC3′) suggests that the Tetrahymena ribozyme undergoes intramolecular transesterification.
For the Candida ribozyme, TES reactions were conducted using 3′-end radiolabeled substrate, (5′GACUCUGCUUAC3′, underlined nucleotide added during the 3′-end labeling procedure), which resulted in the predicted 11-mer product (5′GACUCUCUUAC3′), as well as a high-molecular-weight product band (Figure 5A, lanes I and J). Similar to the Tetrahymena ribozyme, the absence of the predicted hydrolysis product (6 mer, 5′GCUUAC3′) suggests that the Candida ribozyme catalyzes substrate cleavage through intramolecular transesterification.
As a control, TES reactions were conducted with the Pneumocystis ribozyme using 3′-end labeled substrate (5′AUGACUGCUCC3′, underlined nucleotide is due to 3′-end labeling procedure). TES reactions resulted in the expected 12-mer product (5′AUGACUCUCC3′) and the expected high-molecular-weight product band (Figure 3A, lanes I and J) produced in the intramolecular transesterification reaction.
Exogenous guanosine inhibits the TES reaction
The Pneumocystis ribozyme catalyzes the substrate cleavage reaction ∼20-fold faster than either the Tetrahymena or Candida ribozyme (Table 1). This observed decrease in rate for Tetrahymena and Candida could be directly related to the ribozymes ability to utilize their 3′ terminal guanosines for substrate cleavage. For a previously described Tetrahymena ribozyme (24), competition assays using exogenous guanosine were used as a probe for the ribozymes ability to productively bind their terminal guanosine. Using a similar strategy, inhibition of TES product formation could be directly correlated to the ribozymes ability to effectively bind its terminal guanosine into the guanosine-binding site (GBS).
For the Pneumocystis ribozyme, TES reactions were conducted with increasing concentrations of rGTP under optimal TES reaction conditions (Table 1). The results demonstrate that TES product formation is not completely hindered (∼93% hindrance) until an excessively high concentration of rGTP (10 mM) is added (Figure 3C). This indicates that the rPC binds its terminal guanosine much more effectively than the added exogenous guanosine. For the Tetrahymena ribozyme, TES reactions resulted in complete inhibition of TES product formation at much lower rGTP concentrations (0.02 mM, ∼91% hindrance) (Figure 4C). In the case of the Candida ribozyme, TES reactions demonstrate that lower concentrations of rGTP (0.05 mM, ∼93% hindrance) are also effective at completely hindering TES product formation (Figure 5C). These results suggest that the Pneumocystis ribozyme binds its terminal guanosine (G336), via the GBS, more effectively than that for the Tetrahymena (G416) and Candida (G374) ribozymes.
DISCUSSION
In this report, group I intron-derived ribozymes from Tetrahymena and Candida were shown to be capable of catalyzing the excision of internal segments (in this case a single nucleotide) from within their native 5′- and 3′-exon sequences. Furthermore, the Tetrahymena and Candida ribozyme constructs were shown to catalyze the substrate cleavage reaction step through an intramolecular transesterification mechanism, using their 3′ terminal guanosines as nucleophiles. Moreover, both the Tetrahymena and Candida ribozymes were shown to catalyze ribozyme-mediated reverse-splicing reaction, which was not seen for the Pneumocystis ribozyme. Lastly, this is the first report that group I intron-derived ribozymes other than Pneumocystis can catalyze the excision of an internal segment from RNA transcripts via the TES reaction.
The Tetrahymena ribozyme
The Tetrahymena and Pneumocystis ribozymes catalyze the individual steps of the TES reaction under similar optimized magnesium, time and ribozyme conditions (Table 1). The results, however, show that the Tetrahymena ribozyme catalyzes the individual steps of the TES reaction with lower yields (47.5 ± 0.18% versus 68 ± 4.4%) than the Pneumocystis ribozyme. Furthermore, the Pneumocystis ribozyme catalyzes substrate cleavage ∼30-fold faster than the Tetrahymena ribozyme (Table 1). In addition, the Pneumocystis ribozyme catalyzes exon ligation ∼20-fold faster than the Tetrahymena ribozyme.
The decrease in yield of the Tetrahymena ribozyme may be due, at least in part, to its catalysis of reactions other than TES. Previously characterized Tetrahymena ribozymes (12,13), which resemble our Tetrahymena ribozyme construct, can catalyze intramolecular transesterification reactions other than that which occurs in TES, including reverse splicing (12,13). For the reverse-splicing reaction, the ribozyme becomes embedded between the 5′- and 3′-exon regions of its RNA substrate. For the Tetrahymena ribozyme, a high-molecular-weight product band is produced for TES reactions conducted with 5′-end labeled substrates (Figure 5.4A, lane H). This product band was not observed for TES reactions conducted with the Pneumocystis ribozyme, which has allowed an in-depth kinetic analysis of the Pneumocystis substrate cleavage step (26). This high-molecular-weight product band was shown to be the result of ribozyme-mediated reverse splicing (12,13). That two competing reactions (TES and reverse splicing) are simultaneously occurring could explain the lower efficiency of the Tetrahymena ribozyme reactions.
The decrease in rates of the Tetrahymena ribozyme may be due to its ability to utilize its 3′ terminal guanosine as a first-reaction step nucleophile. From the competition studies it appears that the 3′ terminal guanosine of the Pneumocystis ribozyme is more effectively bound (relative to Tetrahymena and Candida) into the GBS of the ribozyme (4) prior to the substrate cleavage step. One reason for this could be that, as seen for similar intramolecular transesterification reactions, the terminal guanosine is predicted to be aligned into the GBS through helix P9.0 (24). However, the Tetrahymena and Candida constructs in this report are not readily able to form simple P9.0 helices. This could be contributing toward the lower binding of the 3′ terminal guanosine and consequently slower rates for Tetrahymena and Candida. Altering the Tetrahymena and Candida ribozymes to resemble their native introns 3′-ends could enhance their ability to effectively utilize their terminal guanosines as intramolecular nucleophiles. Nevertheless, our results demonstrate that all three constructs, in their current incarnations, effectively catalyze the TES reaction.
A previously characterized Tetrahymena ribozyme that lacks both the 5′- and 3′-exon sequences and the RE1 and RE3 sequences is capable of catalyzing a TES-like reaction (27). This highly truncated Tetrahymena ribozyme was shown to be capable of binding pseudoknot structures in trans resulting in the formation of P1 and P10 helices. Unlike the TES reaction, high magnesium concentrations (10–100 mM), high temperatures (55–65°C) and an exogenous guanosine cofactor are required (27). For our studies, the individual ribozymes catalyze the TES reaction in the absence of a guanosine cofactor and require lower magnesium concentrations (10–25 mM) and temperature (44°C).
The Candida ribozyme
The Candida and Pneumocystis ribozymes catalyze the individual steps of the TES reaction under similar optimized time and ribozyme conditions (Table 1). For the Candida ribozyme, however, higher magnesium concentrations are required for optimal TES reactivity. Similar to the Tetrahymena ribozyme, the Candida ribozyme was observed to catalyze the individual steps of the TES reaction with lower yields (50.0 ± 1.5% versus 68 ± 4.4%) than the Pneumocystis ribozyme. Moreover, the Candida ribozyme catalyzes substrate cleavage ∼15-fold slower than the Pneumocystis ribozyme (Table 1). In addition, the Candida ribozyme catalyzes exon ligation ∼10-fold slower than the Candida ribozyme.
The decrease in yield of the Candida ribozyme could be due to the catalysis of a competing reaction, which produces a high-molecular-weight product band for TES reactions conducted with 5′-end radiolabeled substrates (Figure 5.5A, lane H). This high-molecular-weight product band was shown to be the result of ribozyme-mediated reverse splicing (12,13). That both the TES and reverse-splicing reaction are simultaneously occurring could explain the lower yield of the Candida ribozyme reactions. As mentioned for the Tetrahymena ribozyme, the decrease in rates of the Candida ribozyme could partially be due to the ribozyme's ability to effectively utilize its 3′ terminal guanosine as an intramolecular nucleophile.
CONCLUSIONS
Our results demonstrate that group I intron-derived ribozymes, other than Pneumocystis, are inherently capable of catalyzing the TES reaction. Both the Tetrahymena and Candida ribozymes catalyzed the excision of a single nucleotide from within their ribozyme-specific substrates through the TES reaction mechanism. The Pneumocystis ribozyme, however, demonstrated both higher TES product yields and rates compared to the Tetrahymena and Candida ribozymes. For the Tetrahymena and Candida ribozymes, additional cryptic products, including ribozyme-mediated reverse-splicing products were produced. It also appears that the Pneumocystis ribozyme binds its terminal guanosine more effectively than the Tetrahymena and Candida ribozymes. At this stage, it appears that the Pneumocystis ribozyme is the best choice for future TES applications in that it produces higher yields, rates and only a single product. Nevertheless, our results demonstrate that group I introns are multifaceted catalysts.
ACKNOWLEDGEMENTS
This work was supported by grants from the Department of Defense Breast Cancer Research Program DAMD17-03-1-0329 and the Kentucky Lung Cancer Research Program. The authors thank two anonymous reviewers for insightful suggestions. Funding to pay the Open Access publication charges for this article was provided by The University of Kentucky.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cech TR. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Dotson PP, II, Testa SM. Group I intron-derived ribozyme recombination reactions. Rec. Develop. Nucleic Acids Res. 2006;2:307–324. [Google Scholar]
- 3.Bell MA, Johnson AJ, Testa SM. Ribozyme-catalyzed excision of targeted sequences from within RNAs. Biochemistry. 2002;41:15327–15333. doi: 10.1021/bi0267386. [DOI] [PubMed] [Google Scholar]
- 4.Dotson PP, II, Sinha J, Testa SM. A Pneumocystis carinii group I intron-derived ribozyme utilizes an endogenous guanosine as the first reaction step nucleophile in the trans excision-splicing reaction. Biochemistry. 2008;47:4780–4787. doi: 10.1021/bi7020525. [DOI] [PubMed] [Google Scholar]
- 5.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 6.Disney MD, Haidaris CG, Turner DH. Recognition elements for 5′ exon substrate binding to the Candida albicans group I intron. Biochemistry. 2001;40:6507–6519. doi: 10.1021/bi002008r. [DOI] [PubMed] [Google Scholar]
- 7.Testa SM, Haidaris CG, Gigliotti F, Turner DH. A Pneumocystis carinii group I intron ribozyme that does not require 2′ OH groups on its 5′ exon mimic for binding to the catalytic core. Biochemistry. 1997;36:15303–15314. doi: 10.1021/bi9713097. [DOI] [PubMed] [Google Scholar]
- 8.Johnson AJ, Sinha J, Testa SM. Trans insertion-splicing: Ribozyme-catalyzed insertion of targeted sequences into RNAs. Biochemistry. 2005;44:10702–10710. doi: 10.1021/bi0504815. [DOI] [PubMed] [Google Scholar]
- 9.Bell MA, Sinha J, Johnson AJ, Testa SM. Enhancing the second step of the trans excision-splicing reaction of a group I ribozyme by exploiting P9.0 and P10 for intermolecular recognition. Biochemistry. 2004;43:4323–4331. doi: 10.1021/bi035874n. [DOI] [PubMed] [Google Scholar]
- 10.Baum DA, Sinha J, Testa SM. Molecular recognition in a trans excision-splicing ribozyme: non-Watson-Crick base pairs at the 5′ splice site and ωG at the 3′ splice site can play a role in determining the binding register of reaction substrates. Biochemistry. 2005;44:1067–1077. doi: 10.1021/bi0482304. [DOI] [PubMed] [Google Scholar]
- 11.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodson SA, Cech TR. Reverse self-splicing of the Tetrahymena group I intron: Implication for the directionality of splicing and for intron transposition. Cell. 1989;57:335–345. doi: 10.1016/0092-8674(89)90971-9. [DOI] [PubMed] [Google Scholar]
- 13.Roman J, Woodson SA. Integration of the Tetrahymena group I intron into bacterial rRNA by reverse splicing in vivo. Proc. Natl Acad. Sci. USA. 1998;95:2134–2139. doi: 10.1073/pnas.95.5.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue T, Sullivan FX, Cech TR. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J. Mol. Biol. 1986;189:143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- 15.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 16.Zaug AJ, Davila-Aponte JA, Cech TR. Catalysis of RNA cleavage by a ribozyme derived from the group I intron of Anabaena pre-tRNA(Leu) Biochemistry. 1994;33:14935–14947. doi: 10.1021/bi00253a033. [DOI] [PubMed] [Google Scholar]
- 17.Brehm SL, Cech TR. Fate of an intervening sequence ribonucleic acid: Excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry. 1983;22:2390–2397. doi: 10.1021/bi00279a014. [DOI] [PubMed] [Google Scholar]
- 18.Zaug AJ, Grabowski PJ, Cech TR. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983;301:578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- 19.Zaug AJ, Kent JR, Cech TR. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984;224:574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- 20.Zaug AJ, Kent JR, Cech TR. Reactions of the intervening sequence of the Tetrahymena ribosomal ribonucleic acid precursor pH dependence of cyclization and site-specific hydrolysis. Biochemistry. 1985;24:6211–6218. doi: 10.1021/bi00343a027. [DOI] [PubMed] [Google Scholar]
- 21.Zaug AJ, Cech TR. The Tetrahymena intervening sequence ribonucleic acid enzyme is a phosphotransferase and an acid phosphatase. Biochemistry. 1986;25:4478–4482. doi: 10.1021/bi00364a002. [DOI] [PubMed] [Google Scholar]
- 22.Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 23.Chowrira BM, Berzel-Herranz A, Burke JM. Novel RNA polymerization reaction catalyzed by a group I intron-derived ribozyme. EMBO J. 1993;12:3599–3605. doi: 10.1002/j.1460-2075.1993.tb06033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei R, Herschlag D. Mechanistic investigations of a ribozyme derived from the Tetrahymena group I intron: Insights into catalysis and the second step of self-splicing. Biochemistry. 1996;35:5796–5809. doi: 10.1021/bi9527653. [DOI] [PubMed] [Google Scholar]
- 25.Riley CA, Lehman N. Generalized RNA-directed recombination of RNA. Chem. Biol. 2003;10:1233–1243. doi: 10.1016/j.chembiol.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Dotson PP, II, Sinha J, Testa SM. Kinetic characterization of the first reaction step of the ribozyme-catalyzed trans excision-splicing reaction. FEBS J. 2008;275:3110–3122. doi: 10.1111/j.1742-4658.2008.06464.x. [DOI] [PubMed] [Google Scholar]
- 27.Sargueil B, Tanner NK. A shortened form of the Tetrahymena thermophila group I intron can catalyze the complete self-splicing reaction in trans. J. Mol. Biol. 1993;233:629–643. doi: 10.1006/jmbi.1993.1541. [DOI] [PubMed] [Google Scholar]