Abstract
Novel sequencing technologies permit the rapid production of large sequence data sets. These technologies are likely to revolutionize genetics and biomedical research, but a thorough characterization of the ultra-short read output is necessary. We generated and analyzed two Illumina 1G ultra-short read data sets, i.e. 2.8 million 27mer reads from a Beta vulgaris genomic clone and 12.3 million 36mers from the Helicobacter acinonychis genome. We found that error rates range from 0.3% at the beginning of reads to 3.8% at the end of reads. Wrong base calls are frequently preceded by base G. Base substitution error frequencies vary by 10- to 11-fold, with A > C transversion being among the most frequent and C > G transversions among the least frequent substitution errors. Insertions and deletions of single bases occur at very low rates. When simulating re-sequencing we found a 20-fold sequencing coverage to be sufficient to compensate errors by correct reads. The read coverage of the sequenced regions is biased; the highest read density was found in intervals with elevated GC content. High Solexa quality scores are over-optimistic and low scores underestimate the data quality. Our results show different types of biases and ways to detect them. Such biases have implications on the use and interpretation of Solexa data, for de novo sequencing, re-sequencing, the identification of single nucleotide polymorphisms and DNA methylation sites, as well as for transcriptome analysis.
INTRODUCTION
The DNA sequencing field has experienced a major boost with the emergence of novel sequencing technologies. Several systems are currently on the market, including Illumina's Solexa instrument, the Applied Biosystems’ Sequencing by Oligonucleotide Ligation and Detection (SOLiD) technology, and the GS FLX instruments from Roche/454 Life Sciences. The Polony cyclic sequencing by synthesis technology is to be launched (1).
These technologies allow sequence determination much quicker and cheaper than the dideoxy chain terminator method presented by Sanger in 1977 (2). The main difference between Sanger sequencing output and the output of the new technologies is an increased read number, associated with a decrease in the length of individual reads.
To achieve high throughput, the new approaches apply different strategies. 454 Life Sciences has adapted pyrosequencing to a microbead format to sequence 400 000 DNA fragments simultaneously, resulting in a per-run dataset of 100 Mbp with reads averaging 250 bp. SOLiD sequencing also uses templates immobilized onto microbeads. Here, the sequence of the template DNA is decoded by ligation assays involving oligonucleotides labeled with different fluorophores. The SOLiD read length is currently 25–35 bases, and 2–3 Gbp of data can be collected during an 8-day run. Solexa sequencing is based on amplifying single molecules attached to the surface of a flow cell to generate clusters of identical molecules, followed by sequencing using fluorophore-labeled reversible chain terminators. Solexa sequencing proceeds a base at a time and read length depends on the number of sequencing cycles. Current Illumina sequencing instrumentation achieves read lengths of 36 bases. The Solexa flow cell is composed of eight separately loadable lanes. Since each lane has a capacity of about 5 million reads, > 40 million reads can be generated in a run of 3 days, equivalent to > 1.3 Gbp.
The adoption of high-throughput sequencing will revolutionize molecular biology research, similar to the invention of the polymerase chain reaction (PCR) twenty years ago (3). 454 pyrosequencing short (∼100 bp) reads generated on Roche GS20 instruments (now replaced by GS FLX) were successfully used for the de novo sequencing of small genomes and BACs as well as for transcript discovery and characterization (4–9). De novo genomic sequencing succeeded even when ultra-short (27–36 bp) reads generated by Solexa sequencing were employed for a small genome (10). For the human genome, ultra-short reads were applied in studies on chromatin analysis (11,12).
However, working with large data sets of short reads involves difficulties, especially due to wrong base calls. To exploit the full prospects of the novel technologies there is the need to know as much as possible about biases in the output data sets, especially with respect to errors. Previous studies focused on the 454 technology (13) or dealt with the prospects of short read sequencing as such (14). Here, we characterize two Solexa read data sets: 12.3 million 36mer reads (trimmed to 32 bases) from the Helicobacter acinonychis genome and 2.8 million 27mer reads from a Beta vulgaris bacterial artificial chromosome (BAC) clone. We analyze these reads and detect biases with respect to error positions, error rates, erroneous base calls and their neighboring bases and single base insertions or deletions. We determine the compensation of erroneous base calls by correct base calls depending on the sequencing coverage. We analyze read start positions, the read coverage along the target sequence, and dependencies of read coverage and local sequence characteristics. Finally, we assess the reliability of quality values for wrong and correct base calls.
METHODS
Solexa sequencing
Helicobacter acinonychis
DNA was fragmented by nebulization as described in the Solexa protocol (www.illumina.com). Beta vulgaris DNA was sheared for 1 h with a UTR200 sonication device (Hielscher Ultrasonics GmbH) at 100% amplitude and 0.5 cycle mode. Fragmented DNA was further processed as described previously (10). Sequencing was carried out by running 27 or 36 cycles, respectively, on the Illumina 1G sequencing instrument. The Goat module (Firecrest v.1.8.28 and Bustard v.1.8.28 programs) of the Solexa pipeline v.0.2.2.3 (for Helicobacter data set) and v.0.2.2.5 (for Beta data set) were used for image deconvolution and quality value calculation. Parameterization was auto-generated by the pipeline (see Supplementary Data for intensity plots and run parameters, i.e. frequency cross-talk matrix, offsets, phasing). Set up configuration was used as installed by Illumina's technical staff. The Helicobacter data set was collected from three lanes of two flow cells. The Beta data set was generated in a single lane from a further flow cell.
Data analysis
We developed various Perl scripts to extract and process information from ELAND output files (Gerald module v.1.27 of the Solexa pipeline) and to find positions of reads that can be aligned more than once to the reference sequence without mismatches (the positions of those reads are not reported by ELAND). We wrote Perl scripts for the detection of deletions and insertions of single nucleotides in otherwise error-free reads and for the analysis of quality values per base call. Plots were generated with the statistical computing environment R (www.R-project.org) or OpenOffice Calc (www.openoffice.org). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. www.R-project.org) or OpenOffice Calc (www.openoffice.org).
Data availability
Solexa read data are available from the SHARCGS project website at http://sharcgs.molgen.mpg.de.
RESULTS
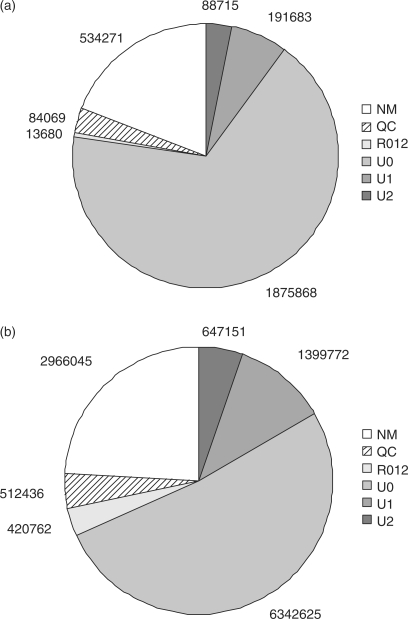
We previously generated 12 288 791 36mer reads from Helicobacter acinonychis on an Illumina 1G sequencing device (10). The Helicobacter genome is 1.55 Mbp in size and has a GC content of 38%. A high-quality reference sequence for Helicobacter is available (GenBank NC_008229) (15). We ran the ELAND software on the read data set (trimmed by the last four bases, because ELAND processes the first 32 bases only) and selected the 8 389 548 32mer reads that ELAND reported to be uniquely matched against the Helicobacter reference sequence with zero, one or two mismatches (labeled U0, U1 or U2, respectively, see Figure 1b). Additionally, we generated a 27mer read data set for the sugar beet (Beta vulgaris) bacterial artificial chromosome (BAC) clone ZR-47B15. The data set consists of 2 788 286 reads, 2 156 266 of which were labeled U0, U1 or U2 in the ELAND output (Figure 1a). The Sanger reference sequence in finished quality of this BAC insert consists of 10 9563 bases with 34.85% GC (Dohm et al., manuscript submitted for publication). For all uniquely matched reads, ELAND reports the match position in the reference sequence as well as the error position(s) in the read.
Figure 1.
Pie charts of the read analysis with ELAND. The ELAND categories are: QC: no matching done because of low quality of the read (more than two positions with quality score = −5), NM, no match found; U0, unique exact match found; U1, unique match with one error; U2, unique match with two errors; R0, multiple exact matches found; R1, multiple matches with one error; R2, multiple matches with two errors. The categories R0, R1, R2 are shown as a single entity. (a) ELAND categorizations for 27mer reads from Beta vulgaris clone ZR-47B15 (2 788 286 in total). (b) ELAND categorizations for 32mer reads from Helicobacter acinonychis (12 288 791 in total, trimmed by the last four base calls of the original 36mer data).
Start positions of reads and read distribution on the target sequence
The preparation of Solexa sequencing libraries involves the fragmentation of the DNA, followed by the adaptor ligation, pre-amplification for material enrichment and amplification within the flow cell prior to sequencing. In order to detect whether the steps preceding sequencing show biases, we analyzed the first bases of a read and the bases that flank the read start position on either side. Of all possible 27mer tuples (Beta) and 32mer tuples (Helicobacter), 99.8 and 98.8% are unique, respectively. We therefore assume that potential biases are representative for the data set.
We calculated the frequency of 2- to 10-base tuples enclosing the starting point for 8 389 548 uniquely matched Helicobacter reads and for 2 156 266 uniquely matched Beta reads relative to the frequency of these tuples in the reference sequences. Since the bases in the reads are subject to errors, we used for both sides the bases of the corresponding region in the reference sequence.
A general sequence bias for the immediate vicinity of the read start position could not be deduced from the two data sets. The results for the Beta data set did not suggest any tendencies (Supplementary Figure 1a). The results for the reads from Helicobacter showed a weak tendency towards T being the most frequent base call to the left and to the right of the read start position (Supplementary Figure 1b). Since two different fragmentation methods were used, sonication for Beta and nebulization for Helicobacter, the results may indicate method-inherent properties.
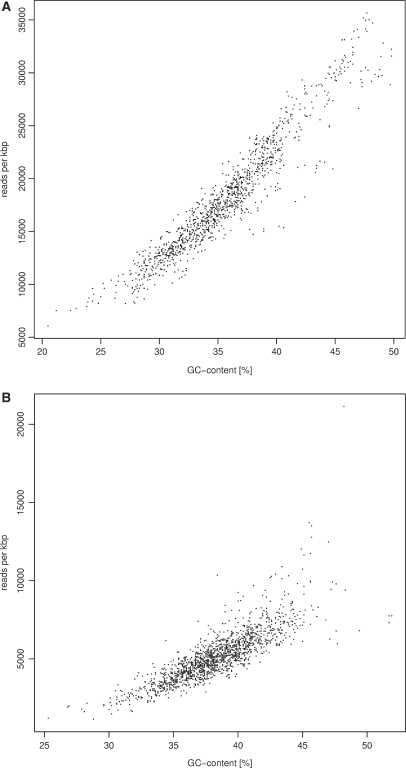
However, by analysing sequence characteristics and number of reads starting in a sliding window of 1 kbp in width, we found a correlation of read coverage and GC content in both data sets (Figure 2). In regions of elevated GC content the number of reads was increased. For instance, windows with a GC content of 40% contain almost twice as many reads as windows with 30% GC in the Beta data set. Thus, while the vicinity of 10 bp was not sufficient to detect a conclusive bias for read starting points, there is a strong preference towards GC-rich regions in 1 kbp sliding windows. Since both templates show the correlation of read coverage and GC content, the shift to GC rich regions seems to be a general feature of the current pre-sequencing procedure. A similar finding was reported by Hillier et al. (16).
Figure 2.
Correlation of the Solexa read coverage and GC content. (a) 27mer reads generated from Beta vulgaris BAC ZR-47B15 (b) 32mer data set from the Helicobacter acinonychis genome. Each data point corresponds to the number of reads recorded for a 1-kbp window (shift of 100 bp in Beta and 1 kbp in Helicobacter).
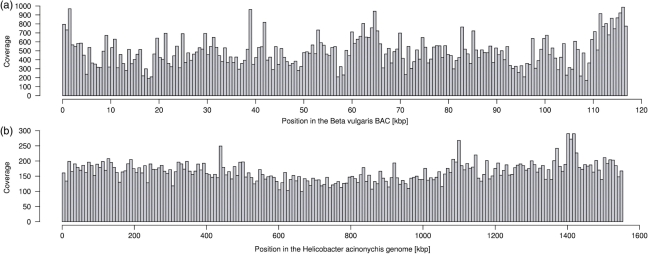
The overall coverage considering matching reads only is 165-fold in the Helicobacter data set (185-fold for 36mer reads) and 465-fold in the Beta data set. The distribution of matching reads along the reference sequences is shown in Figure 3. We calculated the read depth in windows of size 7.77 kbp for Helicobacter (Figure 3a) and of size 0.58 kbp for Beta (Figure 3b). The coverage varied by a factor of 13 and 3.8, respectively, ranging from 49- to 652-fold for Helicobacter and from 238- to 897-fold for Beta (Table 1). We tested whether the distributions shown in Figure 3 are compatible with a uniform distribution of reads across the target sequences. We have applied a χ2-test (goodness of fit) to reject the hypothesis that reads have the same probability to fall into equally sized regions of the target sequence (P < 1e−10 even when dividing target sequences in only five regions).
Figure 3.
Distribution of Solexa reads along the reference sequences considering unique match positions reported by ELAND (zero, one or two mismatch bases) and reads with more than one match position (no mismatch bases) detected with a Perl script. (a) Read distribution along the Beta vulgaris BAC sequence (with cloning vector pBeloBACII). 2 166 892 27mer reads were matched against the finished sequence (enclosed by the cloning vector,∼117 kbp in total). The read coverage was calculated in 200 consecutive 0.58 kbp windows. (b) Read distribution along the 1.55 Mbp Helicobacter genome, based on 8 700 113 32mer reads. The local coverage is shown in 200 consecutive windows of 7.77 kbp.
Table 1.
Proportion of reference sequence and coverage ranges (based on ELAND U0, U1, U2, R0 matched reads and reads with single indels)
| Beta |
Helicobacter |
||
|---|---|---|---|
| Coverage | BAC (%) | Coverage | Genome (%) |
| 200–300 | 4.27 | <100 | 3.53 |
| 300–400 | 23.93 | 100–150 | 26.06 |
| 400–500 | 25.64 | 150–200 | 42.28 |
| 500–600 | 23.93 | 200–250 | 21.49 |
| 600–700 | 12.82 | 250–300 | 4.44 |
| 700–800 | 4.27 | 300–350 | 1.29 |
| 800–900 | 5.13 | >350 | 0.90 |
There is a number of ‘gap’ positions in the target sequences where no read starts from. However, since there are no gaps larger than read length all positions of the target sequence are covered (Supplementary Table 1).
Distribution of error positions along reads
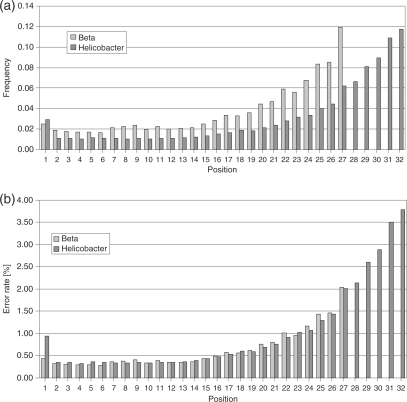
We selected all ELAND U1 and U2 reads, i.e. 28 0173 Beta reads and 2 046 923 Helicobacter reads (cf. Figure 1), to analyze the occurrence of errors per position. We performed two types of calculations. Firstly, we calculated the fraction of wrong base calls at each read position considering wrong base calls only. Secondly, we calculated per-base error rates, i.e. the fraction of wrong base calls per position considering all base calls. The result is shown in Figure 4. The number of occurrences of wrong bases is increased at the first position. Rising from the lowest error rate at the second position, the highest error rate is observed at the last positions of the read [similar observation reported in (16)]: 2.5 and 2.9% of the errors in the data sets of Beta and Helicobacter, respectively, were found at read position 1, and 11.8% of errors were recorded at the last read position (position 27 in the Beta data set and position 32 in the Helicobacter data set, Figure 4a). The per-base error rates range from 0.3% to 3.8% (Figure 4b) resulting in an average error rate of 0.6% for the Beta data set and 1.0% for the Helicobacter data set. Note that only uniquely matched reads with less than three substitution errors are considered.
Figure 4.
Frequency of wrong base calls in Solexa reads depending on the position along the read (27mer reads from Beta vulgaris and 32mer reads from Helicobacter). (a) Error frequency per position calculated from considering wrong base calls only. The highest error frequency is observed at the read 3′ end. (b) Per-base error rates (overall error frequency per position considering all base calls).
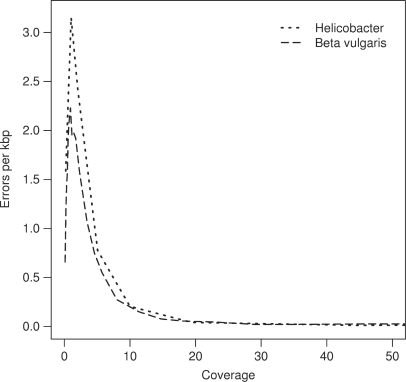
In re-sequencing projects, sequencing errors can be compensated by high-coverage sequencing. In a re-sequencing project, the reads are aligned against a reference sequence. Wherever a mismatch between sequencing data and the reference is observed, a polymorphism is postulated. In order to avoid spurious detection of polymorphisms due to sequencing errors, a consensus between several reads at each position of the reference is common practice. Here, we simulate re-sequencing at different depth by randomly choosing the appropriate number of reads from our two data sets and counting wrong and correct base calls [five (Helicobacter) or ten (Beta) simulations per data point]. An error was considered as compensated when at least one correct base call for the same position existed. A correct base call and the reference sequence hold the majority over one wrong base call, i.e. x wrong base calls at the same position can be compensated by x correct base calls (plus reference sequence). We plotted the dependency of sequencing coverage and error compensation in Figure 5 (range of simulation results: see Supplementary Figure 2). Increasing the sequencing coverage results in a rapid decrease of uncompensated errors. At a coverage of 20-fold the average number of errors per kilo base pair is close to zero and does not decrease any further. However, such estimates are likely to change with improvements of the sequencing technology, as less coverage will be sufficient for reduced error rates.
Figure 5.
Compensation of sequencing errors by deep sequencing in re-sequencing projects. The average number of errors per kbp is shown for different levels of coverage. For coverages below 2, reads are unlikely to overlap and compensation of sequencing errors is rare (thus, sequencing errors accumulate when the coverage is increased). For coverages above 3-fold the number of uncompensated errors drops rapidly with the increase of coverage.
Analysis of reads containing two errors
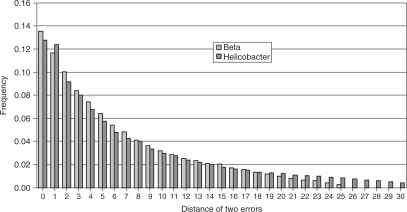
ELAND reported 88 753 reads containing two errors in the Beta data set, corresponding to 4.1% of all uniquely matched reads. In Helicobacter, 647 151 reads contained two errors (7.7% of all uniquely matched reads). We analyzed the distance between erroneous bases and found a preference for small distances between errors (Figure 6). In 25% of reads that contained two errors the erroneous bases were either at adjacent positions or separated by one base. This observation does not contradict the assumption that errors occur independently according to their position-specific probability. The heatmap in Supplementary Figure 3 illustrates the occurrence of two errors relative to the positions in the read. As expected from the per-base error rates, two-error occurrences are concentrated at the 3′ end of reads and are therefore close together. In addition, error pairs also occur with increased frequency at read positions 1 and 2. We provide even stronger evidence for the independence of error positions in two-error reads in Supplementary Figure 4.
Figure 6.
Distance between two errors on a read in the Helicobacter and Beta vulgaris data sets. ‘0’ indicates that the erroneous base calls are next to each other.
Although error positions seem to be independent in reads with two errors, there is evidence that errors accumulate in reads more easily than expected. We deduce this from the ratios of the observed and expected number of reads containing one and two errors respectively: Given the determined error rates per position (for the Helicobacter data set) we expect 3.5 times more correct reads (U0) than reads with one error (U1), but we observe 4.5 times more U0 than U1; we expect 19.8 times more correct reads than reads with two errors (U2), but we observe 9.8 times more U0 than U2. Thus, there are fewer U1 reads than expected and more U2 reads than expected compared to the U0 reads. This tendency is confirmed in the Beta data set (data not shown) and suggests dependencies in the occurrence of errors.
Analysis of error base sequence context
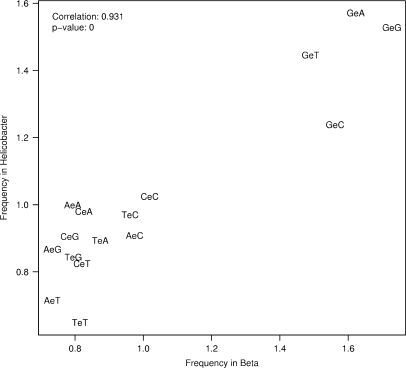
In order to find sequence composition preferences close to wrong base calls, we analyzed the sequence tuples flanking error positions. Since errors at position 1 do not have preceding bases and errors at the last position do not have subsequent bases in the read, we used the corresponding segment of the reference sequence for the analysis. This also avoids analysing wrong base calls in the error-prone read sequences close to the error position under consideration. The sequence composition before the read start is not considered to be responsible for an error at position 1 because this part of the source sequence is not part of the sequenced fragment. However, the bases following the end of the read could have an influence on the base calling. We decided to treat all error positions in the same manner by looking up the flanking bases in the reference sequence. As reference tuples we did not consider all tuples in the reference sequence but all tuples in all uniquely matched reads (taken from the reference sequence and adding 5 bases before and after the corresponding read segment). This is to keep the analysis clean from the read coverage bias towards GC-rich regions of the reference sequence. We calculated the relative frequencies for 3- to 11-base tuples enclosing the error at the middle position and generated sequence logos for Beta and Helicobacter separately. To visualize the general trend we show the 3- and 5-base tuple results in scatterplots with the tuple frequencies for both data sets (Figure 7 and Supplementary Figure 5). All 3-base tuples starting with a G are clearly dominant in both data sets with G-error-G and G-error-A being the top candidates (Figure 7). Error enclosing tuples starting with A or T are underrepresented, and error enclosing tuples starting with C are as frequent as in the reference tuples. The least frequent base after an error is T, being the third base in the three least frequent tuples A-error-T, T-error-T and C-error-T. The trend of G being the most frequent base before an error is preserved and even more emphasized in the scatterplot with 5-base tuples (Supplementary Figure 5). Here, Gs are still the preferred bases before an error, and least frequently we see errors enclosed by Ts. In 35 and 32% of cases (Beta and Helicobacter, respectively), the error position was preceded by G.
Figure 7.
Sequence context of wrong base calls in Solexa reads from Helicobacter acinonychis and Beta vulgaris, considering one base upstream and downstream of the wrong base calls. An ‘e’ indicates the substituted base. The scatterplot shows the correlation of the relative frequencies (relating the frequency of 3-tuples at error positions to the frequency of all 3-tuples in the reads) for the two data sets.
Analysis of base substitution errors in Solexa reads
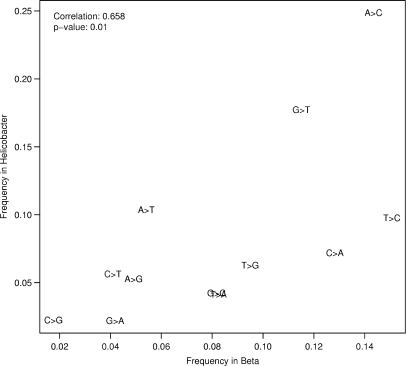
Twelve substitution errors (eight transversions and four transitions) are possible during a base call. We compared the wrong base calls in the reads to the base in the reference sequence and found that base substitution errors in Solexa reads are not equally frequent. Generally, the two data sets show similar tendencies (Figure 8, Table 2). The most frequent base to be changed into is a C, preferentially substituting T or A in the Beta data set and A in the Helicobacter data set (T in Helicobacter as well but at a lower frequency). Consistently for both data sets, C > G transversions are the least frequent substitution errors. The top three types of substitution errors account for >53% of all substitution errors found in the Helicobacter read data set (the transversions A > C, G > T and A > T) and for >42% of all substitution errors found in the Beta data set (the transition T > C and the transversions A > C and C > A).
Figure 8.
Frequency of substitution errors in the Helicobacter acinonychis and Beta vulgaris Solexa read data sets.
Table 2.
Base substitution frequencies in the Beta and Helicobacter read data sets
| From |
|||||
|---|---|---|---|---|---|
| Into | A | C | G | T | Any |
| Beta | |||||
| A | – | 0.13 | 0.04 | 0.08 | 0.25 |
| C | 0.14 | – | 0.08 | 0.15 | 0.38 |
| G | 0.05 | 0.02 | – | 0.09 | 0.16 |
| T | 0.05 | 0.04 | 0.12 | – | 0.21 |
| Any | 0.25 | 0.19 | 0.24 | 0.33 | |
| Helicobacter | |||||
| A | – | 0.07 | 0.02 | 0.04 | 0.14 |
| C | 0.25 | – | 0.04 | 0.10 | 0.39 |
| G | 0.05 | 0.02 | – | 0.06 | 0.14 |
| T | 0.10 | 0.06 | 0.18 | – | 0.34 |
| Any | 0.41 | 0.15 | 0.24 | 0.20 | |
Insertions and deletions in the Solexa read data sets
The ELAND algorithm is limited to the alignment of reads containing up to two substitution errors. In addition to the reads matched by ELAND to the reference sequence there is a substantial amount of unmatched reads (Figure 1). Some of the unmatched reads contain more than two sequencing errors, but another reason for unmatched reads may be the occurrence of insertions and deletions (indels). We implemented a Perl script to find single nucleotide indels within reads without considering additional substitution errors. We observed a very low rate (<0.01%) of indel errors: 323 of 2.8 million Beta reads contain a single nucleotide insertion and 1258 Beta reads contain a single nucleotide deletion; 1215 and 2284 insertion and deletion errors, respectively, were found in the Helicobacter data set. Further inspection of the data revealed that >25% of base insertions occurred in homopolymer tracts of four or more nucleotides. However, no clear trend could be detected for deletions. With respect to the positions within reads where indels occur there is a slight accumulation of such events at internal positions of the reads. No bias for inserted or deleted bases could be detected (data not shown). The reason for detecting this type of error might be sequencing errors in the reference sequences. For Helicobacter, two erroneous insertions in the Sanger sequence were reported (10). Approximately 10% of Illumina reads with one deletion match to these two positions.
Assessment of quality values
The Solexa base caller Bustard reports the quality of each base call by estimating a quality score similar to the phred score based on the image output without considering the reference sequence. More precisely, Bustard estimates the probability P of a base call to be wrong and reports the corresponding quality score Q = −10 log10 (P/(1−P). Thus, a quality score Q = 40 roughly corresponds to an expected error probability of P = 0.01%, and Q = 0 corresponds to an expected error probability of P = 50%. Based on uniquely matched reads reported by ELAND, we have determined 7 201 633 correct and 36 9113 wrong base calls in the Beta data set as well as 70 995 154 correct and 2 694 074 wrong base calls in the Helicobacter data set. We have extracted the corresponding quality scores from Bustard output files and computed observed error rates per quality score. Table 3 shows a comparison of the expected and observed error rates for the base call score quality in our two data sets: Theoretical values underestimate the error probability for high quality values and overestimate the error probability for low quality values.
Table 3.
Observed and expected error rates for base calls of different quality values in the Beta and Helicobacter data sets
| Score | Beta (%) | Helicobacter (%) | Expected (%) |
|---|---|---|---|
| Q = 40 | 1.39 | 0.43 | 0.01 |
| Q = 30 | 3.55 | 1.06 | 0.10 |
| Q = 20 | 5.21 | 1.70 | 0.99 |
| Q = 10 | 9.68 | 4.40 | 9.09 |
| Q = 0 | 39.65 | 28.68 | 50.00 |
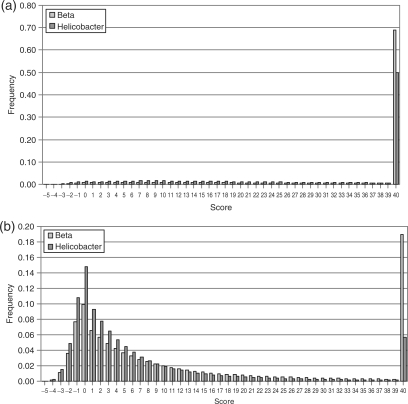
We also collected the quality values for bases reported by ELAND as correct separately from quality values for bases reported as wrong (matching reads only). Figure 9 shows the results in separate histograms. The fraction of the best quality value is increased for correct base calls and low quality values show low fractions as expected. However, there is a substantial amount of high quality values for wrong base calls. Six percent of all wrong base calls in Helicobacter and 19% of all wrong base calls in Beta have Solexa quality scores Q = 40.
Figure 9.
Histograms of base quality values for all correct base calls (a) and all wrong base calls (b) in the Beta and Helicobacter data sets.
DISCUSSION
We have characterized two Solexa read data sets derived from a bacterial genome (Helicobacter acinonychis) and from a Beta vulgaris BAC clone. We looked for systematic biases of read start positions, recorded the error positions and the error frequency along the read length, examined the distribution of reads along the reference sequences, investigated substitution preferences, and assessed the reliability of quality scores. The generalization of our observations may be limited by the fact that the presented data relates to a single Illumina 1G Analyzer. However, since three different flow cells, four different lanes and two different library preparations for two different target sequences are involved we assume that our consistent observations reflect relevant aspects of the current state of Solexa technology.
To explain the observed biases, a comprehensive knowledge of the Solexa technology is necessary. The source DNA is fragmented randomly, and adapter molecules are ligated at both ends of each fragment followed by pre-amplification for enrichment of the material. The DNA fragments are melted, and the single strands are trapped inside the flow cell which is covered by a dense lawn of primers. Subsequent local amplification leads to the formation of clusters of approximately 1000 identical molecules per square micrometer. The base incorporation is started by adding primers, polymerase and the four flourophore-labeled deoxynucleotidetriphosphates. The dNTPs act as reversible terminators, i.e. only a single base is added per molecule in each cycle. The cluster fluorescence is measured to identify which base has been incorporated. A green laser identifies the incorporation of the bases G and T, and a red laser identifies the bases A and C. Two different filters are used to distinguish between G/T and A/C, respectively. After signal detection, the fluorophore and the terminating modification of the nucleotide are removed.
In the context of this work we could not detect a general sequence bias for the immediate vicinity of read start positions, indicating that the fragmentation step is essentially random. Two different methods of fragmentation were used but potential trends for each method were rather weak. However, we did observe a strong correlation between GC richness and read coverage, with the read density being increased in regions of elevated GC content. Uneven coverage of the target genome is well known from Sanger sequencing, but this effect has been attributed to a cloning bias in the underlying plasmid shotgun libraries. Since the propagation of Solexa templates in E. coli is avoided, the cloning procedure cannot be a reason for the read distribution bias. Another reason for biases towards GC-rich sequences could be the different melting behaviour of double-stranded DNA. AT-rich DNA segments denature at lower temperatures than GC-rich DNA. In Serial Analysis of Gene Expression (SAGE) and Massively Parallel Signature Sequencing (MPSS) data sets, the even more dissimilar melting behaviour due to the shortness of the templates (14–21 bases) is supposed to be the reason for the observed bias towards GC-rich sequences (17,18). Since the fragment libraries for Solexa sequencing are larger having sizes of 120–170 bp, denaturation of the DNA is less likely to occur. However, potential denaturation effects are most likely to occur at the adapter-free state of the DNA molecules. Once adapters have been ligated to the fragments, the DNA is no longer sensitive to denaturation. According to the protocol for library generation, we performed a PCR enrichment step. This step might introduce bias as well. However, described PCR-introduced biases have opposite effects, i.e. sub-optimal amplification of GC-rich templates (19).
Solexa sequencing base call errors occur preferentially at the 3′ end of reads. For the accumulation of errors towards the end of the read, we consider the following scenario. All immobilized DNA molecules in a cluster are supposed to give the same signal at a time because each cycle usually adds exactly one base to the growing double strand along the template in a cluster. Whenever single DNA molecules in the cluster are not elongated properly, the overall cluster signal suffers from interference by molecules which are out of phase. Failures in the deprotection (i.e. removal of the terminator group) of incorporated bases can lead to this type of interference. Without deprotection, the next base cannot be added and all bases of following cycles are shifted by one position in this DNA molecule. Thus, with increasing cycle numbers shifts accumulate leading to an increased error rate in later cycles. Additionally, incomplete removal of the fluorophore results in more than one fluorescing base in the following cycle and interferes with signal interpretation as well.
Sequence tuples before an error position are preferentially G-rich. This result suggests that G might be preferentially subject to an incomplete step of deprotection and fluorophore removal.
The way signals are detected offers an explanation for the observed preferences of base substitution errors. The green laser is used to detect G and T at the same time. The brightness of G is enhanced by the use of a filter to distinguish G versus T incorporation. Similarly, A and C are detected by the red laser and distinguished by using different filters. The transversions G > T and A > C are among the most frequent base substitutions in both of our data sets, suggesting that these base call errors arise because of insufficient discrimination of the respective base emission spectra.
The quality of Sanger sequences is affected by the presence of GC-rich sequences as well, but also by polyA or polyT homopolymer runs and by repeats causing secondary structure (20). In a recent survey on the accuracy of 454 pyrosequencing, Huse et al. (13) estimated that 39% of all errors that had passed GS20 quality filtering occur in homopolymer length detection. This is certainly linked to the fact that 454 technology sequences homopolymer tracts in single cycles. In contrast, Solexa sequencing proceeds in a sequential manner, one base at a time. As expected, we have not noted an increased error rate in homopolymer runs of Solexa reads. The Solexa one-by-one sequencing procedure is probably also ensuring that base insertions and deletions in general occur at very low rates.
Each sequencing technology provides base quality values. Sanger sequencing phred scores are calculated from log-transformed probabilities that a base call is incorrect. For example, a phred score of 30 indicates a probability of 0.1% of a wrong base call. In 454 sequencing, quality scores do not provide a measure that a base at a given position is correct, but merely indicate that homopolymer length has been called correctly. It has been found that GS20 reads with average quality scores above 25 had very few errors (13). Solexa scores and phred scores are calculated differently, but scores above 15 have approximately the same meaning. Our observations, however, suggest that the scores determined by the Solexa software underestimate the true error rate by up to 100 times for high quality values and overestimate the true error rate for low quality values.
Our results lead to several implications for analyses with Solexa reads. Even if an excellent Q-value is determined there is a chance for a wrong base call at this position. Thus, during re-sequencing for SNP discovery, variable positions need confirmation, preferably from the opposite strand. Some types of substitution errors occur more frequently than others. Such SNP candidates should be treated with caution, even in case of confirmation. Especially if one or more Gs precede the putative SNP a wrong base call should be considered. However, in re-sequencing projects most sequencing errors can be discerned from bona fide SNPs by applying high coverages (for our data sets: 20-fold and above). In the context of DNA methylation site detection by shotgun bisulfite sequencing (21), the frequency of erroneous detection of C instead of T and vice versa is of particular interest. T > C transitions could be a source for false positives, while C > T substitutions could cause false negative results. For de novo sequencing, systematic substitution errors may confound the ability of filtering correct reads and increase the chance for misassemblies. The read prevalence in GC-rich regions affects all assumptions inferred from the overall read coverage (e.g. the expected maximum number of missing reads in a row). The identification of confirmed SNPs in AT-rich regions may be hampered by poor sequence coverage. Thus, Solexa-based de novo sequencing as well as re-sequencing activities need to calibrate their sequencing output for achieving accordingly high read coverage of AT-rich regions. The bias in read coverage might also impact the estimation of expression levels of transcripts by ultra-short read sequencing. If no compensation is applied the expression levels of GC-rich transcripts may be overestimated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Aleksey Soldatov for comments on the manuscript. Dmitri Parkhomchuk and Peter Marquardt ran the Solexa pipeline. Funding to pay the Open Access publication charges for this article was provided by the Max-Planck-Gesellschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986;51(Pt 1):263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 4.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung F, Haas BJ, Goldberg SMD, May GD, Xiao Y, Town CD. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber AP, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng P, Tan JJS, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, Srinivasan KG, Perbost C, Du L, et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. SHARCGS, a fast and highly accurate short-read assembly algorithm for de novo genomic sequencing. Genome Res. 2007;17:1697–1706. doi: 10.1101/gr.6435207. 10.1101/gr.6435207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barski A, Cuddapah S, Cui K, Roh T, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 13.Huse S, Huber J, Morrison H, Sogin M, Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whiteford N, Haslam N, Weber G, Prügel-Bennett A, Essex JW, Roach PL, Bradley M, Neylon C. An analysis of the feasibility of short read sequencing. Nucleic Acids Res. 2005;33:e171. doi: 10.1093/nar/gni170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, Keller H, Morelli G, Gressmann H, Achtman M, Schuster SC. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2006;2:e120. doi: 10.1371/journal.pgen.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 17.Siddiqui AS, Delaney AD, Schnerch A, Griffith OL, Jones SJM, Marra MA. Sequence biases in large scale gene expression profiling data. Nucleic Acids Res. 2006;34:e83. doi: 10.1093/nar/gkl404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margulies EH, Kardia SL, Innis JW. Identification and prevention of a GC content bias in SAGE libraries. Nucleic Acids Res. 2001;29:E60. doi: 10.1093/nar/29.12.e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutter GL, Boynton KA. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 1995;23:1411–1418. doi: 10.1093/nar/23.8.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley JM. Automated dye-terminator DNA sequencing. In: Adams MD, Fields C, Venter JC, editors. Automated DNA Sequencing and Analysis. 1st edn. London: Academic Press; 1994. [Google Scholar]
- 21.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Solexa read data are available from the SHARCGS project website at http://sharcgs.molgen.mpg.de.