Abstract
‘Whirly’ proteins comprise a plant-specific protein family whose members have been described as DNA-binding proteins that influence nuclear transcription and telomere maintenance, and that associate with nucleoids in chloroplasts and mitochondria. We identified the maize WHY1 ortholog among proteins that coimmunoprecipitate with CRS1, which promotes the splicing of the chloroplast atpF group II intron. ZmWHY1 localizes to the chloroplast stroma and to the thylakoid membrane, to which it is tethered by DNA. Genome-wide coimmunoprecipitation assays showed that ZmWHY1 in chloroplast extract is associated with DNA from throughout the plastid genome and with a subset of plastid RNAs that includes atpF transcripts. Furthermore, ZmWHY1 binds both RNA and DNA in vitro. A severe ZmWhy1 mutant allele conditions albino seedlings lacking plastid ribosomes; these exhibit the altered plastid RNA profile characteristic of ribosome-less plastids. Hypomorphic ZmWhy1 mutants exhibit reduced atpF intron splicing and a reduced content of plastid ribosomes; aberrant 23S rRNA metabolism in these mutants suggests that a defect in the biogenesis of the large ribosomal subunit underlies the ribosome deficiency. However, these mutants contain near normal levels of chloroplast DNA and RNAs, suggesting that ZmWHY1 is not directly required for either DNA replication or for global plastid transcription.
INTRODUCTION
Plant mitochondrial and chloroplast genomes encode ∼50 and ∼100 products, respectively, most of which participate in basal organellar gene expression or energy transduction. Post-transcriptional events play the dominant role in dictating gene product abundance in both organelles (1). In fact, the two organelles house a similar repertoire of RNA-processing pathways that includes RNA editing, group II intron splicing and endonucleolytic processing. Genetic and bioinformatic analyses suggest that many hundreds of nuclear genes encode organelle-localized nucleic acid binding proteins and influence organellar gene expression (2–5), but only a small fraction of such genes has been studied.
The protein that is the focus of this study, ZmWHY1, came to our attention during our characterization of the chloroplast RNA splicing machinery. Nine nucleus-encoded proteins that are necessary for the splicing of various subsets of the ∼20 chloroplast introns in vascular plants have been reported (6–15). One of the first to be characterized, CRS1, is necessary for the splicing of the group II intron in the chloroplast atpF gene (6,9), and binds specifically to that intron in vivo and in vitro (10,11,16). However, the large size of the particles containing CRS1 and atpF intron RNA in vivo, and the fact that CRS1 is not sufficient to promote atpF intron splicing in vitro suggested that additional proteins are involved. We therefore used mass spectrometry to identify proteins that coimmunoprecipitate with CRS1; ZmWHY1 was one such protein.
ZmWHY1 is a member of the ‘Whirly’ protein family, whose orthologs in potato (StWHY1) and Arabidopsis (AtWHY1) were reported to be nuclear transcription factors involved in pathogen-induced transcription (17,18). StWHY1 and AtWHY1 bind single-stranded DNA (ssDNA) in vitro, and StWHY1 adopts a propeller-like structure from which the family acquired its name (17,19). AtWHY1 has also been implicated in telomere binding and maintenance (20). Additional functions for members of the Whirly family were suggested by the fact that GFP fused to each member of the family from Arabidopsis localizes to chloroplasts or mitochondria (21). The copurification of AtWHY1 with a transcriptionally active chloroplast DNA complex (22) and the association of AtWHY2 with mitochondrial nucleoids (23) confirmed that these proteins have organellar functions, but the nature of these functions is not known. Results presented here show that ZmWHY1 plays an essential role in the biogenesis of chloroplasts, that it is associated with DNA from throughout the chloroplast genome and that it interacts in vivo with a subset of chloroplast RNAs that includes the atpF intron. ZmWHY1 enhances atpF intron splicing and influences the biogenesis of the large ribosomal subunit. However, chloroplast DNA and RNAs in ZmWhy1 mutants accumulate to levels similar to those in other mutants with plastid ribosome deficiencies of similar magnitude. These results argue that ZmWHY1 is required neither for chloroplast DNA replication nor directly for global chloroplast transcription.
MATERIALS AND METHODS
Purification of CRS1 ribonucleoproteins and mass spectrometry
Purification of CRS1 ribonucleoprotein particles and mass spectrometry were performed as described for CAF1 and CAF2 particles in (12). The antibody to CRS1 was described previously (11).
Plant material
Our collection of Mu transposon-induced nonphotosynthetic maize mutants (http://chloroplast.uoregon.edu/) was screened by PCR to identify insertions in ZmWHY1, using methods described in (24) and a ZmWhy1-specific primer (5′-CGGCGGCCTTTCTGGAGGA-3′) in conjunction with a Mu terminal inverted repeat primer (5′-GCCTCCATTTCGTCGAATCCCG-3′). The alleles were tested for complementation by crossing phenotypically normal siblings (+/+ or +/−) from ears segregating each allele. Seventy-four ears were recovered, 36 of which segregated chlorophyll-deficient mutants. Other mutants used in this work include iojap (25), hcf7 (26) and crs1 (6). The inbred line B73 (Pioneer HiBred) was used as the source of wild-type tissue for coimmunoprecipitation, sucrose gradient and chloroplast fractionation experiments. Plants were grown in soil in a growth chamber (16 h light, 24°C)/8 h dark, 19°C). Leaf tissue was harvested ∼9 days after planting.
Generation of recombinant ZmWHY1 for antibody production and binding assays
ESTs representing ZmWhy1 were identified as GenBank accessions DV170433 and DV503865; the corresponding cDNAs were obtained from the maize full-length cDNA project (http://www.maizecdna.org/). The complete cDNA sequence was determined and has been entered in GenBank under Accession EU595664. A ZmWHY1 protein fragment (amino acids 86 to 258) with a C-terminal 6x-histidine tag was expressed in Escherichia coli from pET28b (Novagen), purified by nickel affinity chromatography and used for the production of polyclonal antisera in rabbits at the University of Oregon antibody facility. Full-length mature ZmWHY1 (i.e. lacking the transit peptide) for nucleic acid binding assays was generated by PCR amplification of its coding sequence from the cDNA (primers 5′-TATAGGATCCGCCTCCTCCCGTAAG-3′ and 5′-TATAGTCGACTCACCGACGCCATTC-3′), digestion of the product with BamHI and SalI, and cloning into pMAL-TEV. Subsequent steps in expressing and purifying recombinant ZmWHY1 were as described previously for RNC1 (12).
Chloroplast fractionation and protein analysis
Leaf protein extracts were prepared and analyzed as previously described (27). Chloroplast subfractions were those described by Williams and Barkan (24). For RNAse and DNAse treatment of thylakoid membranes, MgCl2 was added to a thylakoid membrane fraction to a concentration of 15 mM. The sample was divided into three 20 μl aliquots: 1 μl RNAse-free RQ1 DNAse (1 U/µl) (Promega, Madison, WI, USA), 1 μl of RNAse A (1 μg/μl), or 1 µl water was added for the DNAse, RNAse, and mock treatments, respectively. Samples were incubated at room temperature for 30 min and then centrifuged at 4°C at 15 000g for 15 min. The pellet was resuspended in 10 mM Tris–HCl pH 7.5, 2 mM EDTA, 0.2 M sucrose, to a volume equivalent to that of the supernatant. The supernatant and pellet fractions were analyzed by SDS–PAGE and immunoblotting. Sucrose gradient sedimentation of stromal extract was performed as described by Jenkins and Barkan (7); aliquots of stroma were treated with either 3 units RQ1 DNAse or 50 μg/ml RNAse A for 30 min at room temperature prior to centrifugation. Antisera to spinach chloroplast RPL2 and MDH were generously provided by A. Subramanian (University of Arizona) and Kathy Newton (University of Missouri), respectively. The other antibodies were generated by us and described previously (28).
Nucleic acid coimmunoprecipitation assays
One hundred microliter aliquots of stromal extract (∼500 µg of protein) were analyzed by RIP-chip, DIP-chip and slot–blot hybridization using methods described in (29), except that stroma used for RIP-chip assays was treated with DNAse prior to immunoprecipitation (10 units RQ1 DNAse at 37°C for 30 min) and again after purification of nucleic acids from the immunoprecipitation. For DIP-chip assays, RNAse A (100 µg/ml final concentration) was added to stroma prior to immunoprecipitation and residual RNA was removed from the recovered nucleic acids by alkali hydrolysis in 200 mM NaOH at 70°C for 30 min.
Analysis of DNA and RNA
DNA extraction from leaf tissue and Southern blot analysis were performed as previously described (30). Leaf RNA was extracted from the middle of the second leaf of 9-day old seedlings, with Tri Reagent (Molecular Research Center, Cincinnati, OH, USA). RNA gel blot hybridizations were performed as previously described (27). The following PCR fragments were used as probes (residue numbers refer to GenBank accession X86563): atpF int/ex2, 35706-36384; atpF int, 36073-35233; ndhA int, 114941-115730; orf99, 86911-88430; petD ex2, 75539-75895; petN, 19081-19415; psbA, 296-1074; rpl16 ex2, 79519-79920; rpl16 int, 80002-80888; rpoB, 23258-24475; rps12 trans, 69307-69420 and 129636-129861; rps12 int1/ex1, 5’, 68793-69460; rps14, 38500-39020; rrn4.5, 102041-102135; rrn5, 102180-102619; rrn16, 95559-96779; rrn23, 98332-98792; trnA mature, 98038-98075 + 98712-98916; trnG mature, 13245-13292 and 13991-14013; trnG int 13293-13990; trnN, 103066-103137; ycf3 int2/ex3, 43820-44873; ycf3 int, 44383-45116. Poisoned primer extension assays to distinguish mature from precursor RNAs were performed as previously described (9) using the following primers and dideoxynucleotide: rrn23, 5′-CGCAAGCCTTTCCTCTTTT-3′ (ddTTP); rpl2, 5′-GGCCGTGCCTAAGGGCATATC-3′ (ddCTP); rps12, 5′-GGTTTTTTGGGGTTGATAG-3′ (ddCTP). Radioactive gels and blots were imaged with a phosphorimager and analyzed using ImageQuant software (GE Healthcare).
Nucleic acid binding assays
Gel mobility shift assays were performed with the same substrates and procedures as described in Watkins et al. (12) except that the binding reactions contained 150 mM NaCl, 5 mM DTT, 50 µg/ml BSA, 25 mM Tris–HCl pH 7.5, 0.1 mg/ml heparin. Filter-binding assays were based on the procedure of Wong and Lohman (31) with modifications (16). The atpF intron RNA substrate for filter-binding assays was transcribed in vitro by T7 RNA from a PCR product generated with the following primers: atpF forward/T7 promoter, 5′-TAATACGACTCACTATAGGGATGAAAAATGTAACCCATTCTT-3′; atpF reverse, 5′-AATGAAAGTAGATTATCTTGC-3′. The RNA, which included atpF exon 1 and the complete intron, was heated in TE to 90°C for 2 min and then placed on ice immediately prior to its addition to binding reactions (300 mM NaCl, 5 mM DTT, 50 µg/ml BSA and 25 mM Tris pH 7.5, 30°C for 30 min).
Chloroplast run-on transcription assay
The chloroplast run-on transcription assay was performed as described by Mullet and Klein (32–34). The radiolabeled products were hybridized to the following synthetic oligonucleotides (10 pmol/slot) that had been applied with a slot–blot manifold to a nylon membrane: rrn16 5′-CCCATTGTAGCACGTGTGTCGCCCAGGGCATAAGGGGCATGATGACTTGG-3′, rrn23 5′-GGACTCTTGGGGAAGATCAGCCTGTTATCCCTAGAGTAACTTTTATCCGA-3′, trnG 5′-CATCTATGTCAGCTTTTCTGTCTGAATGGAACCAAAGCTCTCCGCTTTCTAGATGC-3′ and CFM3 5′-ATACTCGAGCGAAAAACAGGAGGATTAGTAATCTGGCGATCAGGGACTT CTGTTTCTCTGTACCGGGGAGTAGATTATGATGAACC-3′.
RESULTS
Identification of ZmWHY1 in CRS1 coimmunoprecipitates
To find proteins involved in the splicing of the atpF intron we used mass spectrometry to identify proteins that coimmunoprecipitate with the atpF splicing factor CRS1. Stromal extract was initially fractionated on a sucrose gradient, and the fractions that contained the majority of the CRS1 ribonucleoprotein particles (∼600–700 kDa) were used for immunoprecipitation. The immunoprecipitated proteins were separated by SDS–PAGE, and contiguous gel slices containing proteins between ∼20 and ∼120 kDa were used for in-gel trypsin digests and tandem mass spectrometry. Among the proteins identified was a member of the Whirly protein family (Supplementary Table 1, Supplementary Figure 1A) (17,19). The Whirly protein family in vascular plants includes two orthologous groups (Supplementary Figure 1B). The peptides detected in the CRS1 coimmunoprecipitate identified the protein as a member of the orthologous group designated Why1.
Recovery of ZmWhy1 insertion mutants
To elucidate the function of ZmWHY1 we sought insertion mutants in a reverse-genetic screen of our collection of transposon-induced non-photosynthetic maize mutants (http://pml.uoregon.edu/). Two mutant alleles were recovered (Figure 1): the Zmwhy1-1 allele has a MuDR transposon insertion 35-bp downstream of the predicted start codon and conditions an ivory leaf phenotype; the Zmwhy1-2 allele has a Mu1 or Mu1.7 insertion 38-bp upstream of the predicted start codon and conditions a pale green leaf phenotype. The heteroallelic progeny of complementation crosses (Zmwhy1-1/-2) exhibit an intermediate phenotype (Figure 1B). Homozygous mutant plants die after the development of three to four leaves, as is typical of non-photosynthetic maize mutants.
Figure 1.

Mutant alleles of ZmWhy1. (A) Positions of Mu transposon insertions in the ZmWhy1 gene. Protein coding regions are indicated by rectangles, untranslated regions and introns by lines and Mu transposon insertions by triangles. The sequence of each insertion site is shown below, with the nine nucleotides that were duplicated during insertion underlined. The identity of the member of the Mu family is shown for each insertion (why1-2: Mu1/1.7; why1-1: MuDR), and was inferred from polymorphisms in the terminal inverted repeats. (B) Phenotypes of ZmWhy1 mutant seedlings grown for nine days in soil. Seedlings shown are homozygous for either the Zmwhy1-1 or Zmwhy1-2 allele, or are the heteroallelic progeny of a complementation cross. (C) Immunoblot showing loss of ZmWHY1 in mutant leaf tissue. Total leaf extract (10 µg protein, or dilutions as indicated) were analyzed. The same blot stained with Ponceau S is shown below, with the large subunit of Rubisco (RbcL) marked. hcf7 and iojap are pale green and albino maize mutants with weak and severe plastid ribosome deficiencies, respectively (25,26). The apparently higher levels of ZmWHY1 in Zmwhy1-1 mutants relative to Zmwhy1-2 mutants may be an artifact of the fact that samples were loaded on the basis of equal total protein: the abundant photosynthetic enzyme complexes make up the bulk of the protein in the Zmwhy1-2 extract but are missing in the Zmwhy1-1 extract, causing other proteins to appear over-represented. (D) RNA-dependent coimmunoprecipitation of ZmWHY1 with CRS1. Prior to immunoprecipitation, stroma was treated with DNAse or RNAse, or incubated under similar conditions without added nuclease (Mock). The stroma was then subjected to immunoprecipitation with the antibody named at top. Presence of CRS1 in the immunoprecipitation pellets was tested by immunoblot analysis with CRS1 antibody.
A polyclonal antibody was raised to a recombinant fragment of ZmWHY1. This antibody detected a leaf protein whose size is consistent with that anticipated for ZmWHY1 (∼25 kDa) (data not shown) and whose abundance is reduced in ZmWhy1 mutants (Figure 1C), indicating that the detected protein is ZmWHY1. The ZmWHY1 antibody coimmunoprecipitated CRS1 (Figure 1D) from chloroplast extract, confirming that CRS1 and ZmWHY1 associate with one another. This association was disrupted by treatment with ribonuclease A (Figure 1D), indicating it is mediated by RNA. Results described below show that atpF intron RNA, which was shown previously to associate with CRS1 in vivo (10,11), mediates the CRS1/ZmWHY1 interaction.
ZmWHY1 partitions between the chloroplast stroma and thylakoid membrane, to which it is bound in a DNA-dependent manner
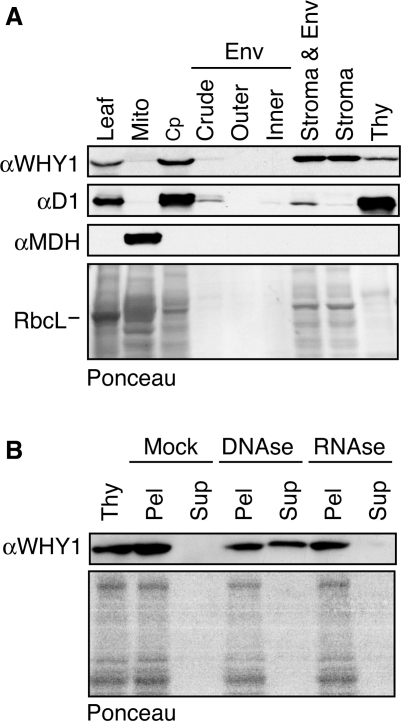
ZmWHY1 was initially recovered from chloroplast stroma and is predicted to localize to chloroplasts by both the TargetP (35) and Predotar (36) algorithms. Immunoblot analysis of proteins from leaf, chloroplasts and mitochondria confirmed that ZmWHY1 is found in chloroplasts and that it is absent, or found at only very low levels, in mitochondria (Figure 2A). Analysis of chloroplast subfractions showed that ZmWHY1 is recovered in both the stromal and thylakoid membrane fractions (Figure 2A); this behavior differs from that of other chloroplast gene expression factors in the same fractionated chloroplast preparation (PPR2, PPR4, RNC1, CAF1, CAF2, CFM2), all of which were found solely in the stromal fraction (8,10,12,15,24).
Figure 2.
Intracellular localization of ZmWHY1. (A) Immunoblots of extracts from leaf and subcellular fractions. The samples in the chloroplast (Cp) and chloroplast subfraction lanes are derived from the same initial number of chloroplasts. The same blot was probed to detect a marker for thylakoid membranes (D1) and mitochondria (MDH). These subcellular fractions are the same as those shown previously for localization of RNC1, where a marker for the envelope membrane fraction was also presented (12). Env; envelope; Mito; mitochondria; Thy; thylakoid membranes. The blot stained with Ponceau S is shown below, with the band corresponding to RbcL marked. (B) DNA-dependent association of ZmWHY1 with thylakoid membranes. The thylakoid membrane fraction was treated with DNAse, RNAse or incubated under similar conditions without added nuclease (Mock). Thylakoid membranes were then pelleted by centrifugation. Pellet (Pel) and supernatant (Sup) fractions were brought to equal volumes, and an equivalent proportion of each fraction was analyzed on an immunoblot probed with ZmWHY1 antibody. The same blot stained with Ponceau S is shown below.
It seemed possible that ZmWHY1 associated with the thylakoid membrane via a DNA tether because chloroplast nucleoids are membrane-associated (37) and AtWHY1 copurified with a chloroplast chromosome preparation (22). In support of this possibility, treatment of the thylakoid membrane fraction with DNAse released a portion of the membrane-associated ZmWHY1 to the soluble fraction (Figure 2B), whereas RNAse treatment had no effect. These results indicate that ZmWHY1 is associated with the thylakoid membrane, at least in part, via an association with chloroplast DNA.
ZmWHY1 is associated with large RNA- and DNA-containing particles
The observations that RNAse and DNAse disrupt ZmWHY1's association with CRS1 and the thylakoid membrane, respectively, suggested that ZmWHY1 associates with both RNA and DNA. To further explore the nature of these interactions, the effects of RNAse or DNAse treatment on the sedimentation properties of ZmWHY1 were investigated (Figure 3). When untreated stroma was sedimented through a sucrose gradient, ZmWHY1 was detected in two peaks (∼400–500 kDa and ∼600–700 kDa) and was also found in pelleted material at the bottom of the gradient. The 600–700 kDa peak coincides with the peak of CRS1 in the same gradient. Treatment of stroma with DNAse reduced the amount of ZmWHY1 in the pellet and in the ∼400–500 kDa peak, but did not reduce its recovery in the 600–700 kDa peak. Conversely, RNAse treatment specifically reduced the recovery of ZmWHY1 in the 600–700 kDa peak. These results together with those described above suggested that ZmWHY1 resides in two types of complexes: one that includes CRS1 and RNA, and the other that includes DNA.
Figure 3.
Sucrose-gradient sedimentation demonstrating that ZmWHY1 is associated with DNA- and RNA-containing particles in chloroplast stroma. Stromal extract was treated with DNAse or RNAse, or incubated under similar conditions without nuclease (Mock), and then sedimented through a sucrose gradient. An equal volume of each gradient fraction was analyzed by probing immunoblots with the antibodies indicated to the left. RPL2, a protein in the large ribosomal subunit, marks the position of ribosomes. Shown below is the blot of the mock-treated fractions stained with Ponceau S, with the RbcL band marked to illustrate the position of Rubisco. The Ponceau S stained blots of experiments involving the DNAse- and RNAse-treated extracts looked similar (data not shown).
Coimmunoprecipitation assays demonstrate that ZmWHY1 associates with a subset of plastid RNAs that includes the atpF intron
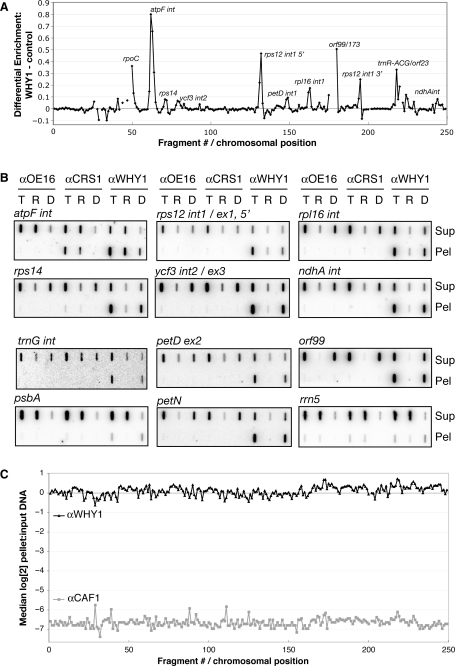
The RNA-dependent association between ZmWHY1 and CRS1 suggested that ZmWHY might associate with CRS1's RNA ligand, the atpF intron. However, the albino phenotype conditioned by the Zmwhy1-1 allele indicated that this could not be ZmWHY1's sole ligand, because mutations in crs1 that completely block atpF intron splicing result in a much less severe chlorophyll deficiency (11). To identify RNAs that associate with ZmWHY in vivo we used a ‘RIP-Chip’ assay (38) as an initial screen: RNAs that coimmunoprecipitate with ZmWHY1 from stromal extract were identified by hybridization to a tiling microarray of the maize chloroplast genome. To ensure that DNA associated with ZmWHY did not contribute to the signal, the extract was treated with DNAse prior to immunoprecipitation, and the nucleic acids recovered from the immunoprecipitation pellet and supernatant were again treated with DNAse. RNAs recovered from the pellet and supernatant were then labeled with red- or green-fluorescing dye, respectively, combined, and hybridized to the microarray. Two replicate immunoprecipitations were analyzed in this manner. To highlight sequences that are enriched in the ZmWHY1 immunoprecipitations, the median enrichment ratio [red(F635)/green (F532)] was plotted according to chromosomal position, after subtracting the median enrichment ratios from control assays (Figure 4A). The results highlight the atpF intron as the major RNA ligand of ZmWHY. The results suggested, in addition, an association between ZmWHY1 and RNAs derived from several other loci (e.g. rps14, rpoC, ycf3, rps12, petD, rpl16, orf99). When the same data were analyzed by considering only the signal in the immunoprecipitation pellets, the results were similar (Supplementary Figure 2A).
Figure 4.
Identification of chloroplast RNAs and DNAs that coimmunoprecipitate with ZmWHY1. (A) RIP-chip data showing coimmunoprecipitation of specific chloroplast RNAs with ZmWHY1. The ratio of signal in the pellet versus the supernatant (F635/F532) for each array fragment is plotted according to chromosomal position. The plot shows the median values for replicate spots across two replicate ZmWHY1 immunoprecipitations after subtracting the corresponding values for two negative control immunoprecipitations (one with OE16 antibody and one without antibody). The same data are plotted using an alternative analysis method in Supplementary Figure 2B; the atpF intron is the most prominent peak in both analyses, but the proportional sizes of other peaks vary depending on the comparison used. (B) Validation of RIP-chip and DIP-chip data by slot–blot hybridization. Stroma was pretreated with DNAse or RNAse or left untreated and then subjected to immunoprecipitation with the antibodies indicated at the top. Nucleic acids purified from the pellets (Pel) and supernatants (Sup) were further treated with DNAse or alkali to remove residual DNA or RNA. The resulting total nucleic acids (T), RNA (R) or DNA (D), were applied to a nylon membrane with a slot blot manifold and hybridized with probes specific for the indicated sequences. Slots contained 1/9th or 1/27th of the nucleic acid recovered from each pellet or supernatant, respectively. (C) DIP-chip data showing genome-wide enrichment of chloroplast DNA in ZmWHY1 immunoprecipitations. Stroma was treated with RNAse prior to immunoprecipitation. Nucleic acids were extracted from the immunoprecipitation pellets and from total input stroma, and subjected to alkali hydrolysis to remove residual RNA prior to analysis by microarray hybridization. The median log2-transformed ratio of fluorescence in the pellet versus the input is plotted for replicate array fragments as a function of chromosomal position.
To validate candidate RNA ligands to emerge from the RIP-chip experiment, RNAs that coimmunoprecipitate with ZmWHY1 were analyzed by slot–blot hybridization using probes corresponding to each RIP-chip peak (Figure 4B). RNAs purified from immunoprecipitations with antibodies to CRS1 and OE16 (a protein that does not bind RNA) were analyzed as controls. As for the RIP-chip assays, the stromal extract was treated with DNAse prior to immunoprecipitation and the nucleic acids recovered from the immunoprecipitation were treated again with DNAse. The results largely recapitulated the RIP-chip data (see lanes ‘R’ in Figure 4B): atpF intron RNA was confirmed to be strongly enriched in ZmWHY1 immunoprecipitations, whereas RNAs from the psbA and petN loci, which did not appear as positives in RIP-chip assays, likewise scored negative in the slot–blot hybridization assay. Coimmunoprecipitation with ZmWHY1 was also confirmed for RNAs from the rps12, ndhA, rpl16, ycf3 and rps14 loci; as predicted by the RIP-chip data, their degree of enrichment was less than that for the atpF intron. However, RNAs from the petD, orf99 and rrn5 loci, which appeared as minor peaks in the RIP-chip data, did not appear to be enriched based on the slot-blot data; the orf99 transcript is of very low abundance, however, so it may be enriched in the pellet at levels that are too low to detect. These issues notwithstanding, the RIP-chip and slot–blot hybridization data together show that ZmWHY1 associates with a subset of RNAs in chloroplast extract, and that the atpF intron is its major RNA ligand.
DNA from throughout the plastid genome coimmunoprecipitates with ZmWHY1
The effects of DNAse-treatment on ZmWHY1's association with the thylakoid membrane (Figure 2B) and on its sedimentation rate (Figure 3) indicated that ZmWHY1 is associated with chloroplast DNA in vivo. To gain insight into which DNA sequences were involved in these interactions, we modified the RIP-chip protocol to detect coimmunoprecipitating DNA (DIP-chip): stromal extract was treated with ribonuclease prior to the immunoprecipitation, and alkali hydrolysis was used to remove residual RNA after the immunoprecipitation. A control immunoprecipitation used antibody to CAF1, a splicing factor that associates with specific chloroplast intron RNAs in vivo (10). Both ZmWHY1 and CAF1 were efficiently immunoprecipitated (Supplementary Figure 2C), but the DIP-chip data were strikingly different (Figure 4C): nearly all of the DNA in the input stromal sample coimmunoprecipitated with ZmWHY1, whereas very little DNA was recovered in CAF1 immunoprecipitations. These results confirm that ZmWHY1 is associated with chloroplast DNA and show further that ZmWHY1 either binds throughout the chloroplast genome, or binds to specific DNA regions and coimmunoprecipitates all other DNA sequences due to their linkage to ZmWHY1-binding sites. Incubation of the extract with various restriction enzymes prior to the immunoprecipitation did not reveal the specific enrichment of any DNA sequences (Supplementary Figure 2B), leading us to favor the interpretation that ZmWHY1 is associated with many sites throughout the chloroplast genome. Nucleic acids recovered from the CAF1 and ZmWHY1 immunoprecipitations were also used as a direct template for PCR (Supplementary Figure 2D). The results support the DIP-Chip data: PCR product was obtained using a variety of chloroplast genome primers from the ZmWHY1 coimmunoprecipitation and not from the CAF1 coimmunoprecipitation.
The enrichment of DNA sequences in ZmWHY1 immunoprecipitations was further confirmed by slot-blot hybridization (Figure 4B). As for the DIP-chip assays, stroma was treated with RNAse prior to the immunoprecipitation, and residual RNA was removed by alkali hydrolysis after the immunoprecipitation (Figure 4B, lanes ‘D’). Antibody to ZmWHY1 coimmunoprecipitated DNA from all sequences tested, whereas DNA was not detected in either the CRS1 or OE16 immunoprecipitations. The DIP-chip, PCR and slot-blot hybridization data provide strong evidence that ZmWHY1 is associated with chloroplast DNA in vivo and that it has many binding sites throughout the genome.
Zm Why1 mutants are deficient for plastid ribosomes
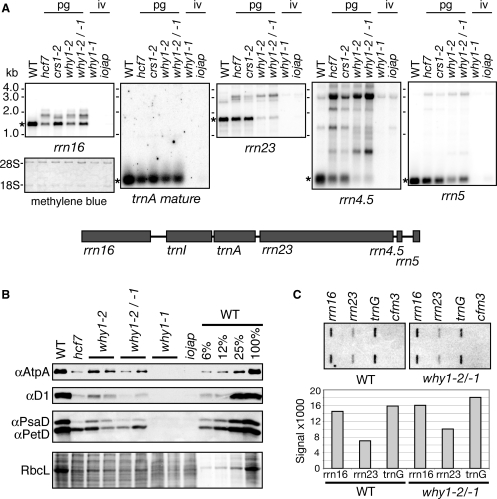
A role for WHY1 in chloroplast gene expression was suggested by the coimmunoprecipitation of ZmWHY1 with CRS1, RNA and DNA, and by the copurification of AtWHY1 with the plastid transcriptionally active chromosome (22). In support of this possibility, core subunits of the chloroplast ATP synthase, photosystem II, photosystem I, the cytochrome b6f complex and Rubisco accumulate to reduced levels in ZmWhy1 mutants (Figure 5B). The protein deficiencies conditioned by the weak allele combinations (Zmwhy1-2/-2 and Zmwhy1-1/-2) resemble those in hcf7mutants, which have a reduced content of chloroplast ribosomes (26). These proteins were not detectable in Zmwhy1-1 homozygotes, as in albino iojap mutants which lack plastid ribosomes (Figure 5B).
Figure 5.
Plastid ribosome deficiency in ZmWhy1 mutants. (A) Total seedling leaf RNA (0.5 μg) was analyzed by RNA gel blot hybridization using probes for the RNAs indicated at the bottom. A map of the plastid rRNA operon is shown below. A cDNA probe was used to detect mature trnA; this lacks intron sequences and therefore hybridizes poorly to unspliced precursor. The probe for 23S rRNA is derived from the 5′ portion of the rrn23 gene and detects just one of the two 23S rRNA fragments found in ribosomes in vivo. The leaf pigmentation conditioned by each mutant allele is indicated: iv: ivory leaves; pg: pale green leaves. The blot used to detect 16S rRNA is shown after staining with methylene blue to illustrate equal loading of cytosolic rRNAs (18S, 28S). Mature RNA forms are indicated with asterisks. (B) Reduced accumulation of photosynthetic enzyme complexes in ZmWhy1 mutants. Immunoblots of leaf extract (5 µg protein or the indicated dilutions) were probed with antibodies to core subunits of photosynthetic enzyme complexes: AtpA (ATP synthase), D1 (photosystem II), PsaD (photosystem I) and PetD (cytochrome b6f complex). The same blot stained with Ponceau S is shown below to illustrate sample loading and the abundance of RbcL. (C) Plastid runon transcription. Chloroplasts prepared from Zmwhy1-1/-2 heteroallelic mutants or their normal siblings (wt) were used for runon transcription assays. RNAs purified from the reactions were hybridized to slot blots harboring oligonucleotides corresponding to the genes indicated at the top. Each probe was present in duplicate. cfm3, a nuclear gene, served as a negative control. The results were quantified with a phosphorimager and plotted on the bar graph below.
The global loss of photosynthetic enzyme complexes in ZmWhy1 mutants suggested an underlying loss of plastid ribosomes. This possibility was confirmed by RNA gel blot hybridizations, which showed a loss of mature 23S, 4.5S and 16S rRNAs in hypomorphic ZmWhy1 mutants, and an increased accumulation of rRNA precursors (Figure 5A). Chloroplast rRNAs were not detectable in plants homozygous for the null Zmwhy1-1 allele, as in albino iojap leaves. Whereas hcf7 mutants show a more severe loss of 16S rRNA than 23S and 4.5S rRNAs, the reverse is true for hypomorphic ZmWhy1 mutants. A dramatic increase in the ratio of 23S rRNA precursors to mature 23S rRNA in these mutants was confirmed with a poisoned-primer extension assay (Supplementary Figure 3C).
Some steps in rRNA processing are dependent upon ribosome assembly in chloroplasts, as in bacteria (15,26,39). The aberrant 23S and 4.5S rRNA processing in ZmWhy1 mutants suggested therefore that ZmWHY1 might promote the expression of a gene needed for the assembly of the large ribosomal subunit (an rRNA or ribosomal protein), with loss of the small ribosomal subunit being a secondary effect. It seemed plausible, for example, that ZmWHY1 might promote processive transcription through the chloroplast rrn operon; this would differentially affect the large ribosomal subunit due to the distal position of the genes encoding its rRNA components (23S, 4.5S and 5S rRNA) in the operon (see map in Figure 5A). However, the results of chloroplast run-on transcriptions assays argue against this possibility (Figure 5C): the ratio of polymerase transit through the 23S gene in comparison to the 16S rRNA gene, and the ratio of rrn operon transcription in comparison to transcription from a different chloroplast locus (trnG-UCC) were similar in wild-type and Zmwhy1-1/-2 mutant chloroplasts. Furthermore, the rRNA components of the large ribosomal subunit were not reproducibly enriched in ZmWHY coimmunoprecipitates (Figure 4A, Supplementary Figure 2B); this suggests that ZmWHY1 does not interact directly with rRNAs or 50S ribosomal subunits, although such interactions cannot be eliminated based on these negative results. Taken together, these results argue that ZmWHY1 directly impacts the expression of a gene encoding a component of the large ribosomal subunit and/or promotes ribosome assembly. Elucidation of its precise role in this process will require further study.
ZmWHY1 promotes atpF intron splicing
The coimmunoprecipitation of ZmWHY1 with the atpF splicing factor CRS1 and with RNA from the atpF locus suggested that ZmWHY1 might be involved in the splicing of atpF pre-mRNA. To test this possibility, atpF RNA from Zmwhy1 mutants was analyzed by RNA gel blot hybridization (Figure 6). To control for pleiotropic effects of weak and severe plastid ribosome deficiencies, RNAs in pale green (hypomorphic) Zmwhy1-2 and Zmwhy1-2/-1 mutants were compared to those in hcf7 mutants, and RNAs in albino (null) Zmwhy1-1 mutants were compared to those in iojap mutants. These comparisons were important because the complete absence of plastid ribosomes results in the failure to splice all chloroplast subgroup IIA introns, including the atpF intron (6,40,41).
Figure 6.
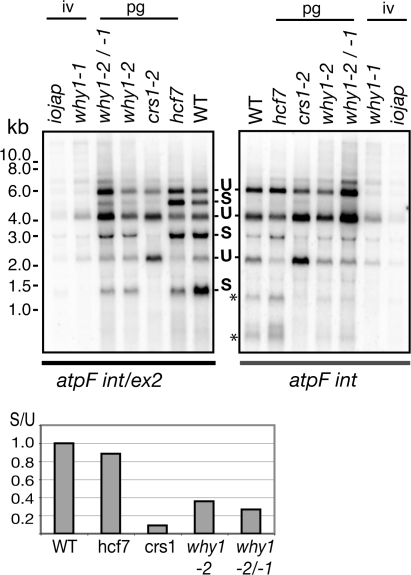
Reduced atpF intron splicing in ZmWhy1 mutants. (A) RNA gel blot analysis of atpF splicing. Total seedling leaf RNA (5 μg) was analyzed by RNA gel blot analysis using a probe including atpF exon 2 and a portion of the atpF intron (atpF int/ex2), or with an intron-specific probe (atpFint). The atpF gene is part of a polycistronic transcription unit that gives rise to a previously characterized population of RNAs (59,60). Spliced (S) and unspliced (U) transcripts are indicated. Asterisks mark bands that we believe correspond to the excised intron and its degradation products. The ratio of spliced to unspliced transcripts was quantified with a phosphorimager, normalized to the wild-type ratio and plotted below using arbitrary units.
The results in Figure 6 show that the ratio of spliced (S) to unspliced (U) atpF transcripts is reduced in hypomorphic ZmWhy1 mutants in comparison to wild-type and hcf7 plants, albeit not as severely as in crs1 mutants. The ratio of excised intron (asterisks) to unspliced RNA is also reduced, supporting the interpretation that ZmWHY1 promotes atpF splicing rather than stabilizing the spliced product. The normal splicing of the atpF intron in hcf7 mutants argues that the partial plastid ribosome deficiency in hypomorphic ZmWhy1 mutants cannot account for their reduced atpF splicing. Furthermore, a different subgroup IIA intron, the rpl2 intron, is spliced normally in the same plants (Supplementary Figure 3B), showing that not all subgroup IIA introns are affected in the hypomorphic ZmWhy1 mutants. These results provide strong evidence that ZmWHY1's association with atpF RNA enhances the splicing of the atpF intron.
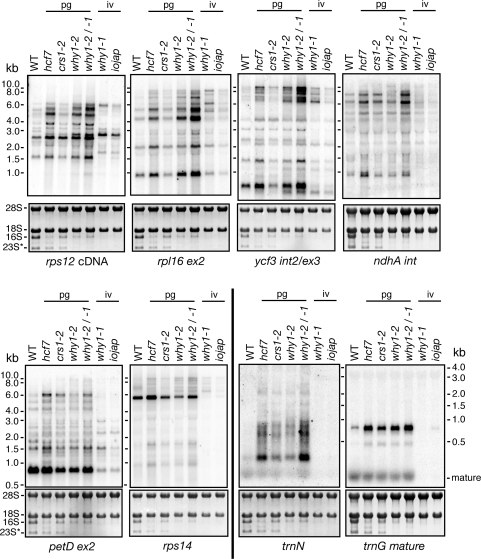
The coimmunoprecipitation data demonstrated an association between ZmWHY1 and RNAs from several loci other than atpF. However, RNA gel blot hybridizations showed that the transcripts from all such genes were qualitatively similar in ZmWhy1 mutants and in the relevant control mutant (Figure 7). The coimmunoprecipitation of ZmWHY1 with RNAs from both loci encoding the trans-spliced group II intron in rps12 was intriguing (Figure 4A), but splicing of this RNA is not disrupted in ZmWhy1 mutants (Supplementary Figure 3B). These results show that ZmWHY1 is not necessary for the normal processing of most chloroplast transcripts.
Figure 7.
Accumulation of plastid RNAs in ZmWhy1 mutants. Total seedling leaf RNA (5 μg) was analyzed by RNA gel blot hybridization using probes specific for the RNAs indicated at bottom. The rps12 probe was a cDNA probe containing exons 1 and 2. The leaf pigmentation conditioned by each mutant allele is indicated: iv: ivory; pg: pale green. The methylene blue-stained blots are shown below, with rRNAs marked. Additional RNAs that were analyzed analogously are shown in Supplementary Figure 3A.
A structural homolog of ZmWHY1 in Trypanosoma brucei is required for mitochondrial RNA editing (42). Several plastid RNAs that are known to be substrates for RNA editing were represented among the RNAs that coimmunoprecipitate with ZmWHY1. Direct sequencing of RT-PCR products demonstrated, however, that the editing of the known edited sites in the petB, rpl20, ycf3 and rps14 transcripts is not disrupted in Zmwhy1-1 and Zmwhy1-2/-1 mutants (data not shown), suggesting that ZmWHY1 is not required for RNA editing.
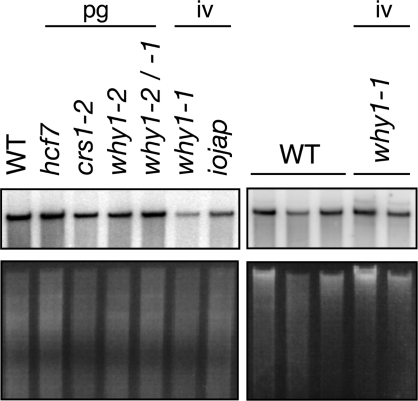
ZmWHY1 is required neither for chloroplast DNA replication nor for global plastid transcription
The association of ZmWHY1 with plastid DNA suggested that it might be involved in chloroplast transcription or DNA replication. However, Southern blot analysis of total leaf DNA showed that plastid DNA levels in ZmWhy1 mutants, although somewhat variable from sample to sample, were generally similar to those in normal and control mutant plants (Figure 8). In addition to the plastid transcripts shown in Figure 7, a variety of other transcripts were examined by RNA gel blot hybridization (Supplementary Figure 3A). In no case was a significant reduction in transcript level detected, indicating that ZmWHY1 is not necessary for global plastid transcription. In fact, a trend is apparent toward increased transcript abundance in ZmWhy1 mutants, but these changes are rather subtle and indirect effects on RNA abundance cannot be excluded.
Figure 8.
Chloroplast DNA levels in ZmWhy1 mutants. Seedling leaf DNA (5 µg) was digested with EcoRI (left), or PvuII (right) and analyzed by DNA gel blot hybridization using a probe from the chloroplast rrn23 gene (top left), or orf99 (top right). The same gels stained with ethidium bromide are shown below. The small fluctuations in relative band intensity may result from small differences in sample loading.
Recombinant ZmWHY1 binds single-stranded RNA and DNA in vitro
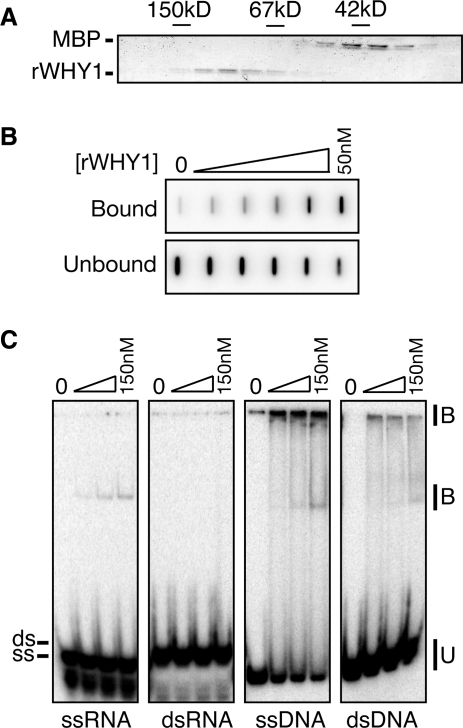
To determine whether ZmWHY1 can directly bind both RNA and DNA, recombinant ZmWHY1 (rWHY1) was generated by expression as a maltose-binding protein (MBP) fusion. rWHY1 was released from the MBP moiety by protease cleavage and further purified on a gel filtration column (Figure 9A). rWHY1 eluted from the sizing column at a position corresponding to a globular protein of ∼100 kDa, consistent with the report that StWHY1 forms a homotetramer (19). Filter-binding assays showed that rWHY1 binds to unspliced atpF RNA in vitro (Figure 9B), but it did not show specificity for this RNA relative to other RNAs of similar size under the conditions tested (data not shown).
Figure 9.
Recombinant ZmWHY1 binds ssRNA and DNA. (A) Elution of recombinant ZmWHY1 from a gel filtration column. MBP–WHY1 was purified by amylose affinity chromatography, cleaved with TEV protease to separate the WHY1 and MBP moieties, and applied to a Superdex 200 column. Column fractions were analyzed by SDS–PAGE and staining with Coomassie blue. The elution position of size markers (alcohol dehydrogenase, 150 kDa; BSA, 67 kDa, MBP, 42 kDa) is shown. The peak WHY1 fractions were pooled and used for in vitro assays. (B) Filter binding assay showing RNA binding activity of ZmWHY1. Assays containing 10 pM radiolabeled atpF intron RNA and increasing ZmWHY1 concentrations (50 nM maximum) were filtered through sandwiched nitrocellulose and nylon membranes. Protein–RNA complexes were captured on the nitrocellulose (bound); unbound RNA was captured on the nylon membrane below. (C) Gel mobility shift assay showing rWHY1's relative affinity for double- and single-stranded RNA and DNA. A 31-mer oligonucleotide in RNA or DNA form was radiolabeled, heated and, either snap cooled (ssRNA, ssDNA) or cooled slowly in the presence of monovalent salts and a two-fold excess of its complement (dsRNA, dsDNA). The substrate (40 pM) was mixed with increasing concentrations of ZmWHY1 (17, 50 and 150 nM). Protein binding is illustrated by the appearance of an upper band and retention at the top of the gel, and by the disappearance of unbound substrate.
To compare the affinity of ZmWHY1 for single-stranded and double-stranded RNA and DNA, gel mobility shift assays were used to detect binding to a synthetic 31-mer oligonucleotide in the context of ssDNA, ssRNA, ds DNA or dsRNA (Figure 9C). ZmWHY1 bound rather weakly to these short oligonucleotides but the results showed, nonetheless, that rWHY1 binds both ssDNA and ssRNA, and binds poorly to dsRNA and dsDNA.
DISCUSSION
Previous reports have attributed diverse functions and intracellular locations to WHY1. WHY1 in dicots has been reported to be a ssDNA-binding protein that functions in the nucleus as both a transcription factor (17,19) and as a negative regulator of telomere length (20). Arabidopsis WHY1 copurified with the ‘transcriptionally active chromosome’ from chloroplasts (22). Our results add another layer to this complex picture. We demonstrate that ZmWHY1 is essential for chloroplast biogenesis, and that it localizes to the chloroplast where it plays multiple roles in gene expression. We also add RNA binding to WHY1's repertoire of biochemical activities and demonstrate that ZmWHY1 is bound to a subset of chloroplast RNAs in chloroplast extract.
Multiple roles of ZmWHY1 in chloroplast biogenesis
ZmWHY was identified among proteins that coimmunoprecipitate with CRS1, which is required for the splicing of the group II intron in the chloroplast atpF pre-mRNA. We showed that ZmWHY1 is associated with atpF intron RNA in vivo and that the coimmunoprecipitation of ZmWHY1 and CRS1 is disrupted by RNAse, indicating that they coimmunoprecipitate due to their association with the same RNA molecule. ZmWHY1's association with atpF RNA is functionally significant, as atpF intron splicing is disrupted in ZmWhy1 mutants. However, the splicing of this intron is more sensitive to a partial loss of CRS1 than to a partial loss of ZmWHY1, suggesting that ZmWHY1 plays an accessory function in atpF splicing but may not be absolutely required.
The atpF splicing defect in ZmWhy1 mutants cannot account for their loss of plastid ribosomes, as the more severe atpF splicing defect in crs1-1 mutants is not accompanied by a substantial plastid ribosome deficiency (11). The specific role of ZmWHY1 in promoting the biogenesis of the plastid translation machinery remains unclear. Although several RNAs with translation-related functions are among the RNAs that coimmunoprecipitate with ZmWHY1, the abundance and processing of these RNAs are similar in ZmWhy1 mutants and in control mutants that exhibit a ribosome-deficiency of similar severity. The specific rRNA deficiencies in ZmWhy1 mutants do suggest, however, that ZmWHY1 is most directly involved in the biogenesis of the large ribosomal subunit: the accumulation and processing of the 23S and 4.5S rRNAs are more sensitive to the partial loss of ZmWhy1 function than are those of 16S rRNA, whereas the reverse is true for hcf7 mutants. Furthermore, in ppr5 mutants, whose primary defect is in the maturation of a specific plastid tRNA, the rRNAs from the two ribosomal subunits are impacted to a similar extent (39). Thus, our results point to the biogenesis of the plastid large ribosomal subunit as one function of ZmWHY1 but definition of its precise role in this process will require additional study. The strong defect in the processing step that separates 23S rRNA from 4.5S rRNA in hypomorphic ZmWhy1 mutants is reminiscent of defects reported for mutations in the DCL, DAL and RNR1 genes in dicots (43–46). Although it is unclear whether any of these genes function directly in 23S/4.5S rRNA processing, it is possible that WHY1 acts in concert with one or more of these proteins.
ZmWHY1 binds both RNA and DNA in vitro and in vivo
We show here that chloroplast DNA coimmunoprecipitates with ZmWHY1 from plastid extract, that a fraction of ZmWHY1 is tethered to the thylakoid membrane in a DNA-dependent fashion, that a fraction of stromal ZmWHY1 is found in DNA-containing particles of ∼400 kDa, and that ZmWHY1 binds ssDNA in vitro. These results are consistent with previous reports that dicot WHY1 binds ssDNA (19,20) and that it copurifies with a chloroplast ‘transcriptionally active chromosome’ (22). Our findings suggest that ZmWHY1 either binds DNA in a sequence non-specific fashion or that it has many binding sites distributed throughout the plastid genome, because DNA sequences from throughout the plastid genome coimmunoprecipitated to a similar extent with ZmWHY1. It remains possible, however, that ZmWHY1 associates with specific DNA regions in vivo, but that these associations were disrupted during lysate preparation. A DNA immunoprecipitation experiment was recently reported for AtWHY2, a mitochondrial-localized Whirly protein (23), with analogous results: DNA sequences from a variety of regions throughout the mitochondrial genome coimmunoprecipitated with AtWHY2, when assayed by PCR.
We demonstrate here that ZmWHY1 interacts not only with DNA, as anticipated by previous reports, but that it also binds RNA in vivo and in vitro. That ZmWHY1 interacts with RNA is, perhaps, not surprising given that a structural homolog of ZmWHY1 has been shown to bind RNAs involved in kinetoplastid RNA editing (42), and that many proteins that bind ssDNA also bind RNA. The atpF intron RNA was the major RNA ligand of ZmWHY1 detected in the RNA coimmunoprecipitation assays. This RNA is not particularly abundant in vivo so its enrichment in ZmWHY1 immunoprecipitations likely reflects a specific interaction in vivo. Although intrinsic specificity for this RNA did not emerge from in vitro binding assays using the entire intron, a high-affinity site within a large RNA such as the atpF intron (∼800 nt) can be masked in vitro due to the overwhelming number of nonspecific sites available for protein binding. Therefore, more detailed studies involving smaller RNA ligands will be required to determine whether ZmWHY1 binds RNA with sequence-specificity or whether it is recruited to the atpF intron via protein–protein interactions.
What is WHY1's DNA-related function in the chloroplast?
The association of ZmyWHY1 with DNA sequences from throughout the chloroplast genome suggests that it participates in transcription and/or DNA metabolism. However, our results argue against a general role in transcription, as all plastid mRNAs examined accumulate in hypomorphic Zmwhy1 mutants to levels that are comparable to those in the relevant control mutants. The results of chloroplast transcription runon experiments argue that the preferential loss of 23S rRNA in these mutants is due to aberrant ribosome assembly rather than to reduced rRNA transcription rates. It remains possible, however, that ZmWHY1 does play a role in chloroplast transcription but that another gene with a partially redundant function serves this purpose in ZmWhy1 mutants.
It is intriguing that ZmWHY1 binds preferentially to DNA in single stranded form because opportunities to interact with ssDNA in vivo are expected to be limited. DNA replication, recombination and repair involve the transient occurrence of ssDNA, and torsional stress can induce DNA unwinding. The Southern blot data showing that plastid DNA levels are no more than minimally decreased in ZmWhy1 null mutants argue against a central role for ZmWHY1 in DNA replication; however participation of ZmWHY1 in DNA recombination or repair remains possible. In fact, the participation of an unrelated ssDNA-binding protein, OSB1, in plant mitochondrial DNA recombination was reported recently (47).
There are several parallels between our findings with ZmWHY1 and the activities reported for the bacterial protein HU. HU is associated with the bacterial nucleoid, binds preferentially to DNA with irregular structural features (e.g. single stranded gaps and bulges), and is involved in DNA recombination and repair (48,49). Despite its high conservation in bacteria and the presence of an HU homolog in a plastid genome in red algae (50), HU homologs are not encoded in the nuclear or plastid genomes of vascular plants (50,51). Thus, alternative proteins have presumably been recruited in vascular plants to fulfill the functions performed by HU in the chloroplast's cyanobacterial ancestor. The nucleoid-associated protein sulfite reductase has been suggested to be one such protein (51–53), and perhaps WHY1 is another. HU influences global transcription patterns through its effect on nucleoid architecture, and mediates the formation of DNA loops that repress transcription from specific genes (54–56). HU is also an RNA-binding protein, and functions in vivo to repress the translation of the E. coli rpoS mRNA (57,58). Like HU, ZmWHY1 interacts globally with plastid DNA, but specifically with certain plastid RNAs, and binds preferentially to nucleic acids with single-stranded character. The abundance of several chloroplast mRNAs is increased in ZmWhy1 mutants, consistent with a global repressive role for ZmWHY1 in transcription. This possibility is in accord with the recent report that over-expression of AtWHY2 in Arabidopsis causes a reduction in the levels of several mitochondrial RNAs (23). Although its role in DNA metabolism remains uncertain, our results demonstrate that description of WHY1 as a chloroplast transcription factor is, at best, an over-simplification of the complex roles played by this interesting protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Rosalind Williams-Carrier for her participation in the purification of CRS1-associated proteins for mass spectrometry, Susan Belcher for maintaining and propagating the mutant lines, and Tiffany Kroeger and Amy Cooke for identifying ZmWhy1 mutants. We are also grateful to Christian Schmitz-Linneweber for overseeing the sequencing of ZmWhy1 cDNAs. This work was supported by grants to A.B. (DBI-0421799 and MCB-0314597) and K.J.v.W. (DBI-0211935) from the National Science Foundation. Funding to pay the Open Access publication charges for this article was provided by NSF grant DBI-0421799.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marchfelder A, Binder S. Plastid and plant mitochondrial RNA processing and RNA stability. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles. Kluwer Academic Publishers: Dordrecht, The Netherlands; 2004. pp. 261–294. [Google Scholar]
- 2.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2193. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins K. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13:55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nickelsen J. Chloroplast RNA-binding proteins. Curr. Genet. 2003;43:392–399. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 5.Schwacke R, Fischer K, Ketelsen B, Krupinska K, Krause K. Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics. 2007;277:631–646. doi: 10.1007/s00438-007-0214-4. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins B, Kulhanek D, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins B, Barkan A. Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 2001;20:872–879. doi: 10.1093/emboj/20.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asakura Y, Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19:3864–3875. doi: 10.1105/tpc.107.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostheimer G, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA. 2001;7:1227–1238. doi: 10.1017/s1355838201010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watkins K, Kroeger T, Cooke A, Williams-Carrier R, Friso G, Belcher S, Wijk Kv, Barkan A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19:2606–2623. doi: 10.1105/tpc.107.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asakura Y, Bayraktar O, Barkan A. RNA. 2008. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falcon de Longevialle A, Hendrickson L, Taylor N, Delannoy E, Lurin C, Badger M, Millar AH, Small I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03581.x. (in press) [DOI] [PubMed] [Google Scholar]
- 15.Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 Pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostersetzer O, Watkins K, Cooke A, Barkan A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desveaux D, Subramaniam R, Despres C, Mess JN, Levesque C, Fobert PR, Dangl JL, Brisson N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 18.Desveaux D, Despres C, Joyeux A, Subramaniam R, Brisson N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell. 2000;12:1477–1489. doi: 10.1105/tpc.12.8.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desveaux D, Allard J, Brisson N, Sygusch J. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat. Struct. Biol. 2002;9:512–517. doi: 10.1038/nsb814. [DOI] [PubMed] [Google Scholar]
- 20.Yoo HH, Kwon C, Lee MM, Chung IK. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 2007;49:442–451. doi: 10.1111/j.1365-313X.2006.02974.x. [DOI] [PubMed] [Google Scholar]
- 21.Krause K, Kilbienski I, Mulisch M, Rodiger A, Schafer A, Krupinska K. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005;579:3707–3712. doi: 10.1016/j.febslet.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 22.Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marechal A, Parent JS, Sabar M, Veronneau-Lafortune F, Abou-Rached C, Brisson N. Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function. BMC Plant Biol. 2008;8:42. doi: 10.1186/1471-2229-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams P, Barkan A. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 2003;36:675–686. doi: 10.1046/j.1365-313x.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- 25.Walbot V, Coe EH. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl Acad. Sci. USA. 1979;76:2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 1998;297:38–57. [Google Scholar]
- 28.Voelker R, Barkan A. Nuclear genes required for post-translational steps in the biogensis of the chloroplast cytochrome b6f complex. Molec. Gen. Genet. 1995;249:507–514. doi: 10.1007/BF00290576. [DOI] [PubMed] [Google Scholar]
- 29.Barkan A. Genome-wide analysis of RNA-protein interactions in plants. In: Belostotsky D, editor. Plant Systems Biology, Series: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2008. [DOI] [PubMed] [Google Scholar]
- 30.Voelker R, Mendel-Hartvig J, Barkan A. Transposon-disruption of a maize nuclear gene, tha1, encoding a chloroplast SecA homolog: in vivo role of cp-SecA in thylakoid protein targeting. Genetics. 1997;145:467–478. doi: 10.1093/genetics/145.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong I, Lohman T. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullet J, Klein R. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapp JC, Baumgartner BJ, Mullet J. Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes: transcription rates and mRNA levels vary over 300-fold; predicted mRNA stabilities vary 30-fold. J. Biol. Chem. 1992;267:21404–21411. [PubMed] [Google Scholar]
- 34.Klein R, Mullet J. Light-induced transcription of chloroplast genes. J. Biol. Chem. 1990;265:1895–1902. [PubMed] [Google Scholar]
- 35.Emanuelsson O, Heijne Gv. Prediction of organellar targeting signals. Biochim. Biophys. Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- 36.Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 37.Sato N, Terasawa K, Miyajima K, Kabeya Y. Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 2003;232:217–262. doi: 10.1016/s0074-7696(03)32006-6. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.00563-08. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess WR, Hoch B, Zeltz P, Huebschmann T, Koessel H, Boerner T. Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell. 1994;6:1455–1465. doi: 10.1105/tpc.6.10.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel J, Boerner T, Hess W. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 43.Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D. RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellaoui M, Keddie JS, Gruissem W. DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Mol. Biol. 2003;53:531–543. doi: 10.1023/B:PLAN.0000019061.79773.06. [DOI] [PubMed] [Google Scholar]
- 45.Bellaoui M, Gruissem W. Altered expression of the Arabidopsis ortholog of DCL affects normal plant development. Planta. 2004;219:819–826. doi: 10.1007/s00425-004-1295-5. [DOI] [PubMed] [Google Scholar]
- 46.Bisanz C, Begot L, Carol P, Perez P, Bligny M, Pesey H, Gallois JL, Lerbs-Mache S, Mache R. The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 2003;51:651–663. doi: 10.1023/a:1022557825768. [DOI] [PubMed] [Google Scholar]
- 47.Zaegel V, Guermann B, Le Ret M, Andres C, Meyer D, Erhardt M, Canaday J, Gualberto JM, Imbault P. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006;18:3548–3563. doi: 10.1105/tpc.106.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 49.Kamashev D, Balandina A, Mazur AK, Arimondo PB, Rouviere-Yaniv J. HU binds and folds single-stranded DNA. Nucleic Acids Res. 2008;36:1026–1036. doi: 10.1093/nar/gkm667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi T, Takahara M, Miyagishima SY, Kuroiwa H, Sasaki N, Ohta N, Matsuzaki M, Kuroiwa T. Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell. 2002;14:1579–1589. doi: 10.1105/tpc.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato N. Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 2001;6:151–55. doi: 10.1016/s1360-1385(01)01888-x. [DOI] [PubMed] [Google Scholar]
- 52.Sato N, Nakayama M, Hase T. The 70-kDa major DNA-compacting protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett. 2001;487:347–350. doi: 10.1016/s0014-5793(00)02342-5. [DOI] [PubMed] [Google Scholar]
- 53.Sekine K, Fujiwara M, Nakayama M, Takao T, Hase T, Sato N. DNA binding and partial nucleoid localization of the chloroplast stromal enzyme ferredoxin:sulfite reductase. FEBS J. 2007;274:2054–2069. doi: 10.1111/j.1742-4658.2007.05748.x. [DOI] [PubMed] [Google Scholar]
- 54.Lia G, Bensimon D, Croquette V, Allemand JF, Dunlap D, Lewis DE, Adhya S, Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kar S, Edgar R, Adhya S. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl Acad. Sci. USA. 2005;102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis DE, Geanacopoulos M, Adhya S. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol. Microbiol. 1999;31:451–461. doi: 10.1046/j.1365-2958.1999.01186.x. [DOI] [PubMed] [Google Scholar]
- 57.Balandina A, Kamashev D, Rouviere-Yaniv J. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 2002;277:27622–27628. doi: 10.1074/jbc.M201978200. [DOI] [PubMed] [Google Scholar]
- 58.Balandina A, Claret L, Hengge-Aronis R, Rouviere-Yaniv J. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 2001;39:1069–1079. doi: 10.1046/j.1365-2958.2001.02305.x. [DOI] [PubMed] [Google Scholar]
- 59.Barkan A. Tissue-dependent plastid RNA splicing in maize: Transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell. 1989;1:437–445. doi: 10.1105/tpc.1.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stahl DJ, Rodermel SR, Bogorad L, Subramanian AR. Co-transcription pattern of an introgressed operon in the maize chloroplast genome comprising four ATP synthase subunit genes and the ribosomal rps2. Plant Molec. Biol. 1993;21:1069–1076. doi: 10.1007/BF00023603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.